Abstract
High-affinity iron acquisition in Bacillus subtilis is mediated via the bacillibactin catechole siderophore pathway. Three of the four essential pathway steps, bacillibactin synthesis, Fe-bacillibactin uptake, and Fe-bacillibactin hydrolysis have been characterized previously. The functional and regulatory components for bacillibactin secretion, the second step of the siderophore pathway, remained unknown. In this study, the screening of a B. subtilis exporter mutant library led to the identification of the YmfE major facilitator superfamily (MFS)-type transporter as a target for bacillibactin export. Analysis of iron-limited ymfE mutant cultures displayed an eightfold reduced bacillibactin secretion and, on the other hand, a 25-fold increased secretion of the bacillibactin precursor 2,3-dihydroxybenzoate. Investigation of the regulatory aspect revealed that bacillibactin secretion is, in contrast to all other components of the pathway, independent of the ferric uptake repressor Fur. Indeed, the MerR-type transcriptional regulator Mta was found to activate both bacillibactin secretion and ymfE gene expression, exposing Mta as an additional regulatory member of the bacillibactin pathway.
Siderophores are secreted by a plethora of microorganisms for high-affinity iron acquisition in iron-limited habitats. They represent important virulence factors of many pathogens, especially of those that colonize mammalian hosts. The elucidation of siderophore pathways has become an emerging strategy to define novel antibiotic targets (22, 26). In this light, it is evident that the cellular components of siderophore secretion, in contrast to siderophore biosynthesis, Fe-siderophore uptake, and iron release, are rarely investigated. Only a few siderophore secretion systems, mainly in gram-negative bacteria, have been described. In most cases, they are represented by transporters of the major facilitator superfamily (MFS) and are transcriptionally regulated by the ferric uptake repressor Fur. These exporters comprise the proteins EntS in Escherichia coli (11), YhcA in Erwinia chrysanthemi (10), LbtB in Legionella pneumophila (1), PvsC in Vibrio parahaemolyticus (34), and CsbX in Azotobacter vinelandii (24), which are used for secretion of enterobactin, achromobactin, legiobactin, vibrioferrin, and protochelin-like siderophores, respectively. The MFS-type alcaligin exporter AlcS in Bordetella pertussis and Bordetella bronchiseptica is an example of a siderophore secretion system that is regulated independent of Fur and that is constitutively expressed and acts as part of a regulatory circuit that depends on the intracellular siderophore-sensing regulator AlcR (6). A further type of a siderophore efflux system is the MexA-MexB-OprM system of the resistance-nodulation-cell division superfamily in Pseudomonas aeruginosa, which was suggested to be involved in pyoverdine secretion (19, 25). The only known siderophore exporter in gram-positive bacteria is the ABC-type exporter ExiT for exochelin secretion in Mycobacterium smegmatis, which is regulated by the Fur analog IdeR (38). Recently, another ABC-type of secretion system, IroC, was identified as an exporter of enterobactin and its glucosylated derivative salmochelin in Salmonella enterica serovar Typhimurium (9). In addition to transport across the cytoplasmic membrane, the TolC outer membrane factor was identified as a requirement for enterobactin secretion across the outer membrane in E. coli (5).
The gram-positive model bacterium Bacillus subtilis uses the (2,3-dihydroxybenzoate-glycine-threonine)3 triscatecholate trilactone siderophore bacillibactin (BB) for high-affinity iron acquisition. BB is synthesized by the nonribosomal peptide synthetase DhbEBF that condenses and subsequently cyclotrimerizes the precursors 2,3-dihydroxybenzoate (DHB), glycine, and threonine to yield the functional siderophore (20). The dhbEBF genes are cotranscribed with the genes dhbA (coding for a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase) and dhbC (coding for an isochorismate synthase) that, together with the isochorismate lyase activity of dhbB, permit the synthesis of DHB using chorismate as a primary metabolism precursor (27, 28). After BB secretion, which has not been characterized, iron charging takes place in the extracellular environment, and Fe-BB is shuffled back into the cell via the ABC-type transporter FeuABC-YusV (21, 23). For the matter of intracellular iron release, Fe-BB is subsequently hydrolyzed by the Fe-BB esterase BesA (YuiI) (21). The main regulator of the BB pathway is represented by Fur, which represses BB synthesis, Fe-BB uptake, and hydrolysis under nonlimiting conditions of iron availability (3). Additionally, the feuABC genes for Fe-BB uptake are positively regulated by the BB transport regulator Btr (YbbB) that is activated upon intracellular sensing of (Fe)-BB (12).
This study focused on the investigation of the remaining essential BB pathway step, siderophore secretion, and presents new members of the pathway that were found to be involved in this process. The MFS-type exporter YmfE was identified during a screening of B. subtilis exporter single mutants for the loss of BB efflux. The lack of functional YmfE led to a severe defect in BB secretion and diminished growth in iron-limited medium. Furthermore, it was shown that BB secretion is not dependent on extracellular levels of iron and, hence, Fur regulation. The multidrug-efflux activator Mta was found to affect both BB secretion and ymfE expression, revealing Mta as a new regulatory component of the BB pathway. The identification of functional and regulatory components for BB secretion allows us to describe for the first time a closed, high-affinity iron acquisition pathway in B. subtilis.
MATERIALS AND METHODS
Strains and general methods.
