Abstract
The genome of the halophilic archaeon Halobacterium sp. strain NRC-1 encodes homologs of the eukaryotic Mre11 and Rad50 proteins, which are involved in the recognition and end processing of DNA double-strand breaks in the homologous recombination repair pathway. We have analyzed the phenotype of Halobacterium deletion mutants lacking mre11 and/or rad50 after exposure to UV-C radiation, an alkylating agent (N-methyl-N′-nitro-N-nitrosoguanidine), and γ radiation, none of which resulted in a decrease in survival of the mutant strains compared to that of the background strain. However, a decreased rate of repair of DNA double-strand breaks in strains lacking the mre11 gene was observed using pulsed-field gel electrophoresis. These observations led to the hypothesis that Mre11 is essential for the repair of DNA double-strand breaks in Halobacterium, whereas Rad50 is dispensable. This is the first identification of a Rad50-independent function for the Mre11 protein, and it represents a shift in the Archaea away from the eukaryotic model of homologous recombination repair of DNA double-strand breaks.
Maintaining genome integrity through the repair of DNA double-strand breaks (DSBs) is critical to cell survival. It is particularly essential following exposure to ionizing radiation, which has been shown to produce extensive DNA fragmentation (11, 14). Multiple pathways for repairing DNA DSBs have been identified, including synthesis-dependent strand annealing both with and without extended DNA synthesis (31, 45), nonhomologous end joining, and homologous recombination (HR), working in a coordinated manner on the various damage types (36, 37). Although the pathways utilized depend on the protein complement of the organism, the cell cycle, and the number of genome copies present, some elements are shared between pathways across the three domains of life. The involvement of a crossover structure and a recombinase such as RecA (Bacteria), RadA (Archaea), or Rad51 (Eukarya) is common to both HR and extended-synthesis-dependent strand annealing, and the processing of double-stranded DNA ends to produce a 3′ overhang is found in HR, synthesis-dependent strand annealing, and extended-synthesis-dependent strand annealing (45).
The process of HR repair of DNA DSBs in both the Bacteria and Eukarya has been extensively studied (see reviews in references 24, 25, 38, 41, and 44). The first step in this pathway is the recognition of the DSB and the resection of the 5′ strand to produce a 3′-OH overhang that can be recognized by the recombinase. In the Bacteria, this is done primarily by the RecBCD complex, although there are redundant pathways for RecBCD activities, including the use of the RecFOR complex for recombinase loading along with the RecQ helicase and RecJ nuclease for DNA end processing (44).
The Mre11-Rad50 complex in Eukarya performs DSB recognition (see reviews in references 2, 15, 38, and 39). Rad50 has an ATP-dependent DNA binding activity, and coiled-coil domains of Rad50 are separated by a zinc hook found to be required for the repair of DSBs by HR (43). Mre11 is a nuclease with double-stranded DNA exonuclease and single-stranded DNA endonuclease activities as well as a helicase. Homodimers of both Mre11 and Rad50 interact to form a complex referred to hereafter as the MR complex. Bacterial genomes can also encode a structural homolog of the MR complex, namely, the SbcCD complex (24). SbcCD has been shown to cleave hairpin DNA, which can block stalled replication forks, prior to homologous recombination rescue of the fork (8). Deletions of sbcC together with sbcB, encoding an ExoI 3′-to-5′ exonuclease, have been shown to complement ΔrecBC in Escherichia coli by shunting repair from the RecBCD pathway into the RecFOR pathway (24). In Bacillus subtilis and Deinococcus radiodurans, the deletion of sbcC results in an increased sensitivity of the cells to ionizing radiation (28). Deletions of sbcC and/or sbcD in D. radiodurans also result in delayed repair kinetics of DNA DSBs following γ irradiation (5).
The MR complex in eukaryotes also includes the Nbs1 (human) (Xrs2 in Saccharomyces cerevisiae) protein. Xrs2 binds DNA and Mre11 and aids in the localization of the complex to the DSB as well as the stimulation of the Mre11 exonuclease (38). Yeast MR complex mutants are sensitive to ionizing radiation and exposure to alkylating agents (38) and have a slow-growth phenotype (35). Mre11 has been shown to be required for complex formation with Rad50 and Xrs2 (38). The structure and activity of archaeal Rad50 and Mre11 were examined in Pyrococcus furiosus, a hyperthermophilic archaeon (18, 19). The data confirmed the structural conservation of MR complex homologs through all three domains of life. In thermophilic archaea, the genes encoding Rad50 and Mre11 are also found in an operon with genes coding for a 5′-to-3′ nuclease (nurA) (9, 40) and helicase (herA [mla]) (10, 27). Interestingly, only genes encoding Rad50 and Mre11 are present in the genome of the mesophilic archaeon Halobacterium sp. strain NRC-1 (Halobacterium).
