Abstract
The Irr protein is a global regulator of iron homeostasis in Bradyrhizobium japonicum, and a subset of genes within the Irr regulon are negatively controlled under iron limitation. However, repressor function, high-affinity DNA binding in vitro, or promoter occupancy in vivo of Irr for a negatively regulated gene has not been demonstrated. Here, we show that the blr7895 and bll6680 genes are negatively regulated by Irr as determined by derepression of transcript levels in iron-limited cells of an irr mutant strain. Electrophoretic gel mobility shift analysis showed that a component in extracts of wild-type cells grown under iron limitation bound the iron control elements (ICE) within the promoters of blr7895 and bll6680 identified previously (G. Rudolph, G. Semini, F. Hauser, A. Lindemann, M. Friberg, H. Hennecke, and H. M. Fischer, J. Bacteriol. 188:733-744, 2006). Binding was not observed with extracts of cells from the parent strain grown under high iron conditions or with those from an irr mutant. Furthermore, gel mobility supershift experiments identified Irr as a component of the binding complex. Purified recombinant Irr bound to ICE DNA with high affinity in the presence of divalent metal, with Kd values of 7 to 19 nM, consistent with a physiological role for Irr as a transcriptional regulator. In addition, in vitro transcription initiated from the blr7895 promoter was inhibited by Irr. Whole-cell cross-linking and immunoprecipitation experiments showed that Irr occupies the promoters of blr7895 and bll6680 in vivo in an iron-dependent manner. The findings demonstrate that Irr is a transcriptional repressor that binds DNA with high affinity.
Regulation of iron-dependent gene expression in Alphaproteobacteria differs substantially from that in Escherichia coli or other well-studied model organisms. The iron response regulator (Irr) is found in most Alphaproteobacteria species, and many within this taxonomic group contain RirA as well (6, 23, 24). Irr has been studied most intensely in Bradyrhizobium japonicum, where it was initially identified in a screen for mutants defective in the control of heme biosynthesis (11). Irr is a conditionally stable protein that degrades in response to cell exposure to iron (18). Thus, unlike other Fur family metalloregulators, Irr is absent in the presence of the regulatory metal. Also, unlike Fur, Irr does not respond to iron directly but, rather, forms a complex with the heme biosynthetic enzyme ferrochelatase and responds to the status of heme at the site of synthesis (19). Irr binds heme directly (13, 18), which triggers its degradation by a mechanism that involves the redox activity of heme and oxidation of the protein (25, 26).
Although the regulatory input information for Irr is the status of heme biosynthesis, it is now clear that Irr is a global regulator of iron homeostasis and metabolism (21, 22, 27). B. japonicum senses iron through the status of heme in an Irr-dependent manner to control iron homeostasis (27). A role for Irr as both a positive and a negative global regulator is based on several observations. Irr was initially discovered as a positive effector of ferric iron transport as well as a negative regulator of heme biosynthesis (11), and similar roles have been described for Brucella abortus Irr (13, 14). Microarray analysis of a B. japonicum irr mutant shows that Irr affects the expression of many iron-regulated genes, and a heme-defective strain cannot maintain normal iron homeostasis (27). In addition, several iron-regulated genes in Rhizobium leguminosarum are derepressed in an irr strain (22). A cis-acting DNA element called an iron control element (ICE) was found in the promoters of the divergently transcribed genes hmuR and hmuT and shown to be necessary for the activation of those genes under iron limitation (15). This element was found to bind Irr in a yeast one-hybrid screen (21), and ICE-like motifs are found upstream of many genes in B. japonicum and other Alphaproteobacteria species (20, 21).
In the present study, we are interested in addressing negative regulation by B. japonicum Irr. Specifically, we wanted to determine whether Irr functions as a repressor and to assess Irr binding to the promoters of negatively controlled genes in vitro and in vivo. Among the 17 genes negatively regulated by Irr in microarray analysis (27), only two have an ICE-like motif (20, 21), and high-affinity binding by Irr to those elements has not been demonstrated. The dependence of hemB expression on Irr is well documented (11); however, this gene lacks an ICE motif, and binding of Irr to the hemB promoter has not been shown. The ICE motifs within the promoters of the blr7895 and bll6680 genes are required for repression under iron limitation and can recruit an Irr fusion in a yeast one-hybrid screen. However, the affinity of Irr for those DNA elements is very weak in vitro (21). In fact, in vitro binding studies of Irr from several organisms all require micromolar concentrations of protein to detect DNA binding (3, 14, 21, 27). Herein, we show that Irr is a transcriptional repressor with high affinity for target DNA and that Irr occupies the promoters of target genes in vivo.