The bacterial strains used in this study are listed in Table 1. Strains in liquid culture were grown under agitation (250 rpm) at 37°C. Strains on LB plates were incubated for growth at 37°C and then stored at room temperature. Culture plate medium was prepared by adding 1.2% (wt/vol) Agar No. 1 to the growth medium, which was then heated at 121°C and 1.5 × 105 Pa for 30 min. Antibiotics for selection of B. subtilis strains were used at the following concentrations: erythromycin, 1 μg/ml; lincomycin, 25 μg/ml; spectinomycin, 200 μg/ml; and kanamycin, 10 μg/ml. DNA preparations and transformations were carried out as described previously (16, 29). For growth under conditions of iron depletion, Belitsky minimal medium without citrate and iron salt was used (33). Stock solutions were stored in polyethylene tubes, and cultures were grown in polycarbonate flasks to avoid iron contamination during growth. The absolute iron content of this medium is about 0.1 μM as determined previously by inductively coupled plasma mass spectrometry (21). For iron repletion controls, 200 μM FeCl3 was added. To restore BB synthesis in strain BMM100 and controls, 3 mM DHB was added.
TABLE 1.
Strains used in this study
| Strain | Genotype | Functional category of deleted/disrupted gene(s) | Source or reference |
|---|---|---|---|
| ATCC 21332 | Wild type | 21 | |
| JJM405 | ΔdhbF2::kan | Dimodular nonribosomal peptide synthetase for BB assembly (complete second module deleted) | 20 |
| BMM100 | ΔdhbC::erm | Isochorismate synthase | 21 |
| BMM141 | ymfE::pMUTIN | MFS-type transport | This studya |
| BMM142 | mta::pMUTIN | Transcriptional activator of multidrug-efflux transporters; disruption leaves residues 1 to 99 of the N terminal DNA binding domain of Mta (N-Mta) under native control | This study |
| BMM143 | Δmta::spc | Transcriptional activator of multidrug-efflux transporters | This study |
| BMM150 | ΔywjA::erm | ABC-type transport (Fur target) | This study |
| BMM161 | yttB::pMUTIN | MFS-type transport | This study |
| BMM162 | yubD::pMUTIN | MFS-type transport | This study |
| BMM163 | ygaD::pMUTIN | ABC-type transport | This study |
| BMM164 | yvmA::pMUTIN | MFS-type transport | This study |
| BMM165 | yvkA::pMUTIN | MFS-type transport | This study |
| BMM166 | yvaE::pMUTIN | DMT-type transport | This study |
| BMM167 | ywfF::pMUTIN | MFS-type transport | This study |
| BMM168 | yusP::pMUTIN | MFS-type transport | This study |
| BMM169 | mrpF::pMUTIN | Efflux for pH homeostasis | This study |
| BMM170 | ykuC::pMUTIN | MFS-type transport | This study |
| BMM171 | ykkD::pMUTIN | DMT-type transport | This study |
| BMM172 | ykkC::pMUTIN | DMT-type transport | This study |
| BMM173 | yeaB::pMUTIN | CDF-type transport | This study |
| BMM174 | yerP::pMUTIN | RND-type transport | This study |
| BMM175 | yvqJ::pMUTIN | Macrolide efflux | This study |
| BMM176 | ebrA::pMUTIN | DMT-type transport | This study |
| BMM177 | ebrB::pMUTIN | DMT-type transport | This study |
| BMM178 | ymfD::pMUTIN | MFS-type transport | This study |
| BMM179 | yitG::pMUTIN | MFS-type transport | This study |
| BMM180 | yitZ::pMUTIN | MFS-type transport | This study |
| BMM181 | tatAC::pMUTIN | Twin arginine-dependent transport | This study |
| BMM182 | tatAD::pMUTIN | Twin arginine-dependent transport | This study |
| BMM183 | tatAY::pMUTIN | Twin arginine-dependent transport | This study |
| BMM184 | tatAC::pMUTIN | Twin arginine-dependent transport | This study |
| BMM185 | tatCY::pMUTIN | Twin arginine-dependent transport | This study |
| BMM186 | yfmI::pMUTIN | Macrolide efflux | This study |
| BMM187 | ywjA::pMUTIN | ABC-type transport (Fur target) | This study |
| BMM188 | yhcA::pMUTIN | MFS-type transport | This study |
| BMM189 | yfiS::pMUTIN | MFS-type transport | This study |
| BMM190 | ybbB::pMUTIN | Transcriptional activator for Fe-BB uptake (Fur target) | This study |
| BMM191 | yfiB::pMUTIN | ABC-type transport | This study |
| BMM192 | yfiC::pMUTIN | ABC-type transport | This study |
| BMM193 | blt::pMUTIN | MFS-type transport | This study |
| BMM194 | yceJ::pMUTIN | MFS-type transport | This study |
| BMM195 | ycnB::pMUTIN | MFS-type transport | This study |
| BMM196 | yfiU::pMUTIN | MFS-type transport | This study |
| BMM197 | yfkF::pMUTIN | MFS-type transport | This study |
| BMM198 | yfkL::pMUTIN | MFS-type transport | This study |
| BMM199 | yhjO::pMUTIN | MFS-type transport | This study |
| BMM200 | yqjV::pMUTIN | MFS-type transport | This study |
| BMM201 | yuxJ::pMUTIN | MFS-type transport | This study |
| BMM202 | ycbK::pMUTIN | DMT-type transport | This study |
| BMM203 | yjbB::pMUTIN | Macrolide efflux | This study |
| BMM204 | yrhP::pMUTIN | Putative lysine exporter | This study |
| BMM205 | yheH::pMUTIN | ABC-type transport | This study |
| BMM206 | yknU::pMUTIN | ABC-type transport | This study |
| BMM207 | yknV::pMUTIN | ABC-type transport | This study |
| BMM208 | yvcC::pMUTIN | ABC-type transport | This study |
| BMM209 | yhbJ::pMUTIN | Unknown type of multidrug efflux | This study |
| BMM210 | ywfA::pMUTIN | MFS-type transport | This study |
| BMM211 | ywcC::pMUTIN | Putative transcriptional regulator | This study |
| BMM212 | ysbA::pMUTIN | Putative transcriptional regulator | This study |
| BMM213 | yncC::pMUTIN | MFS-type transport | This study |
| BMM214 | ywbG::pMUTIN | Predicted exporter | This study |
All of the donor strains used in this study for transformation into the ATCC 21332 background were obtained from the BFA mutant library (18).