Halobacterium is an extreme halophile growing optimally in 4 M NaCl (12). Intracellular salt, 4 M KCl (16), acts as a counterbalance to the high external salt concentration. The exceptional ability of Halobacterium to survive high levels of ionizing radiation has been attributed to its adaptation to hypersaline environments characterized by high levels of solar radiation and periodic desiccation (23, 42). Halobacterium has up to 25 copies of its genome during log-phase growth and 15 copies during stationary phase (7), potentially providing additional templates for homologous recombination-repair. The free-radical-scavenging capability of membrane pigments, specifically bacterioruberin, has been shown to provide Halobacterium with protection against cellular damages by ionizing radiation (23, 34). Whole-genome transcriptional analysis suggested that HR is the major pathway for the repair of DSBs in Halobacterium (42), and homologs of eukaryotic HR proteins have been identified in its genome (30). The Halobacterium genome lacks homologs for genes encoding the Ku70 and Ku80 proteins involved in the Ku-mediated nonhomologous end-joining pathway for the repair of DNA DSBs, although 36% of the Halobacterium genome open reading frames were reported to be unrelated to any known proteins (30), allowing the possibility of currently unknown repair proteins.
Here, Halobacterium is used as a model system for archaea in a genetic approach to investigate the cellular roles of Rad50 and Mre11 in the repair of DNA DSBs. Our phenotypic analysis of rad50, mre11, and mre11-rad50 knockout mutants did not show increased sensitivity to ionizing radiation, UV, or the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). This is in contrast to previous studies with yeast Rad50 mutants that are highly sensitive to γ radiation and MNNG, which produce DSBs. We also show a delay in the repair of DSBs with mre11 and rad50-mre11 mutants, raising interesting questions about the role of the MR complex in this archaeon.
METHODS AND MATERIALS
Cultures and growth conditions.
Halobacterium sp. strain NRC-1 cultures were grown in standard GN101 medium (250 g/liter NaCl, 20 g/liter MgSO4·7H2O, 2 g/liter KCl, 3 g/liter Na citrate, and 10 g/liter Oxoid peptone [pH 7.2] with the addition of 1 ml/liter Trace Elements solution [31.5 mg/liter FeSO4·7H2O, 4.4 mg/liter ZnSO4·7H2O, 3.3 mg/liter MnSO4·H2O, 0.1 mg/liter CuSO4·5H2O]) (17) at 42°C with shaking at 220 rpm (Gyromax 737; Amerex Instruments, Lafyatte, CA). Tryptophan and uracil dropout media were made using yeast synthetic dropout medium supplements (without tryptophan and without uracil) and yeast nitrogen base (without amino acids) (both from Sigma, St. Louis, MO) suspended in a basal salt solution (250 g/liter NaCl, 20 g/liter MgSO4·7H2O, 2 g/liter KCl, 3 g/liter Na citrate [pH 7]) with the addition of 1 ml/liter Trace Elements solution (31.5 mg/liter FeSO4·7H2O, 4.4 mg/liter ZnSO4·7H2O, 3.3 mg/liter MnSO4·H2O, 0.1 mg/liter CuSO4·5H2O) and 20 g/liter agar. When specified, uracil was added to a final concentration of 50 μg/ml, tryptophan was added to a final concentration of 50 μg/ml, and 5-fluoroorotic acid was added to a final concentration of 0.3 mg/ml.
Targeted gene deletion.