MATERIALS AND METHODS
Strains and media.
Bradyrhizobium japonicum LO was the parent strain used in the present work. Strain LODTM5 is a mutant derivative of LO that contains a transposon, Tn5, within the irr gene (11). B. japonicum strains were routinely grown at 29°C in glycerol-salts-yeast extract (GSY) medium as described previously (9). Strain LODTM5 was grown in medium supplemented with 50 μg/ml kanamycin and 50 μg/ml streptomycin. For the iron experiments, modified GSY medium, which contains 0.5 g/liter yeast extract instead of 1 g/liter, was used, and either no exogenous iron was added for low-iron medium or 12 μM FeCl3·6H2O was added for high-iron medium. The actual iron concentration of the unsupplemented medium was 0.3 μM, as determined with a Perkin-Elmer model 1100B atomic absorption spectrometer.
RNA isolation and quantitative real-time PCR analysis.
We determined the expression levels of blr7895, bll6680, and gapA by quantitative real-time PCR with iQ Sybr green supermix (Bio-Rad) by using an iCycler thermal cycler (Bio-Rad) as described in detail elsewhere (27). The standard curve method was employed for relative quantitation, and gapA was a housekeeping gene control. Genomic DNA from parent strain LO was used as the PCR template to generate a standard curve for each gene. Relative starting quantities of mRNAs for blr7895, bll6680, and gapA were calculated from corresponding standard curves. The quantities of blr7895 and bll6680 were then normalized to the quantity of gapA for each condition. The results are based on the average of triplicate assays, and the standard deviation is shown as an error bar in the graphed results.
EMSA.
Electrophoretic mobility shift assays (EMSA) were used to determine the binding of DNA to purified Irr or to components in cell extracts as described previously (8). A 0.1 nM DNA probe was used for Kd determination experiments, and DNA probes at other concentrations were used as indicated in the figure legends. The negative-control DNA corresponds to a sequence found in the multiple-cloning site of pBluescript SK+. The test DNA probes were 42 bp in length and contained nucleotide sequences corresponding to the 21-bp ICE (underlined residues of sequences below) flanked by DNA sequences found in the multiple-cloning site region of pBluescript SK+. The sequences of the probes (only one strand of each double-stranded DNA) are as follows: 5′-CAGGAATTCGATAATTTAGAATCATTCTAAACTGACCTCGAC-3′ (ICE of blr7895 [ICE7895]), 5′-CAGGAATTCGATAATTTAGAACGCTTCTAAATGGACCTCGAC-3′ (ICE of bll6680 [ICE6680]), and 5′-CAGGAATTCGATATCAAGCTTATCGATACCGTCGACCTCGAC-3′ (control).
Following incubation, EMSA reactions were analyzed as autoradiograms of 6% nondenaturing polyacrylamide gels. Autoradiograms were scanned using a GS-700 densitometer (Bio-Rad, Hercules, PA), and signal intensities were detected and quantified using Quantity One software (Bio-Rad, Hercules, PA). To determine the dissociation binding constant (Kd), binding reaction mixtures were titrated with various concentrations of Irr. Bound and unbound DNAs were quantified by comparing relative signal intensities and analyzed using GraphPad Prism (GraphPad Software Inc., San Diego, CA).
In vitro transcription analysis.
Transcription of DNA initiated from the blr7895 promoter by purified RNA polymerase was carried out as described by Beck et al. (4). A 4 nM DNA template was incubated with no or 200 nM Irr in in vitro transcription buffer (40 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.1 mM EDTA [pH 8.0], 0.1 mg bovine serum albumin ml−1, 150 mM KCl, 0.04 mM K3PO4, 0.1 mM dithiothreitol) in a 9-μl volume for 30 min at 37°C. A total of 0.5 μg of RNA polymerase was added (1-μl volume) and incubated for an additional 10 min. The reaction was started by adding 10 μl of a solution containing a 2 mM concentration of each nucleoside triphosphate (1 mM final concentration), 1 μCi of [α-32P]UTP, and RNasin and incubated for 30 min at 37°C. The reaction was terminated on ice. An equal volume of 2× RNA loading buffer (Ambion) was added, followed by heating at 95°C for 3 min, run on an 8% urea acrylamide gel, and exposed by autoradiography. A commercial RNA ladder (Fermentas) was radiolabeled using T4 RNA ligase and used to estimate molecular mass.