Construction of deletion mutants.
Deletion mutants were generated according to the PCR synthesis method of marker cassettes with long flanking homology regions (37). All PCRs were carried out with a Platinum Pfx DNA polymerase possessing proofreading activity. In a first round of PCR, chromosomal DNA of B. subtilis ATCC 21332 was used as a template to amplify the up- and downstream regions of the target gene. The primers for generation of the ΔywjA mutant were the pair 5′-CGATGCGGCAGGAAGCATGAAG-3′ and 5′-GCAAGTCAGCACGAACACGAACCCTTACAGAACACCTGAAAACAGGCG-3′ and the pair 5′-CTATTTTTAATAGTTATCTATTATTTAACGGGAGGAAAGACCTGATCGAAGCCGGAGGC-3′ and 5′-CATGAAAAATATCAAGTCTCTGAAAGTAGCC-3′ (sites generating complementary 3′ ends to the up- and downstream flanks of the pMUTIN erythromycin resistance cassette are underlined). The primer for generation of the Δmta mutant were the pair 5′-CAGCACCCGGCCAAGAAC-3′ and 5′-CTCTTGCCAGTCACGTTACGTTATTAGAACGCCTGATATCTCCGCCAC-3′ and the pair 5′-GCATAGTTAAGCCAGCCCCGCATCAGGAAAACCCCCGGC-3′ and 5′-AGAAGGTGTTCAGGTCAGAGTGC-3′ (sites generating complementary 3′ ends to the up- and downstream flanks of the pUS19 spectinomycin resistance cassette are underlined). The resulting PCR products were used in a second round of PCR generating a fusion construct of the homologous genomic regions and the resistance marker that was used as a template. Transformants were selected on antibiotic-containing LB plates. Chromosomal DNA of the mutants was isolated, and the deletions were confirmed by PCR using the distal fusion construct primers.
Siderophore secretion assay.
A chrome azurol sulfonate-hexadecyltrimethylammonium bromide (CAS-HDTMA) stock solution was prepared according to published protocols (30). The CAS-HDTMA solution was mixed 1:10 with Belitsky minimal medium containing 1.2% agar-agar. Siderophore secretion is indicated by bright halo formation around the cell colonies. To test halo formation of B. subtilis wild type (WT) and mutants, strains were first grown on LB plates over night at 37°C. From these plates, cells were spotted onto CAS agar plates that were incubated for 20 h at 30°C and then at room temperature. After 48 h, the plates were scanned for equal comparison of the secretion phenotypes.
Mutant library screening.
Chromosomal DNA of B. subtilis Functional Analysis (BFA) mutant strains carrying single gene disruptions in the B. subtilis 168 background (trpC2 sfp0) was isolated and used for transformation into B. subtilis ATCC 21332 (sfp+). Transformants were selected on erythromycin/lincomycin-containing plates. At least two transformants from each strain were used for BB secretion analysis. For this purpose, the transformants were grown in parallel with WT on CAS agar containing 0.5 mM isopropyl-β-d-thiogalactopyranoside to avoid polar effects (18). To eliminate false positives, the transformants were screened in parallel on blood agar plates (BD) for surfactin secretion as a marker for a functional nonribosomal peptide synthetase system. Additionally, all strains were tested for tryptophan prototrophy in Belitsky minimal medium to avoid selection of the B. subtilis 168 donor strain that occasionally survived in spore form on the transformation plates. Target gene disruption in the ymfE mutant was confirmed by PCR with the primers 5′-ATGGTCGCTGGCTTTATACGGTC-3′ and 5′-GACCGTCCCTAAGTTTTTTTCTTC-3′. Disruption of the mta gene in the Mta mutant consisting of the N-terminal binding domain of Mta (N-Mta) was checked using the primers 5′-ATGAAATATCAAGTTAAACAAGTGGCG-3′ and 5′-CGGCCGGGGGTTTTCCTGATG-3′, and the presence of the N-terminal part of the gene (downstream of its native promoter) was confirmed using the primers 5′-CTTTTATACACTATTTGTGAGAAG-3′ and 5′-TTGAATCATCTCATCCATTCTTTG-3′.
Analytical chromatography.