Targeted gene deletions were constructed using the protocol described previously by Peck et al. (32) and plasmid pAK07, modified from plasmid pNBK07 (from M. P. Krebs, Illinois State University, Normal, IL) to include the trpA gene for tryptophan biosynthesis under the control of the constitutively expressed ferrodoxin (fdx) promoter. Changes to the protocol are as follows: the gene knockout construct for each gene was composed of 500 bp upstream and downstream of the target gene flanking the trpA gene and fdx promoter, as described previously (1). Strain AK071, auxotrophic for both tryptophan and uracil biosynthesis (ΔtrpA Δura3), was used in place of strain ΑΚ07 (Δura3) to allow the use of uracil and tryptophan dropout media. Uracil dropout medium was used as the selective agent for selection for uracil prototrophy following transformation with plasmid pAK07 bearing knockout gene constructs and the ura3 marker for uracil biosynthesis. Tryptophan dropout medium supplemented with 5-fluoroorotic acid (Sigma, St. Louis, MO) was subsequently used to select intramolecular recombinants that lost the plasmid and acquired tryptophan prototrophy. Deletions were confirmed by Southern hybridization using probes within the deleted region and up- or downstream of the deleted region. In addition, the production of a transcript for the mre11 and rad50 genes was analyzed to confirm that the disruption in the first gene in the operon did not affect the transcription of the second gene. Total RNA was extracted using the Stratagene (La Jolla, CA) Absolute RNA kit, followed by treatment with RQ1 RNase-free DNase (Promega, Madison, WI) and the production of cDNA using the TaqMan reverse transcription (RT) kit (Applied Biosystems, Foster City, CA). FastTaq PCR reagents (Roche Applied Sciences, Indianapolis, IN) were used to eliminate multiple PCR products due to the high GC content of the Halobacterium genome. PCR products were analyzed by agarose gel electrophoresis. Northern blot hybridization and RT-PCR analysis were used to confirm the absence of rad50 mRNA in strain AK073. A 500-bp DNA probe within the gene product (Fig. 1) was used for the Northern blot hybridization.
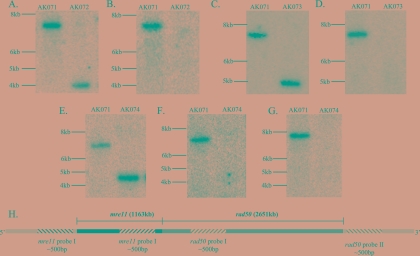
FIG. 1.
Southern hybridization blots showing deletions of the mre11, rad50, and mre11-rad50 genes (A to G) and schematic of probe locations (H). Probes were designed to hybridize to the regions 500 bp immediately upstream of the mre11 coding region (mre11 probe I) (A), 500 bp immediately downstream of the rad50 coding region (rad50 probe I) (C and E), and within the coding regions of mre11 (mre11 probe II) (B and G) and rad50 (rad50 probe II) (D and F).
UV-C and γ-irradiation survival.
UV-C (254 nm) and γ-irradiation survival assays were conducted and quantified as described previously by Baliga et al. (4) and Kottemann et al. (23), respectively. Recovery after UV-C irradiation was conducted in the dark to limit photoreactivation repair. Cells were plated within 30 min following irradiation with both UV and γ. Survival was measured by calculating the ratio of the total number of viable cells after irradiation to the total number of viable cells in the control sample.
MNNG survival.
Cultures were grown to an optical density at 600 nm (OD600) of 0.8 in standard GN101 medium at 42°C with shaking prior to being divided into aliquots and diluted to and OD600 of 0.2 in GN101 medium with the addition of 50 μg/ml and 100 μg/ml MNNG to a final volume of 5 ml. Optical density values were recorded after approximately 24 h of incubation at 42°C; control cultures were grown in standard GN101 medium. At least three independent experiments were carried out for each strain and MNNG concentration.
Growth curves and temperature-dependent growth assays.
Cultures of each deletion mutant strain and background strain AK071 were grown to an OD600 of 0.6 (log phase, 2 × 108cells/ml), diluted back to an OD600 of 0.05 in GN101 medium supplemented with tryptophan and uracil, and incubated at 42°C with shaking. OD600 readings were taken at intervals and recorded. At least three independent experiments were carried out for each strain.
γ-Irradiation time course of recovery PFGE analysis.
Cultures of Halobacterium background and mutant strains were grown in GN101 medium with uracil plus tryptophan to an OD600 of 0.6, concentrated by centrifugation, and irradiated using a 60Co γ source (University of Maryland College Park γ Test Facility) to a final dose of 2.5 kGy (dose rate of 3 to 13 kGy/h). Cultures were brought back to a full volume in GN101 medium with uracil plus tryptophan and incubated at 42°C with shaking for 12 h. Samples were taken at each of the following time points: preirradiation, 0 h, 2 h, 4 h, 8 h, and 12 h. Cells were pelleted by centrifugation at 8,000 × g for 5 min and resuspended in room-temperature BSS (250 g/liter NaCl, 20 g/liter MgSO4, 2 g/liter KCl, 3 g/liter sodium citrate) prior to being embedded into InCert agarose plugs (0.8% final concentration prepared in 3:1 BSS-H2O; Bio-Rad, Hercules, CA) at a final cell concentration of 1 × 109 cells/ml. Plugs were lysed in proteinase K solution (0.25 M EDTA [pH 8], 1% N-lauryl sarkosine, and 0.5 mg/ml proteinase K) at 54°C for 1 to 2 days. Plug washes consisted of two washes for 1 h in 20 ml 1× Tris-EDTA (TE) buffer at room temperature, two washes for 1 h in 20 ml 0.5× TE buffer at room temperature, and four washes for 24 h in 0.5× TE buffer at 4°C. Plugs were stored in 5 ml 0.5× TE buffer at 4°C after wash steps. Halobacterium genomic DNA plugs were analyzed using a CHEF DR-III electrophoresis system (Bio-Rad, Hercules, CA) with1% pulsed-field gel electrophoresis (PFGE)-certified agarose (Bio-Rad, Hercules, CA) gels and 0.25× Tris-borate-EDTA in both the running and gel buffers. Run conditions were 6 V/cm, 10- to 60-s switching times, and a 120° included angle for 24 h at 14°C.