In vivo cross-linking and immunoprecipitation.
Two-hundred-milliliter cultures of parent strain LO or irr strain LODTM5 were grown under low- or high-iron conditions to mid-log phase. For cross-linking, formaldehyde was added to 1% (vol/vol), and the cells were gently rocked in a flask at room temperature for 10 min. To quench the cross-linking, glycine was added to a final concentration of 10 mg/ml, and the cells were shaken gently at 4°C for 30 min. Cells were collected by centrifugation, and the pellets were resuspended and washed twice with phosphate-buffered saline (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl [pH 7.4]). Washed cells were resuspended in 2 ml lysis buffer (100 mM Tris [pH 8.0], 300 mM NaCl, and 10 mM EDTA) and lysed with a French pressure cell as described previously (17). The lysis also sheared the DNA. The cell extracts were aliquoted into 700-μl volumes and frozen at −80°C until further use. Agarose beads, preblocked with salmon sperm DNA and bovine serum albumin (USB, Cleveland, OH), were washed with lysis buffer and resuspended to a 33% slurry. To preclear the lysate, 70 μl of the bead slurry was added to 700 μl of cell lysate and rotated at room temperature for 1 h to mix, followed by centrifugation for 5 min at 3,000 × g. The supernatant was divided into two 300-μl samples. Three hundred microliters of lysis buffer plus 0.5% Triton X-100 was added to each sample. Two microliters of serum containing anti-Irr polyclonal antibodies was added to one sample, and nothing was added to the other sample, which served as a negative control. Both samples were rotated at 4°C overnight. Fifty microliters of washed-bead slurry was added to both samples and rotated at room temperature for 1 h. The samples were then centrifuged as before, and the supernatant of the negative control was saved to use as input DNA. The protein-DNA complex was washed and eluted from the beads per the manufacturer's instructions. All steps were carried out at room temperature. The DNA was un-cross-linked from the protein by adding NaCl to the eluted samples to 0.2 M, and the samples were incubated at 65°C overnight. DNA was purified using the Qiagen PCR purification kit and eluted in 30 μl of elution buffer. DNA was then diluted 1:10,000 or 1:5,000 in water, and PCR was performed with primers against blr7895 and bll6680, respectively. Primers targeted a 100- to 250-base-pair region in the promoters of target genes. PCR was carried out with 25-μl volumes and run for 32 cycles with an annealing temperature of 56°C and an extension time of 1 min.
RESULTS
Iron-dependent regulation of blr7895 and bll6680 is lost in an irr mutant strain.
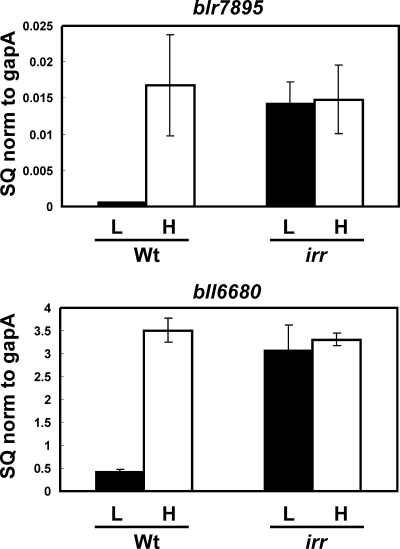
The bll6680 gene encodes a putative bacterioferritin, and blr7895 encodes an unknown protein predicted to have a ferritin-like fold at the N terminus. Each gene contains an ICE in their promoter regions that are involved in regulation by iron (21). These elements bind Irr in a yeast one-hybrid screen and also weakly in vitro (21), strongly suggesting that blr7895 and bll6680 are regulated by Irr. To address this directly, we measured transcripts of each gene in wild-type B. japonicum cells and in an irr mutant by quantitative real-time PCR (Fig. 1). Both genes were strongly regulated by iron in the parent strain, with high transcript levels found in cells grown under iron-replete conditions and very low levels in iron-limited cells. These observations agree with a previous report using primer extension analysis and lacZ reporter fusions (21). In the irr mutant strain, iron-dependent regulation of blr7895 and bll6680 was lost, with high levels of expression in cells grown in either high- or low-iron medium (Fig. 1). This indicates that Irr normally negatively regulates the expression of these genes in iron-limited cells, which is derepressed in the irr strain. Despite this control, Irr was reported to bind ICE motif DNA in vitro only weakly (21). One possibility was that maximal binding required an additional component in cells that was missing in the recombinant protein. This was addressed in several ways, as described below.