Culture supernatant samples yielded from 20-ml cell cultures (each strain grown in triplicates) after 10 h of growth were analyzed by high-performance liquid chromatography coupled with mass spectrometry (HPLC-MS). Analysis was performed with a Macherey-Nagel 125/2 Nucleodur 100-3 C18 column (using a flow rate of 0.3 ml/min, column temperature of 45°C, and a linear gradient from 10 to 95% water-methanol-0.05% formic acid over 20 min), which was coupled with an electrospray ionization-quadrupole mass spectrometer. After background subtraction, peak area integration of the obtained UV signals (at 215 nm) resulted in the relative quantification of secreted BB and DHB amounts, which were normalized to the culture optical densities (ODs). For control analysis of the intracellular fractions, the cell pellets (fresh weight ranging from 2 to 6 mg) were washed twice with Tris-EDTA buffer, and the cells were disrupted by sonication. After the cell debris was removed by centrifugation, the lysate was extracted three times with ethyl acetate; the pellet that remained after evaporation was dissolved in 300 μl of 20% methanol, and samples were subjected to HPLC-MS analysis. The threshold for UV detection of BB was at a concentration of ∼0.1 μM.
Transcriptional analysis.
B. subtilis strains were cultivated in iron-limited Belitsky minimal medium. An iron-deficient preculture was inoculated into fresh medium to an initial OD at 600 nm (OD600) of 0.05, and cells were harvested at an OD600 of 0.35. Total RNA was isolated from these cell cultures according to the acidic phenol method (17). The RNA was quantified by measuring the UV absorption at 260 nm and subsequent multiplication with the RNA-specific factor 40. The ratio of RNA to protein (260 nm/280 nm) was above 1.65 in all samples. Quality of the RNA was further checked by comparing the 16S and 23S rRNA bands after denaturing gel electrophoresis. One and two micrograms of total RNA from each strain were dotted onto a nylon membrane using a dot blot apparatus and hybridized after UV cross-linking with a UTP-11-digoxigenin-labeled antisense RNA probe specific for ymfE mRNA. The riboprobe was synthesized by in vitro transcription using T7 RNA polymerase and a PCR product of the ymfE gene containing a T7 promoter extension that was introduced by the primer pair 5′-CGGATTACTGGCAATCCCAC-3′ and 5′-CTAATACGACTCACTATAGGGAGTAGCATCTACAGTAAAGAGCAC-3′ (T7 promoter sequence is underlined). After hybridization and washing, the filters were treated with a digoxigenin-specific antibody fragment conjugated with an alkaline phosphatase (Roche) and AttoPhos (Amersham Biosciences) as an enhanced chemifluorescence substrate. The hybridization signals were detected with a Storm860 fluorescence imager, and relative signal quantification was done with ImageQuant software.
RESULTS
BB secretion is Fur independent.
Since the majority of siderophore exporters are transcriptionally regulated by the global Fur repressor, it was initially examined whether the BB secretion system is among the genes of the B. subtilis Fur regulon, which has been extensively studied by transcriptomic, proteomic, and motif search analyses (3, 23). A functional database search of the B. subtilis Fur regulon led to the identification of only one Fur-regulated exporter, which is the ABC-type secretion system YwjA. To investigate the possible role of ywjA in BB secretion, a ywjA deletion mutant was constructed and assayed for BB secretion. The cells were applied onto CAS agar plates, on which BB secretion can be detected and estimated semiquantitatively by comparing the emerging halo zones around the cell colonies. However, no differences in BB secretion in the ΔywjA and WT strains were observed in this assay (Fig. 1A). A subsequent HPLC-MS analysis of secreted BB amounts did not show significant differences in BB secretion of ΔywjA and WT in iron-limited liquid cell cultures (Fig. 1B). Thus, it was concluded that YwjA is not essential for BB export.
FIG. 1.
(A) CAS agar phenotypes of the WT and ΔywjA mutant. (B) HPLC traces of BB secreted in iron-limited cultures of WT and ΔywjA mutant strains after 10 hours of growth. UV detection was done at 215 nm, and compound identity was confirmed by subsequent MS analysis. After background subtraction, peaks were integrated for quantitative determination of BB secretion.
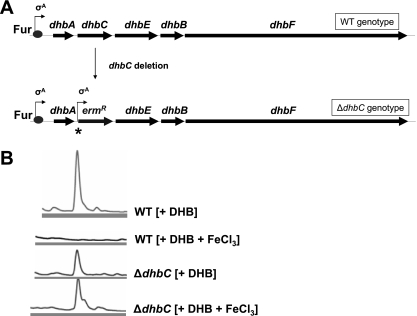
Next, it was generally examined whether BB secretion is dependent on Fur regulation. To investigate this issue, the levels of BB secretion under conditions of iron depletion and iron repletion were compared. As an essential requirement for this analysis, BB biosynthesis was uncoupled from iron-dependent expression. This was achieved by using a deletion mutant of the dhbC gene in which the downstream BB assembly genes dhbEBF are set under the control of a constitutive σA promoter provided by the introduced erythromycin resistance marker cassette (Fig. 2A). Since it is possible to restore BB synthesis in the DHB-deficient ΔdhbC mutant by supplementing the cultures with DHB (21), the ΔdhbC mutant was used to investigate iron-dependent secretion of constitutively synthesized BB. For this purpose, the WT (as control) and ΔdhbC mutant were grown in iron-limited minimal medium in the presence of 3 mM DHB with or without the addition of 200 μM FeCl3. The supernatants were analyzed after 10 h of growth by HPLC-MS (Fig. 2B). As expected, secretion of BB in the WT culture was strictly iron dependent. In contrast, the ΔdhbC mutant showed a completely iron-independent BB secretion. To test if the detected amounts of BB in the culture supernatants partially resulted from cell lysis during growth, the intracellular fractions of the cultures were analyzed for their BB contents by HPLC-MS. With the same UV light detection method applied, there was no intracellular BB found in the HPLC trace (data not shown) according to a previous study (21), which excluded substantial BB release by cell lysis. These data led to the conclusion that, if BB biosynthesis is released, BB secretion occurs independently of iron availability and, thus, is not subject to Fur regulation.