RESULTS
Targeted gene deletion of mre11 and rad50.
To determine whether the Rad50 and Mre11 proteins encoded in the genome of Halobacterium were involved in the repair of DNA DSBs, we carried out targeted gene deletions of the rad50 and mre11 genes (single and double deletions) using a modified gene deletion system (see Materials and Methods). For this study, a strain of Halobacterium deficient in the biosynthesis of both tryptophan and uracil (AK071 [ΔtrpA Δura3]) was constructed for use as a background strain for targeted gene deletions, as were strains AK07 (Δura3), AK072 (Δmre11 ΔtrpA Δura3), AK073 (Δrad50 ΔtrpA Δura3), and AK074 (Δmre11 Δrad50 ΔtrpA Δura3). AK071, a double auxotroph, allowed the selection of recombinants on a high-salt dropout medium that we developed for this study, thereby reducing the number of colonies to screen for the mutant genotypes. The mre11 and rad50 genes are located together in an operon on the Halobacterium main chromosome, with the mre11 coding region positioned upstream of the rad50 coding region.
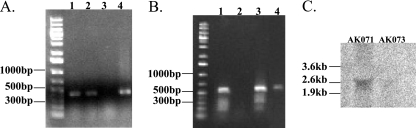
The presence of multiple genome copies in each cell of Halobacterium complicates the construction of targeted gene deletion mutant strains but does not prevent achieving full gene deletion (see references 4, 22, 32, and 42 for examples). In this study, the deletion of all copies of the rad50 gene was achieved after transformation with the appropriate deletion construct. In contrast, copies of both the wild-type gene and the deletion construct were detected by PCR in the clones targeted for mre11 gene deletion after transformation with the appropriate deletion construct. Repeated transformations of those hybrid clones with the same deletion construct eventually resulted in the complete deletion of the mre11 gene. Double mutant strain AK074 (Δmre11 Δrad50) was constructed using strain AK073 (Δrad50). Genotypes of mutant strains AK072, AK073, and AK074 were confirmed by Southern hybridization (Fig. 1) after initial screening by PCR. RT-PCR was used to confirm that single-gene deletions of mre11 or rad50 did not interfere with the transcription of the remaining gene in the operon (Fig. 2A and B). The absence of any transcriptional product from rad50 was further confirmed for AK073 by Northern hybridization and RT-PCR analysis (Fig. 2B and C).
FIG. 2.
mRNA analysis for mre11, rad50, and mre11-rad50 genes in the background strain, deletion mutants, and wild-type Halobacterium. (A and B) RT-PCR analysis showing the presence or absence of mRNA transcripts in the AK071 background strain (lane 1), AK073 (Δrad50) (lane 2), AK072 (Δmre11) (lane 3), and wild-type Halobacterium (lane 4). A 500-bp fragment of the transcript from mre11 was identified in AK071, AK073, and the wild type but was lacking in AK072, as expected (A). A 500-bp fragment of the transcript from rad50 was identified in AK071, AK072, and the wild type but was lacking in AK073, as expected (B). (C) Northern hybridization blot showing the absence of mRNA transcription in AK073 (Δrad50) compared to the observed mRNA in AK071 (background strain). DNA probes were designed to hybridize to a 500-bp region within the rad50 transcript.
Growth and survival of mre11 and rad50 mutants.