FIG. 1.
Iron-dependent expression of the blr7895 and bll6680 genes in parent strain LO (Wt) or irr mutant strain LODTM5 (irr). mRNAs from cells grown in medium supplemented with no added iron (L) or with 12 μM FeCl3 (H) were analyzed by quantitative real-time PCR. The data are expressed as the relative starting quantity (SQ) of the respective mRNAs normalized to the housekeeping gene gapA. The data are expressed as the average of the results of three replicates ± the standard deviation. Error bars indicate the standard errors of the mean.
Identification of Irr as a component in cell extracts that binds to ICE.
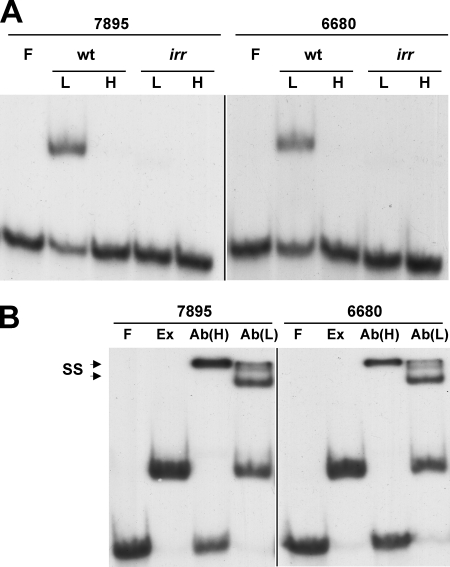
EMSA were carried out using extracts from cells grown in low- or high-iron medium (0.1 μg) and radiolabeled DNA containing the ICE motif sequence in the promoter of blr7895 or bll6680 (Fig. 2A). A complex was formed with either ICE DNA in extracts of wild-type cells grown under low-iron conditions as discerned by the mobility shift of the labeled DNA. Formation of this complex was iron dependent, since it was not observed with extracts from cells grown in iron-replete medium. EMSA using upstream regions of the blr7895 or bll6680 gene that did not include the ICE did not show complex formation (data not shown). No complexes were formed with ICE DNA using extracts from cells of the irr strain grown in low- or high-iron medium, showing that complex formation was Irr dependent (Fig. 2A). This observation is consistent with a lack of gel shift in extracts of wild-type cells grown in iron-replete medium, because Irr is not expressed in those cells (11).
FIG. 2.
Identification of Irr as a component in extracts of wild-type cells grown under iron limitation that binds to ICE DNA. (A) Extracts prepared from cells of parent strain LO (wt) or irr strain LODTM5 (irr) grown in medium supplemented with no iron (lanes L) or with 12 μM FeCl3 (lanes H) were incubated with 0.1 nM radiolabeled ICE DNA from blr7895 or bll6680. EMSA was carried out with free DNA probe (F) or DNA with 0.1 μg extract protein. Complexes were resolved on 6% nondenaturing gels and visualized by autoradiography. (B) Electrophoretic mobility supershift assays were carried out using free DNA probe (lanes F), 5 μg extract protein from wild-type cells grown in low-iron medium (lanes Ex), and extract plus anti-Irr antibodies at 1:100 [lanes Ab(H)] or 1:1,000 [lanes Ab(L)] dilutions in the binding reaction mixtures. The complexes were resolved and visualized as described for panel A. SS denotes the supershifted species in the presence of antibody.