FIG. 2.
(A) Scheme of genotype alteration after dhbC deletion. The dhbC gene was replaced by an erythromycin resistance cassette that is transcribed without terminator from a constitutive σA promoter (indicated by the star). (B) HPLC traces of secreted BB in iron-limited cultures of WT and ΔdhbC mutant strains after 10 hours of growth. The addition of DHB (3.0 mM final concentration) and FeCl3 (200 μM final concentration for iron repletion) is indicated in brackets. Detection was done at 215 nm, and compound identity was confirmed by subsequent MS analysis.
YmfE is the major component of BB secretion.
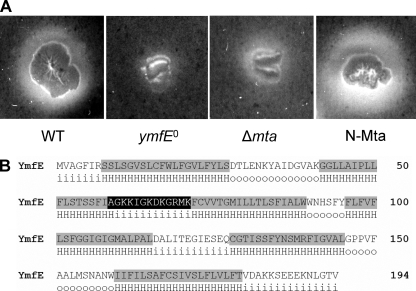
In order to identify the BB exporter and its regulatory component(s), 56 mutants of secretion systems (mainly from the MFS and ABC types) and transcriptional regulators were used in total to screen for effects on BB secretion (Table 1). The mutants were grown on CAS plates in parallel with the WT, and three mutants showed significant differences in their BB secretion phenotypes. The mutant of the transcriptional regulator YbbB (Btr) showed strongly increased halo formation (data not shown). According to the recently described function of Btr as an Fe-BB uptake activator (12), this observation was explained by extracellular (Fe-)BB accumulation due to blocked uptake of the iron-charged siderophore. Interestingly, a similar increase in halo formation was observed with the mutant of the transcriptional regulator Mta (Fig. 3A), which will be further examined below. In contrast, the mutant of the predicted MFS-type exporter YmfE displayed drastically decreased halo formation (Fig. 3A), indicating that YmfE is a functional component for BB secretion.
FIG. 3.
(A) CAS agar phenotypes of ymfE, Δmta, and N-Mta mutants in comparison with the WT after 2 days of incubation at 30°C. (B) Amino acid sequence of YmfE with the membrane-spanning domains (shaded in gray) predicted by HMMTOP (35, 36). The amino acids of the conserved motif between transmembrane regions two and three are shaded in black. H, hydrophobic α-helical regions; i, in (intracellular loop); o, out (extracellular loop).
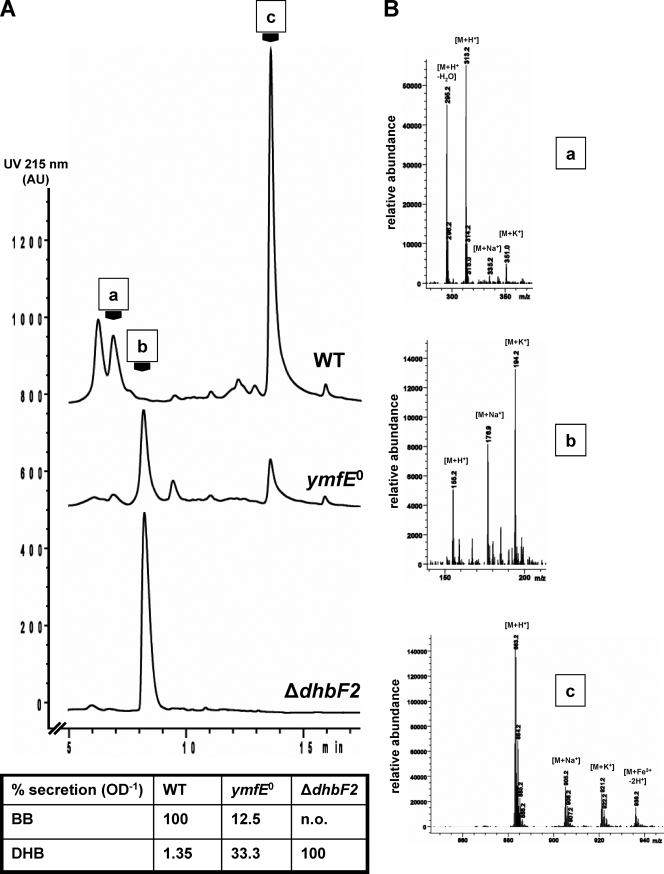
To examine the growth of the ymfE mutant and to quantify the secreted amounts of BB, the strain was grown in minimal medium under iron depletion and repletion conditions in parallel with the WT and the BB biosynthesis mutant ΔdhbF2 that lacks a major part of the nonribosomal peptide synthetase (Fig. 4). While growth of the ymfE mutant was not significantly affected under iron repletion, the mutant showed a markedly diminished growth rate during iron starvation, which was similar to that of the ΔdhbF2 mutant. Samples of the culture supernatants were used for subsequent HPLC-MS analysis to determine the BB and DHB contents (Fig. 5). The relative quantification of the compounds showed that BB secretion was about eightfold reduced in the ymfE mutant compared to the WT. In contrast, secretion of the BB precursor DHB was about 25-fold enhanced, which was still below the level in the BB-deficient ΔdhbF2 mutant. Thus, the reduction of BB secretion in the ymfE mutant to about 10% under iron-limited growth conditions revealed YmfE to be the major BB secretion component of B. subtilis.