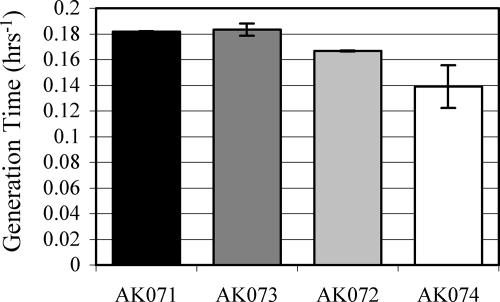
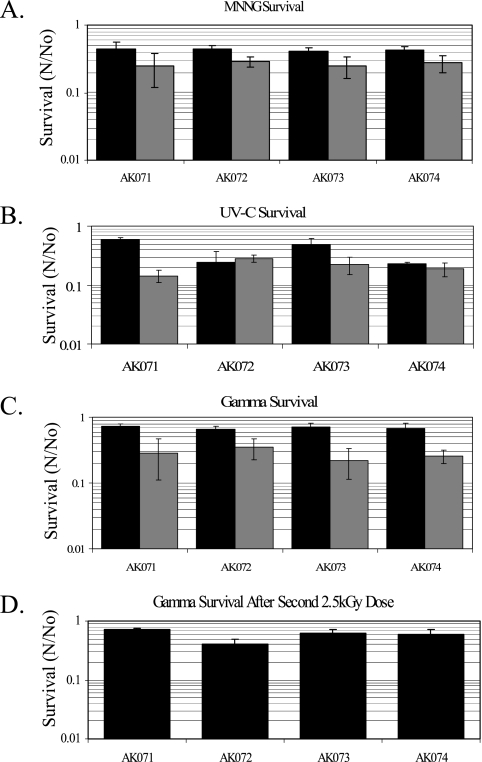
Phenotypic characterization of mutant strains AK072, AK073, and AK074 revealed no temperature-dependent growth defects (data not shown) but did show a slightly slower, albeit reproducible, growth rate under standard culturing conditions compared to that of the background strain (Fig. 3). The mutant strains showed the same level of resistance to the alkylating agent MNNG as the background strain (Fig. 4A) (P value of >0.05, indicating that the change was not statistically significant). In contrast, a decrease in survival was observed in both strains AK072 and AK074 (Δmre11 and Δmre11 Δrad50, respectively) after exposure to 200 J/m2 of UV-C (254 nm) radiation (P values were 0.0414 and 0.0002, respectively), which was eliminated at the higher dose of 350 J/m2, compared to the survival of background strain AK071 (Fig. 4B). Allowing the cells to recover in liquid medium for up to 1 h did not alter the survival of these mutant strains compared to strain AK071, eliminating the possibility of plating as a compounding stress. Surprisingly, no differential survival in any of the mutants was observed after exposure to 2.5 kGy of γ radiation, even after a second 2.5-kGy dose following a 4-h incubation at 42°C with shaking to allow time for the minimal repair of DSBs (Fig. 4C and D) (P > 0.05). Sensitivity to ionizing radiation is the hallmark of mutations of Mre11 and Rad50 in eukaryotic systems (38). For all treatments, the levels of survival of the background strain were comparable to those previously reported for wild-type Halobacterium strains (4, 23).
FIG. 3.
Generation time for strains AK071 (background), AK072 (Δmre11), AK073 (Δrad50), and AK074 (Δmre11Δrad50). Data shown are the averages of at least two replicates, with standard errors.
FIG. 4.
Survival of strains AK071 (background), AK072 (Δmre11), AK073 (Δrad50), and AK074 (Δmre11-Δrad50) after exposure to 50 μg/ml (black) and 100 μg/ml (gray) MNNG (A), 200 J/m2 (black) and 350 J/m2 (gray) UV-C radiation with recovery in the dark (B), γ radiation at doses of 2.5 kGy (black) and 5 kGy (gray) (C), and γ radiation at a dose of 2.5 kGy, followed by a 4-h incubation under standard culturing conditions and followed by a second dose of 2.5 kGy (D). Survival was calculated as the average ratio (N/No) of surviving CFU from treated (N) compared to untreated (No) cultures. Data shown are the averages of at least three replicates, with standard errors shown.
DNA DSB repair in mre11 and rad50 mutants.
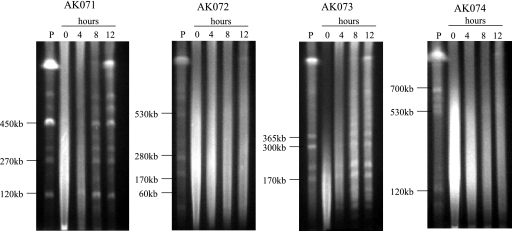
The ability to repair DNA DSBs was assayed in each of the mutant strains after exposure to 2.5 kGy of γ radiation, which represents nearly 80% survival in wild-type Halobacterium strains (23). Samples were taken both prior to irradiation and over a time course of recovery under standard culturing conditions. Agarose plugs containing 1 × 109 cells/ml were made and analyzed by PFGE. In the background strain and strain AK073 (Δrad50), repair of chromosomal fragmentation after exposure to ionizing radiation was initiated within 4 h and completed within 12 h (Fig. 5). In contrast, the recovery process for strains AK072 and AK074 (Δmre11 and Δmre11 Δrad50, respectively) took longer; those mutants showed no evidence of repair until the 12-h time point. Interestingly, mutant strains AK072 and AK074 did not show a decrease in cell survival after exposure to 2.5 kGy of γ radiation, implying that the repair of DNA DSBs is completed, albeit at a reduced rate, and that the inhibitory effect on the kinetics of radiation-induced DSB repair resulted from the absence of Mre11 proteins in the cells.