The dependence on Irr for complex formation means either that Irr is present in the complex or that it regulates the expression of another protein that is found there. Therefore, electrophoretic mobility supershift analysis was carried out using antibodies directed against Irr and extracts from wild-type cells grown under iron limitation (Fig. 2B). At a 1:1,000 antibody titer, a supershift doublet was observed with a mobility slower than that formed in the absence of the antibody. At a 1:100 antibody titer, a supershift was observed as well, but a band corresponding to unbound DNA was also observed, suggesting interference by the antibody in the Irr-DNA complex. Antibody alone did not result in a mobility shift of the labeled DNA (data not shown). The findings indicate that Irr is a component of the binding complex.
Purified recombinant Irr binds ICE DNA with high affinity in the presence of divalent metal.
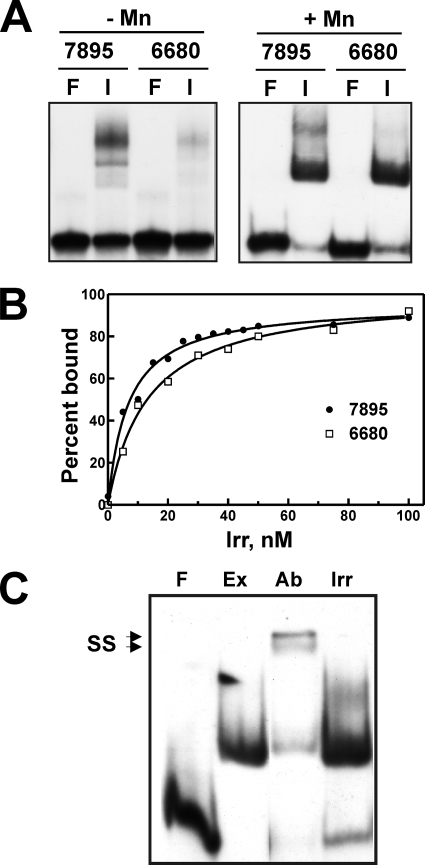
The data presented thus far show that Irr is a component in cell extracts that binds to ICE DNA. We wanted to determine whether Irr binds directly to the DNA by EMSA using purified recombinant DNA. Fur family proteins typically have dissociation binding constant (Kd) values in the 5 to 50 nM range (1, 2, 5, 8, 10, 16). However, less than 50% of ICE DNA was bound in the presence of 1.5 μM Irr in a previous report (21) We carried out EMSA by using purified Irr and also found very weak binding to ICE DNA (Fig. 3A). In these initial experiments, less than half of the 0.25 μM labeled DNA was bound in the presence of 100 nM Irr (Fig. 3A, left panel). Divalent metals bind to Fur family proteins and have both structural and regulatory roles. Mn2+ is often added to buffers in binding reactions for Fur family proteins. We found that the addition of 100 μM MnCl2 resulted in stronger binding of Irr to ICE DNA than that in the absence of metal (Fig. 3A). Irr did not bind to nonspecific DNA (data not shown). Zn2+ also improved binding to a similar extent, but Cu2+, Ni2+, and Co2+ did not (data not shown). The binding affinity of Irr for ICE DNA was quantified as described previously (8) by measuring Irr-DNA complex formation as a function of the Irr concentration (Fig. 3B). The Kd value of Irr for ICE7895 and ICE6680 DNA was 7.1 and 19.3 nM, respectively, in the presence of Mn2+. Thus, Irr binds ICE DNA with high affinity under those conditions.
FIG. 3.
Binding of purified recombinant Irr to ICE DNA in vitro. (A) EMSA was carried out using ICE DNA corresponding to the blr7895 or bll6680 promoters. Binding reactions were carried out using 0.2 nM radiolabeled ICE DNA either without protein (lanes F) or with 200 nM Irr (lanes I). In addition, the reaction, running, and gel buffers were supplemented with 100 μM MnCl2, denoted as +Mn in the right panel. The complexes were resolved and visualized as described in the legend to Fig. 2. (B) Determination of Irr affinity for ICE DNA. EMSA was carried out using 0.1 nM radiolabeled ICE7895 or ICE6680 DNA with various concentrations of purified Irr. The signal intensities of bound and unbound DNA on the autoradiograms were quantified and are expressed as percent bound DNA. (C) Comparison of the mobilities of ICE complexes in purified Irr and in extracts of wild-type cells grown under iron limitation. EMSA was carried out as described in the legend to Fig. 2. Lanes: F, free DNA probe; Ex, probe with 5 μg protein extract; Ab, probe with extract plus 1:1,000 dilution of anti-Irr antibody; Irr, purified Irr.