FIG. 4.
Iron-dependent growth analysis in Belitsky minimal medium. Growth curves of the WT (⋄/⧫), ymfE mutant (○/•), and ΔdhbF2 mutant (□/▪) strains. Growth was without (open symbols) or with (filled symbols) 200 μM FeCl3.
FIG. 5.
HPLC-MS analysis and relative quantification of secreted BB and DHB amounts in the WT, ymfE mutant, and ΔdhbF2 mutant cultures. (A) The UV absorption chromatograms of the HPLC traces are shown. UV peaks a, b, and c correspond to the detected masses of DHB-Gly-Thr ([M+H+] = 313), DHB ([M+H+] = 155), and BB ([M+H+] = 883) shown in panel B. The UV peak in front of peak a (retention time, 6 min) corresponds to DHB-Gly (itoic acid) with [M+H+] = 212 (mass spectrum not shown). The table gives the relative amounts of secreted BB and DHB per OD unit of the culture. The integrated UV peak areas of BB and DHB were set to 100% in the WT and ΔdhbF2, respectively. n.o., not observed. (B) Mass spectra corresponding to UV peaks a, b, and c.
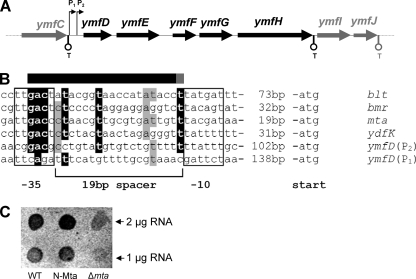
A domain architecture analysis of the 194-amino-acid protein YmfE revealed the presence of six membrane-spanning domains (Fig. 3B). The conserved 13-amino-acid motif of MFS-type transporters that is suggested to be involved in substrate channeling (32) is present at the typical position between the second and third transmembrane domains. A DNA pattern analysis revealed that the nearest transcription terminators to ymfE are located immediately downstream of the genes ymfC and ymfH, suggesting that ymfE is cotranscribed in a pentacistronic operon that is controlled by a promoter region upstream of ymfD, which will be further examined below (Fig. 6A). Incidentally, a further putative MFS-type exporter, ymfD, is located upstream of ymfE. However, since the ymfD mutant was not affected in halo formation on CAS agar, the idea that ymfD is involved in BB secretion was excluded. The genes ymfF, ymfG, and ymfH are predicted to encode peptidases with unknown specificity.
FIG. 6.
(A) Scheme of the putative ymfDEFGH operon structure. Predicted transcription terminators (T) are indicated. The two potential promoters upstream of ymfD are indicated with P1 and P2. (B) Alignment of Mta-dependent promoter sequences of genes blt, bmr, mta, and ydfK (4) with potential MerR-type promoters P1 and P2 located upstream of ymfD. Sequence parts showing a conservation higher than 80% are printed in white letters on black, and parts showing conservation higher than 60% are shaded gray. The bar above the sequence alignment indicates the essential part of the Mta binding site (black) and the part where Mta-binding is partially known (gray) (4). (C) Transcriptional analysis of Mta-dependent ymfE expression. Total RNA (1 and 2 μg) isolated from WT, Δmta, and N-Mta mutant was applied to a nylon membrane by dot blotting and hybridized with a digoxigenin-UTP-labeled antisense riboprobe specific for the ymfE transcript. Hybridization signals were detected by using a digoxigenin-specific antibody fragment conjugated with alkaline phosphatase and AttoPhos as a chemifluorescence substrate.
The Mta multidrug-efflux regulator affects BB secretion.
As mentioned above, the mutant of the gene mta, which codes for a MerR-type multidrug-efflux activator (4), showed an increased accumulation of secreted BB in the CAS plate assay. This led to a closer investigation of the original mutant construction properties of this strain. The pMUTIN construct that was used for integration into mta disrupts the gene after the 99th amino acid codon, thus leaving the dimerizing N-terminal DNA-binding domain (N-Mta), which covers the first ∼100 amino acids of the protein, essentially intact (13). In a previous study, it was shown that the N-terminal domain of Mta is sufficient to activate the expression of Mta-regulated export genes, which leads to elevated secretion of their corresponding substrates (4). Thus, in accordance with the phenotype observed in this study, it was assumed that the N-terminal binding domain of Mta and, thus, Mta itself are involved in the regulation of BB secretion. To verify this hypothesis, a mutant was constructed in which the complete mta gene was deleted. This Δmta mutant was assayed for BB secretion and showed, in contrast to the N-Mta phenotype, reduced secretion of BB on CAS agar plates (Fig. 3A). To quantify BB secretion in the examined mutants, the strains were grown in parallel with the WT in iron-limited minimal medium, and the extracellular culture fractions were analyzed by HPLC-MS. The BB amounts in the culture supernatants of the Δmta and N-Mta strains were 0.55-fold and 2.5-fold, respectively, in comparison to those detected in the WT culture. Altogether, these findings indicated that Mta is an activator of BB secretion.