FIG. 5.
PFGE time course of recovery after exposure to 2.5 kGy of γ radiation. Samples were taken preirradiation (P) and immediately following irradiation (0) and every 4 h during the recovery up to 12 h and were embedded in InCert agarose plugs at a final density of 1 × 109cells/ml. Plugs were digested with XbaI prior to gel electrophoresis. Images were taken from one of three independent replicates of this experiment.
DISCUSSION
Reactive oxygen species, whether produced by desiccation, exposure to ionizing radiation, or the auto-oxidation of dehydrogenases involved in electron transport as part of aerobic respiration, can oxidize DNA base and sugar moieties and generate DNA DSBs (reviewed in references 20 and 21). However, DSB production is not limited to the actions of free-radical species. Base excision repair of oxidized DNA bases found in clusters (6, 13) can lead to the formation of cytotoxic DSBs. Homologous chromosomes serve as templates for recombination repair, leading to the repair of DSBs and yielding advantages to microorganisms with a multiplicity of genomic material.
In yeast, a major pathway for the repair of DSBs is HR, which requires the MR complex (38). This MR complex plays an enzymatic role in DNA end processing and a structural role in DNA end joining (3). In the Archaea, homologs of Mre11 and Rad50 have been biochemically characterized in the hyperthermophile P. furiosus (18, 19), but little is known about the in vivo functions of those proteins. Here, we investigated the cellular role of Rad50 and Mre11 in the radiation-resistant halophile Halobacterium sp. strain NRC-1 through phenotypic analysis of in-frame deletion mutants following DNA-damaging treatments.
Similarly to findings in the yeast mre11 mutant, we found a slight growth defect in Halobacterium mutant strains lacking Δmre11 under standard culturing conditions (35). However, and in contrast to yeast mutants, none of the Halobacterium mutant strains tested displayed increased sensitivity to ionizing radiation or MNNG (alkylating agent) compared to that of the background strain. Sensitivity to ionizing radiation is the defining characteristic of MR complex mutants in yeast, along with sensitivity to alkylating agents (reviewed in reference 38). Studies with bacteria also showed increased sensitivity to ionizing radiation for sbcC and sbcD mutants in B. subtilis and in the radiation-resistant bacterium D. radiodurans (5, 28). Both strains lacking Δmre11 (AK072 and AK074) showed decreased rates of survival after 200 J/m2 of UV-C irradiation compared to the background strain. UV-C irradiation induces mostly bulky lesions in the form of cyclobutane pyrimidine dimers and 6-4 photoproducts, resulting in the production of single-stranded DNA breaks. At 350 J/m2, the survival of all mutant strains closely approximated that of the background strain. This may indicate a minor role for Mre11 in the repair of single-strand breaks at low doses of UV irradiation either as a nuclease or as a damage sensor (33).
Using PFGE analysis, we found that Halobacterium Δmre11 and Δmre11 Δrad50 mutant strains (AK072 and AK074) displayed extensive delay in the repair of DNA DSBs. The deletion of genes encoding the structural homologs of the MR complex, sbcC and sbcD, in D. radiodurans revealed a similar delay in the reconstitution of intact chromosomes as well as delayed growth following γ irradiation (5). In both the background (AK071) and Δrad50 (AK073) Halobacterium strains, the initiation of the repair of chromosomal fragmentation was observed 4 h after exposure to 2.5 kGy of γ radiation, whereas strains lacking the mre11 gene (AK072 and AK074) did not begin to display the repair of DSBs until the 12-h time point. Although the completion of DNA repair was not observed within the 12-h time scale of this experiment, exposure to a second 2.5-kGy dose of γ radiation 4 h after an initial dose failed to result in a significant reduction in the survival of the Δmre11 and Δmre11 Δrad50 mutant strains compared to that of the background strain. These results indicate that despite a reduced rate of homologous recombination repair of DNA DSBs in mutant strains lacking the mre11 gene, a sufficient reduction in chromosomal fragmentation occurred after 4 h of liquid recovery to allow the survival of the strains. The presence of multiple genome copies in Halobacterium cells, resulting in an increase in substrate for recombination repair of DSBs, and the extended recovery period (10 days) during growth on solid medium may have enabled survival in strains lacking the mre11 gene.