Demonstration of strong binding of Irr to target DNA using purified components in vitro suggests that an additional protein is not necessary to form a DNA complex that includes Irr. We compared the mobilities of radiolabeled ICE7895 DNA and purified Irr or with the mobilities of extracts of wild-type cells grown in low-iron medium on the same nondenaturing gel in the presence of Mn2+ (Fig. 3C). The mobility of the complex found in cell extracts was the same as that of the Irr-DNA complex, supporting the conclusion that Irr was the only protein component in the cell extract complex.
Irr represses transcription from the blr7895 promoter in vitro.
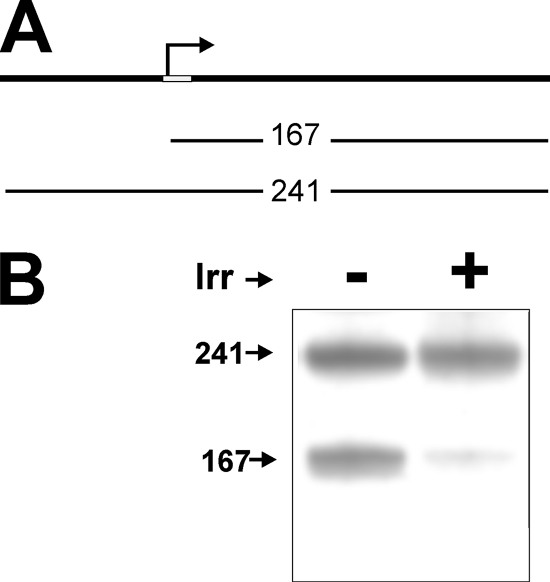
blr7895 and bll6680 are expressed at low levels in iron-limited cells, where Irr accumulates to high levels (Fig. 1) (11). In addition, these genes are expressed at a high level and independently of iron in an irr mutant strain (Fig. 1). Thus, Irr appears to act as a repressor. We addressed this possibility in vitro by examining the ability of purified recombinant Irr to repress transcription from the blr7895 gene promoter. A 241-bp linear double-stranded DNA fragment that corresponds to a region from 77 bp upstream of the blr7895 gene transcription start site, as determined previously (21), to 167 bp downstream of it was used as a template (Fig. 4A). Transcription of the DNA template by using purified B. japonicum RNA polymerase (gift from H. Hennecke) yielded the expected transcript of ∼170 bp in size. Another RNA corresponding to the full-length DNA, ∼240 bp in size, was also observed (Fig. 4B). The larger message may be due to “breathing” at the end of the double-stranded DNA template, allowing transcription initiation from an end, which can serve as a control. In the presence of Irr, abundance of the smaller transcript was drastically diminished, whereas the larger RNA was unaffected. Thus, Irr can repress transcription from the blr7895 promoter in vitro.
FIG. 4.
Inhibition of in vitro transcription by Irr. (A) A linear 241-bp DNA fragment that includes the blr7895 promoter region and is 167 bp downstream of the transcription start site was used as a template. The bent arrow denotes the transcription start site, and the white rectangle represents the ICE. (B) Transcription was carried out using a 4 nM DNA template and 0.5 μg purified B. japonicum RNA polymerase holoenzyme in the absence (−) or presence (+) of 200 nM Irr. The radiolabeled RNA products were separated by electrophoresis and visualized by autoradiography.
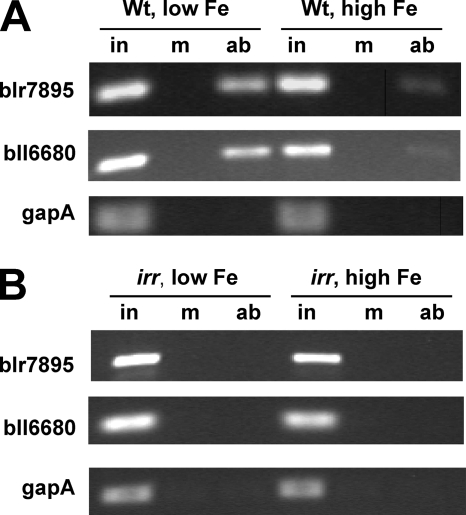
Irr occupies the blr7895 and bll6680 gene promoters in vivo in an iron-dependent manner.