Mta induces expression of ymfE.
Subsequently, the possibility of Mta-dependent expression of ymfE was investigated. The intergenic region of ymfC and ymfD contains sequences of two potential MerR-type promoters, P1 and P2, indicated by the typical 19-bp-long spacer between the −35 and −10 consensus elements (Fig. 6B). Sequence alignments with known Mta-dependent MerR-type promoters reveal that the spacer region of the proximal promoter P2 in particular shows significant similarity with conserved Mta binding motifs that are typically found within this region (4). To elucidate possible effects on ymfE expression in Δmta and N-Mta, the strains were, in parallel with WT, cultured in iron-limited minimal medium and then transcriptionally analyzed by incubating the isolated total RNA with a digoxigenin-11-UTP-labeled antisense RNA probe specific for ymfE mRNA (Fig. 6C). The transcription of ymfE was found to be approximately threefold downregulated in Δmta, while the N-Mta strain showed approximately twofold induction of ymfE expression. The results of the transcriptional analysis thus correlate with the observed enhanced BB secretion in this strain, demonstrating that Mta is involved in ymfE expression as a positive regulatory factor.
DISCUSSION
Secretion of the BB siderophore is the last unidentified step of high-affinity iron acquisition in B. subtilis. The present study shows that YmfE represents the major functional component that is involved in this process. YmfE is predicted to be an MFS-type efflux with six membrane-spanning domains, and, thus, it is not a typical member of the MFS. In general, MFS-type secretion systems are known to possess 12 or 14 membrane-spanning α-helices within a single folding unit, which are organized as two bundles of six (plus two) that form a hydrophobic cavity within the plane of the bilayer (15, 32). Thus, it is likely that YmfE has to adopt an oligomeric state (possibly as a homo- or heterodimer) to gain a similar domain arrangement for substrate recognition and translocation. At this point, it cannot be ruled out that more functional components would be needed to form the functional transport system.
Disruption of the ymfE gene resulted in a reduction of BB secretion to about 10% of the WT level. In the pathway of the structurally related E. coli siderophore enterobactin, deletion of the EntS exporter resulted in a 50% reduction of enterobactin export, and, thus, the presence of at least one more enterobactin-specific export system has been suggested (11). Since the remaining BB export capacity in the absence of YmfE is comparatively low, compensating exporters might be represented by efflux systems with low substrate specificity, such as multidrug transporters, which are highly abundant in Bacillus spp. These additional transport activities might be part of a compensatory strategy that helps to prevent intracellular accumulation of nonloaded BB. Similar to the B. subtilis ymfE mutant, the E. coli entS mutant showed increased efflux of siderophore precursors. It was suggested that blocked siderophore secretion results in inefficient siderophore biosynthesis (due to equilibrium displacement or possibly due to abolished direct interaction of the synthesis machinery and the export system) and hence leads to an increased release of biosynthesis precursors and/or intermediates (11, 22). Export systems for these precursors have not been described so far.
The fact that BB secretion in B. subtilis was found to be Fur independent is neither the rule nor an exception concerning the regulation of siderophore secretion systems. Most of the known siderophore export systems are Fur regulated; however, the alcaligin exporter in Bordetella spp. is constitutively expressed (6). Interestingly, both in the BB and the alcaligin pathways, positive regulators (AlcR in Bordetella and Btr in B. subtilis) were found to bind the intracellular siderophores as inducers for transcriptional activation of siderophore pathway genes (7, 12). Both deletion and overexpression of the alcaligin exporter AlcS revealed alterations in the intracellular alcaligin pool, which led to imbalanced induction of the AlcR-regulated systems (6). Since Fur derepression is a relatively strong transcriptional response (2), the maintenance of appropriate intracellular siderophore levels in such finely tuned inducer-dependent systems might have been accomplished by providing siderophore export with alternative regulation mechanisms. This study demonstrates that the MerR-type multidrug-efflux activator Mta is involved in transcriptional activation of the BB secretion system. Thus, in addition to Fur and Btr, a third family of transcriptional regulators has to be added to the regulatory circuit of the BB pathway. In general, MerR proteins possess an N-terminal domain that contains the conserved DNA-binding motif and the dimerization region (8, 31) and a C-terminal modulation domain, which largely varies in size and is essential for binding of the coactivating ligand(s) (4, 14). The native ligand of Mta has not been identified yet, and further studies will address this aspect with regard to the involvement of the regulator in BB secretion. Mta was previously reported to autoregulate its expression and to activate three further export systems (4). Two of them, Bmr and Blt, are well-characterized multidrug-efflux systems with broad substrate specificity. The third one, YdfK, is an uncharacterized membrane protein with unknown function. However, both Bmr and Blt and putatively also YdfK are regulated by further “target-specific” MerR-type regulators (4), and the functional reason for their additional Mta-mediated activation has not been elucidated so far. It is now conceivable that Mta-dependent regulation of these other export systems is also linked to BB secretion, possibly as part of the compensating strategy. Concurrently, since the present results do not exclude the possibility that Mta is the exclusive regulator of YmfE, the regulation of BB secretion may be further ramified. However, YmfE and Mta were shown to be the main functional and regulatory components of BB secretion, respectively, and their addition to the previously identified components for BB utilization leads to the first closed BB pathway model in B. subtilis (Fig. 7). Ongoing studies will focus on the molecular details of Mta-dependent ymfE activation with respect to ligand sensing, promoter binding, and possible cotranscription of adjacent genes and their functions. Other studies will also investigate whether additional components are involved in secretion of BB as well as its precursors and, in the same way, its degradation products of intracellular hydrolysis.