The absence of phenotype in mutants lacking the rad50 gene after exposure to a range of DNA-damaging conditions, combined with the decreased rate of DNA DSB repair in Δmre11 mutants observed using PFGE analysis, suggests that there is a decoupling of the archaeal Mre11-Rad50 complex, permitting the recombinational repair of DNA DSBs in Halobacterium in the absence of Rad50. This represents a departure from both bacterial (SbcC and SbcD) and eukaryotic (Mre11 and Rad50) homologs that do not function independently of their role as a complex. Recently, data for Sulfolobus acidocaldarius (33) has lent strength to the hypothesis that Mre11 and Rad50 may have independent functions outside the MR complex in archaea. Using immunodetection methods, Quaiser et al. (33) previously found that Rad50 was bound to DNA under normal growth conditions. Upon chromosomal fragmentation by γ radiation, both the recombinase RadA and the Mre11 protein were recruited to the DNA, suggesting a possible role for Mre11 as an inducible damage sensor (33).
Archaeal Mre11 and Rad50 are able to form a complex, as demonstrated for P. furiosus (18, 19), but the formation of the MR complex is not required for the repair of DNA DSBs in Halobacterium. The absence of Rad50 had no observable effect on the repair of DNA DSBs, whereas the loss of Mre11 was more difficult for cells to compensate for, due to the loss of either the nuclease activity or the DNA damage-sensing activity of the Mre11 protein. In yeast, the deletion of mre11 or rad50 can be complemented by the overexpression of the ExoI 5′-to-3′ exonuclease (26). Conversely, the deletion of both mre11 and exoI increases the radiation-sensitive phenotype compared to that of the yeast mre11 single mutant (29). This suggests an emphasis on the nuclease function of the MR complex in eukaryotes, which may explain the importance of archaeal Mre11 in the repair of DNA DSBs. The development of more effective genetic tools for hyperthermophilic archaea will greatly improve our knowledge of the MR complex through the investigation of NurA activity, a 5′-to-3′ exonuclease associated with the MR complex in the Crenarchaeota (33), and of other nucleases that may compensate for the absence of Mre11.
Taken together, the in vivo results presented here, using the powerful genetic tools available for halophilic archaea, suggest an essential role for Mre11 in the repair of DNA DSBs in Halobacterium. Mre11 is hypothesized to act as a sensor for DNA DSBs and as a nuclease for the generation of single-stranded templates for recombinase activity. Data presented in this study suggest the presence of an alternative nuclease or repair pathway that operates in the absence of Mre11, albeit at a reduced rate of repair, to permit cell survival even after repeated rounds of γ irradiation. The activity of Rad50 and the formation of the MR complex are not required for the repair of DNA DSBs, representing a difference in the roles of the MR complex in the archaea compared to those of the MR complex in the eukaryotic model. The in vivo role of archaeal Rad50 and the identification of alternative nucleases for DNA DSB end processing require further investigation.
Acknowledgments
This work was supported by grants from NASA (NNG05GN58G) and AFOSR (FA95500710158) to J.D.