To determine whether Irr binds directly to target gene promoters in vivo, we carried out cross-linking and immunoprecipitation (IP) experiments (also called chromatin IP or ChIP). In these experiments, cells grown in low- or high-iron medium were treated with formaldehyde to cross-link Irr to its in vivo targets. Irr was then immunoprecipitated from cells that were previously lysed, and the DNA was sheared. After the cross-linking was reversed, DNA that coprecipitated with Irr was analyzed by PCR using primers that amplified the promoter regions of blr7895 and bll6680 (Fig. 5). Products were observed for each promoter when wild-type cells grown under iron limitation were used, but very little was observed when cells grown in high-iron medium were used (Fig. 5A). gapA, which is not regulated by Irr, was used as a negative control. As an additional control, the cross-linking IP experiments were carried out with the irr strain grown in low- or high-iron medium (Fig. 5B). The findings show that Irr occupies the promoters of blr7895 and bll6680 in vivo, which further supports the conclusion that Irr is a direct regulator of those genes.
FIG. 5.
Detection of Irr at the blr7895 and bll6680 gene promoters in vivo by cross-linking IP. Cross-linking of parent strain LO (Wt) (A) or irr strain LODTM5 (irr) (B) cells grown in low- or high-iron medium followed by cell breakage and co-IP was carried out as described in Materials and Methods. The primers used amplify the promoter region of blr7895, bll6680, and gapA. Lanes: in, input DNA; m, mock IP in the absence of antibody; ab, IP using anti-Irr antibodies. gapA is a control for a gene not regulated by Irr.
DISCUSSION
The Irr protein was initially discovered in a screen for a negative regulator of heme biosynthesis and was subsequently shown to be a global regulator of iron homeostasis in B. japonicum. In the present study, we establish that Irr is a transcriptional repressor and binds to target DNA with high affinity. We also show that Irr occupies the promoters of two Irr-regulated genes in an iron-dependent manner in vivo, which further supports the conclusion that Irr directly regulates those genes. These findings do not rule out the possibility of indirect control of some genes by Irr, resulting in downregulation under iron limitation, but they clearly demonstrate a direct repressor function for Irr. The presence of Irr was sufficient to repress transcription initiated from the blr7895 promoter in vitro, indicating that an auxiliary protein is not needed for that activity. Consistent with this, Irr was likely the only protein component in cell extracts that bound to ICE DNA in EMSA, as discerned by comparison of the mobility of DNA in the presence of pure protein and that in extracts (Fig. 3C).
Rather than an auxiliary protein, high-affinity binding of Irr to DNA requires a divalent metal in vitro. Metals have both structural and regulatory roles in Fur family proteins. Fur and PerR have a structural zinc atom that is present in purified recombinant protein preparations that require denaturation in the presence of chelators to remove them (2, 12). Recombinant Irr does not contain zinc or manganese as purified from E. coli (unpublished data). Unlike other Fur family proteins, Irr functions in the absence of the regulatory metal, and the degradation response to iron is an indirect consequence of heme. Thus, the role for divalent metal is not yet known, but it is likely to have a structural function rather than a regulatory role since Irr does not respond to Mn2+ or Zn2+ in vivo (11). The Mn2+ concentration used in the in vitro experiments is within the physiological range reported for bacterial cells (7).
The identification of the ICE motif as an Irr target by Rudolph et al. (21) provides an important tool for studying DNA-Irr interactions that was not previously available. It provides a means to predict Irr target genes (20, 21) and has allowed us to optimize Irr activity in vitro. Nevertheless, some caveats regarding this element should be noted. Among the large number of genes identified in a bioinformatics search to contain a putative upstream ICE motif (21), fewer than 15% (24 of 172) are regulated by iron, based on microarray data (27), and even fewer (11 of 172) by Irr. These discrepancies are likely due to both the imprecision of predictive informatics and the false negatives from the microarrays. A separate bioinformatics analysis (20) identified many fewer genes with putative ICE motifs than Irr-regulated genes identified in the microarray studies. Several factors could contribute to this, including the fact that not all Irr-regulated genes are likely to be direct targets of the protein and that Irr may have DNA targets dissimilar from the ICE motif. In support of the latter possibility, a cis-acting element was identified for Bartonella quintana Irr that is dissimilar to an ICE motif (3).