FIG. 7.
Current model of the BB pathway. Functional components for biosynthesis, secretion, uptake, and hydrolysis are indicated in rectangular boxes. Regulatory components are shown in star-shaped boxes. Positive or negative regulation is indicated at the arrows with plus or minus sign, respectively. Iron or ligand sensing (s) is also indicated. Dashed arrows and question marks in the case of Mta ligand sensing and degradation or export of BB hydrolysis products indicate that these processes are to be analyzed. NRPS, nonribosomal peptide sythetase.
Acknowledgments
We thank the group of E. Bremer (Marburg) for help with chemiluminescence detection. We are indebted to O. P. Kuipers (Groningen), H. Yoshikawa (Tokyo), M. Hecker (Greifswald), H. Westers (Groningen), and U. Gerth (Greifswald) for kindly providing BFA mutant strains.
This work was supported by EC grant LSHG-CT-2004-503468 and the DFG.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 1881351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 1845826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 451613-1629. [DOI] [PubMed] [Google Scholar]
- 4.Baranova, N. N., A. Danchin, and A. A. Neyfakh. 1999. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug-efflux transporters. Mol. Microbiol. 311549-1559. [DOI] [PubMed] [Google Scholar]
- 5.Bleuel, C., C. Grosse, N. Taudte, J. Scherer, D. Wesenberg, G. J. Krauss, D. H. Nies, and G. Grass. 2005. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 1876701-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 1873650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caguiat, J. J., A. L. Watson, and A. O. Summers. 1999. Cd(II)-responsive and constitutive mutants implicate a novel domain in MerR. J. Bacteriol. 1813462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouch, M. L., M. Castor, J. E. Karlinsey, T. Kalhorn, and F. C. Fang. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67971-983. [DOI] [PubMed] [Google Scholar]
- 10.Franza, T., B. Mahe, and D. Expert. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55261-275. [DOI] [PubMed] [Google Scholar]
- 11.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 441225-1234. [DOI] [PubMed] [Google Scholar]
- 12.Gaballa, A., and J. D. Helmann. 2007. Substrate induction of siderophore transport in Bacillus subtilis mediated by a novel one-component regulator. Mol. Microbiol. 66164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 27647178-47184. [DOI] [PubMed] [Google Scholar]
- 14.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2000. Crystallization and preliminary X-ray diffraction studies on the DNA-binding domain of the multidrug transporter activation protein (MtaN) from Bacillus subtilis. Acta Crystallogr. D 561456-1458. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, C. F. 2007. Multiple molecular mechanisms for multidrug resistance transporters. Nature 446749-757. [DOI] [PubMed] [Google Scholar]
- 16.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204305-320. [DOI] [PubMed] [Google Scholar]
- 17.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 1791153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, K., et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 391948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 2767209-7217. [DOI] [PubMed] [Google Scholar]
- 21.Miethke, M., O. Klotz, U. Linne, J. J. May, C. L. Beckering, and M. A. Marahiel. 2006. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 611413-1427. [DOI] [PubMed] [Google Scholar]
- 22.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 1883664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page, W. J., E. Kwon, A. S. Cornish, and A. E. Tindale. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol. Lett. 228211-216. [DOI] [PubMed] [Google Scholar]
- 25.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10529-544. [DOI] [PubMed] [Google Scholar]
- 26.Quadri, L. E. 2007. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infect. Disord. Drug Targets 7230-237. [DOI] [PubMed] [Google Scholar]
- 27.Rowland, B. M., T. H. Grossman, M. S. Osburne, and H. W. Taber. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178119-123. [DOI] [PubMed] [Google Scholar]
- 28.Rowland, B. M., and H. W. Taber. 1996. Duplicate isochorismate synthase genes of Bacillus subtilis: regulation and involvement in the biosyntheses of menaquinone and 2,3-dihydroxybenzoate. J. Bacteriol. 178854-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 16047-56. [DOI] [PubMed] [Google Scholar]
- 31.Shewchuk, L. M., J. D. Helmann, W. Ross, S. J. Park, A. O. Summers, and C. T. Walsh. 1989. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry 282340-2344. [DOI] [PubMed] [Google Scholar]
- 32.Stergiopoulos, I., L.-H. Zwiers, and M. A. De Waard. 2002. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 108719-734. [Google Scholar]
- 33.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 1392041-2045. [DOI] [PubMed] [Google Scholar]
- 34.Tanabe, T., H. Nakao, T. Kuroda, T. Tsuchiya, and S. Yamamoto. 2006. Involvement of the Vibrio parahaemolyticus pvsC gene in export of the siderophore vibrioferrin. Microbiol. Immunol. 50871-876. [PubMed] [Google Scholar]
- 35.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17849-850. [DOI] [PubMed] [Google Scholar]
- 36.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283489-506. [DOI] [PubMed] [Google Scholar]
- 37.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12259-265. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, W., J. E. Arceneaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two nonribosomal peptide synthetase genes. Mol. Microbiol. 29629-639. [DOI] [PubMed] [Google Scholar]