We thank Russell Rosenblatt for his technical support.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assenmacher, N., and K. P. Hopfner. 2004. MRE11/RAD50/NBS1: complex activities. Chromosoma 113157-166. [DOI] [PubMed] [Google Scholar]
- 3.Aylon, Y., and M. Kupiec. 2004. DSB repair: the yeast paradigm. DNA Repair (Amsterdam) 3797-815. [DOI] [PubMed] [Google Scholar]
- 4.Baliga, N. S., S. J. Bjork, R. Bonneau, M. Pan, C. Iloanusi, M. C. Kottemann, L. Hood, and J. DiRuggiero. 2004. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 141025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentchikou, E., P. Servant, G. Coste, and S. Sommer. 2007. Additive effects of SbcCD and PolX deficiencies in the in vivo repair of DNA double-strand breaks in Deinococcus radiodurans. J. Bacteriol. 1894784-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaisdell, J. O., and S. S. Wallace. 2001. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl. Acad. Sci. USA 987426-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breuert, S., T. Allers, G. Spohn, and J. Soppa. 2006. Regulated polyploidy in halophilic archaea. PLoS ONE 1e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly, J. C., L. A. Kirkham, and D. R. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 957969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantinesco, F., P. Forterre, and C. Elie. 2002. NurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 3537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Constantinesco, F., P. Forterre, E. V. Koonin, L. Aravind, and C. Elie. 2004. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 321439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly, M. J., L. Ouyang, P. Fuchs, and K. W. Minton. 1994. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J. Bacteriol. 1763508-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DasSarma, S., and E. Fleischmann. 1995. Halophilic Archaea: an overview, p. 3-11. In F. Robb, A. Place, K. Sowers, H. Schreier, S. DasSarma, and E. Fleischmann (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Dianov, G. L., P. O'Neill, and D. T. Goodhead. 2001. Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. Bioessays 23745-749. [DOI] [PubMed] [Google Scholar]
- 14.DiRuggiero, J., N. Santangelo, Z. Nackerdien, J. Ravel, and F. T. Robb. 1997. Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1794643-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudas, A., and M. Chovanec. 2004. DNA double-strand break repair by homologous recombination. Mutat. Res. 566131-167. [DOI] [PubMed] [Google Scholar]
- 16.Engel, M. B., and H. R. Catchpole. 2005. A microprobe analysis of inorganic elements in Halobacterium salinarum. Cell Biol. Int. 29616-622. [DOI] [PubMed] [Google Scholar]
- 17.Hackett, N. R., and S. DasSarma. 1989. Characterization of the small endogenous plasmid of Halobacterium strain SB3 and its use in transformation of H. halobium. Can. J. Microbiol. 3586-91. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105473-485. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner, K. P., A. Karcher, D. Shin, C. Fairley, J. A. Tainer, and J. P. Carney. 2000. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 1826036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57395-418. [DOI] [PubMed] [Google Scholar]
- 21.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 2401302-1309. [DOI] [PubMed] [Google Scholar]
- 22.Kaur, A., M. Pan, M. Meislin, M. T. Facciotti, R. El Gewely, and N. S. Baliga. 2006. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 16841-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kottemann, M., A. Kish, C. Iloanusi, S. Bjork, and J. DiRuggiero. 2005. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9219-227. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38233-271. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 16049-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manzan, A., G. Pfeiffer, M. L. Hefferin, C. E. Lang, J. P. Carney, and K. P. Hopfner. 2004. MlaA, a hexameric ATPase linked to the Mre11 complex in archaeal genomes. EMBO Rep. 554-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascarenhas, J., H. Sanchez, S. Tadesse, D. Kidane, M. Krisnamurthy, J. C. Alonso, and P. L. Graumann. 2006. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol. Biol. 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 1591423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 9712176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35667-676. [DOI] [PubMed] [Google Scholar]
- 33.Quaiser, A., F. Constantinesco, M. F. White, P. Forterre, and C. Elie. 2008. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following gamma irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol. Biol. 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahmohammadi, H. R., E. Asgarani, H. Terato, T. Saito, Y. Ohyama, K. Gekko, O. Yamamoto, and H. Ide. 1998. Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents. J. Radiat. Res. (Tokyo) 39251-262. [DOI] [PubMed] [Google Scholar]
- 35.Shor, E., S. Gangloff, M. Wagner, J. Weinstein, G. Price, and R. Rothstein. 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slupphaug, G., B. Kavli, and H. E. Krokan. 2003. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 531231-251. [DOI] [PubMed] [Google Scholar]
- 37.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 192929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Bosch, M., R. T. Bree, and N. F. Lowndes. 2003. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, T., S. Zhang, S. Zhu, D. Sheng, J. Ni, and Y. Shen. 2008. Physical and functional interaction between archaeal single-stranded DNA-binding protein and the 5′-3′ nuclease NurA. Biochem. Biophys. Res. Commun. 367523-529. [DOI] [PubMed] [Google Scholar]
- 41.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4435-445. [DOI] [PubMed] [Google Scholar]
- 42.Whitehead, K., A. Kish, M. Pan, A. Kaur, D. J. Reiss, N. King, L. Hohmann, J. DiRuggiero, and N. S. Baliga. 2006. An integrated systems approach for understanding cellular responses to gamma radiation. Mol. Syst. Biol. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiltzius, J. J., M. Hohl, J. C. Fleming, and J. H. Petrini. 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12403-407. [DOI] [PubMed] [Google Scholar]
- 44.Wyman, C., D. Ristic, and R. Kanaar. 2004. Homologous recombination-mediated double-strand break repair. DNA Repair (Amsterdam) 3827-833. [DOI] [PubMed] [Google Scholar]
- 45.Zahradka, K., D. Slade, A. Bailone, S. Sommer, D. Averbeck, M. Petranovic, A. B. Lindner, and M. Radman. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443569-573. [DOI] [PubMed] [Google Scholar]