Acknowledgments
We thank H. Hennecke for the gift of purified B. japonicum RNA polymerase.
This work was supported by National Institutes of Health grant R01 GM067966 to M.R.O.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Ahn, B. E., J. Cha, E. J. Lee, A. R. Han, C. J. Thompson, and J. H. Roe. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 591848-1858. [DOI] [PubMed] [Google Scholar]
- 2.Althaus, E. W., C. E. Outten, K. E. Olson, H. Cao, and T. V. O'Halloran. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 386555-6569. [DOI] [PubMed] [Google Scholar]
- 3.Battisti, J. M., L. S. Smitherman, K. N. Sappington, N. L. Parrow, R. Raghavan, and M. F. Minnick. 2007. Transcriptional regulation of the heme binding protein gene family of Bartonella quintana is accomplished by a novel promoter element and iron response regulator. Infect. Immun. 754373-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, C., R. Marty, S. Klausli, H. Hennecke, and M. Gottfert. 1997. Dissection of the transcription machinery for housekeeping genes of Bradyrhizobium japonicum. J. Bacteriol. 179364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellini, P., and A. M. Hemmings. 2006. In vitro characterization of a bacterial manganese uptake regulator of the fur superfamily. Biochemistry 452686-2698. [DOI] [PubMed] [Google Scholar]
- 6.Chao, T. C., J. Buhrmester, N. Hansmeier, A. Pühler, and S. Weidner. 2005. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl. Environ. Microbiol. 715969-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300931-936. [DOI] [PubMed] [Google Scholar]
- 8.Friedman, Y. E., and M. R. O'Brian. 2003. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J. Biol. Chem. 27838395-38401. [DOI] [PubMed] [Google Scholar]
- 9.Frustaci, J. M., I. Sangwan, and M. R. O'Brian. 1991. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J. Bacteriol. 1731145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuangthong, M., and J. D. Helmann. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 1856348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 27321669-21674. [DOI] [PubMed] [Google Scholar]
- 12.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41849-859. [DOI] [PubMed] [Google Scholar]
- 13.Martínez, M., R. A. Ugalde, and M. Almirón. 2005. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 1513427-3433. [DOI] [PubMed] [Google Scholar]
- 14.Martínez, M., R. A. Ugalde, and M. Almirón. 2006. Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 1522591-2598. [DOI] [PubMed] [Google Scholar]
- 15.Nienaber, A., H. Hennecke, and H. M. Fischer. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41787-800. [DOI] [PubMed] [Google Scholar]
- 16.Platero, R., V. de Lorenzo, B. Garat, and E. Fabiano. 2007. Sinorhizobium meliloti Fur-like (Mur) protein binds a Fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl. Environ. Microbiol. 734832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puri, S., and M. R. O'Brian. 2006. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 1886476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 9613056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi, Z., and M. R. O'Brian. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9155-162. [DOI] [PubMed] [Google Scholar]
- 20.Rodionov, D. A., M. S. Gelfand, J. D. Todd, A. R. Curson, and A. W. Johnston. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-Proteobacteria. PLoS Comput. Biol. 2e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph, G., G. Semini, F. Hauser, A. Lindemann, M. Friberg, H. Hennecke, and H. M. Fischer. 2006. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J. Bacteriol. 188733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todd, J. D., G. Sawers, D. A. Rodionov, and A. W. Johnston. 2006. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol. Genet. Genomics 275564-577. [DOI] [PubMed] [Google Scholar]
- 23.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. Johnston. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 1484059-4071. [DOI] [PubMed] [Google Scholar]
- 24.Viguier, C., P. O. Cuiv, P. Clarke, and M. O'Connell. 2005. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol. Lett. 246235-242. [DOI] [PubMed] [Google Scholar]
- 25.Yang, J., K. Ishimori, and M. R. O'Brian. 2005. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J. Biol. Chem. 2807671-7676. [DOI] [PubMed] [Google Scholar]
- 26.Yang, J., H. R. Panek, and M. R. O'Brian. 2006. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol. Microbiol. 60209-218. [DOI] [PubMed] [Google Scholar]
- 27.Yang, J., I. Sangwan, A. Lindemann, F. Hauser, H. Hennecke, H. M. Fischer, and M. R. O'Brian. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]