Abstract
Microcin E492 is a channel-forming bacteriocin that is found in two forms, namely, a posttranslationally modified form obtained by the covalent linkage of salmochelin-like molecules to serine 84 and an unmodified form. The production of modified microcin E492 requires the synthesis of enterochelin, which is subsequently glycosylated by MceC and converted into salmochelin. mceC mutants produced inactive microcin E492, and this phenotype was reversed either by complementation with iroB from Salmonella enterica or by the addition of exogenous salmochelin. Cyclic salmochelin uptake by Escherichia coli occurred mainly through the outer membrane catecholate siderophore receptor Fiu. The production of inactive microcin E492 by mutants in entB and entC was reverted by the addition of the end product of the respective mutated pathway (2,3-dihydroxybenzoic acid and enterochelin/salmochelin, respectively), while mutants in entF did not produce active microcin E492 in the presence of enterochelin or salmochelin. The EntF adenylation domain was the only domain required for this microcin E492 maturation step. Inactivation of the enzymatic activity of this domain by site-directed mutagenesis did not prevent the synthesis of active microcin E492 in the presence of salmochelin, indicating that the adenylation activity is not essential for the function of EntF at this stage of microcin E492 maturation.
Microcin E492 (MccE492) is a channel-forming bacteriocin that is produced and excreted by Klebsiella pneumoniae RYC492 (13, 26). The genetic determinants involved in the production of active MccE492 have been cloned and expressed in Escherichia coli (45). Recombinant MccE492 has the same biochemical and electrophysiological properties as MccE492 isolated from Klebsiella. This bacteriocin exhibits characteristics that make its study very attractive: it behaves as a toxin on several malignant human cell lines through induction of apoptosis (22); it forms amyloid-like fibrils, a structure that modulates MccE492 activity (5); and it undergoes posttranslational modification by the attachment of salmochelin derivatives to its C terminus (43) through a process named enzymatic tailoring (33). Maturation requires the action of MceC, -D, -I, and -J, which are proteins encoded by the MccE492 system (24) whose activities in vitro were recently characterized (33). MceI presents identity with an acyltransferase, MceD has identity with an esterase, MceJ does not present homology with any protein of known function, and MceC is homologous to IroB (24). IroB is a glycosyltransferase found in Salmonella enterica (4) and in pathogenicity islands from uropathogenic E. coli strains (14) that glycosylates enterochelin to produce salmochelin (16, 21, 35). The production of salmochelin seems to be a defense strategy of virulent strains to restore an iron acquisition system in response to enterochelin sequestration by mammalian siderocalin (discussed in reference 17).
MccE492 uptake occurs through the outer membrane proteins FepA, Fiu, and Cir (7, 35, 42), which are the receptors for the ferric form of enterochelin and its hydrolysis products (20). MccE492 uptake is inhibited by enterochelin and its linear dimer and trimer derivatives, although ferric enterochelin has no effect on antibacterial activity (34, 42).
Other microcins, such as M and H47, have maturation genes equivalent to those present in the MccE492 system (7). Therefore, it is presumed that these bacteriocins are also modified in the C-terminal amino acid by salmochelin. This assumption is supported by the fact that there is a functional relationship in the maturation process of microcins H47, I47, and E492, as determined by heterologous complementation of antibacterial activity (36). The modification of the C-terminal amino acid by salmochelin is called a “Trojan horse strategy,” because the modification would facilitate the uptake of these bacteriocins by the ferric catecholate receptors located in the outer membranes of the target cells (17).
MccE492 was described as an unmodified bacteriocin (37), until Thomas et al. (43) communicated that under certain growth conditions cells carrying the MccE492 system produced an important fraction of modified MccE492 along with a fraction of unmodified bacteriocin. MccE492 preparations obtained under such conditions had four to eight times more antibacterial activity than preparations obtained under conditions in which the modified form was not detected (43). On the other hand, mutants in the maturation genes (mceC, -I, and -J) that participate in the modification of MccE492 produce an inactive microcin (24), meaning that either modification is required for antibacterial activity or the maturation gene products also participate in an unknown form in the maturation process of MccE492. Therefore, it is necessary to establish to what extent the lack of posttranslational modification affects MccE492 antibacterial activity and to determine whether there is a contribution of host factors to the production of active MccE492.
The aim of this work was to determine how mutants impaired in the synthesis of salmochelin affect the production of active MccE492 in vivo. To this end, several mutants in the synthesis of enterochelin were used as hosts of the MccE492 system. The pathway of enterochelin/salmochelin synthesis in vivo was disconnected from the modification of MccE492 by salmochelin, as demonstrated by providing this substrate exogenously. The biological assay developed for this purpose showed that salmochelin can be exported and imported in nonpathogenic strains of E. coli and that Fiu is the main receptor for salmochelin uptake. Additionally, MceC plays no role in the production of active MccE492 apart from the synthesis of salmochelin. Strikingly, we found that EntF has a dual function for the production of active MccE492. It is indispensable for the production of active MccE492 in a step that is upstream of, and not related to, enterochelin synthesis. Moreover, the participation of EntF at this stage is restricted only to its adenylation domain, but the adenylation activity necessary for the synthesis of enterochelin is not essential for the production of active MccE492.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmids and strains used in this work are described in Table 1. The following strains and plasmids were kindly donated by M. Fishbach, from the laboratory of Chris Walsh at Harvard Medical School: E. coli ER1100A, ER1300H, and K-12 entB and plasmids pER311, pER307A, and pER304A. E. coli H5311 was kindly provided by K. Hantke.
TABLE 1.
Genotypes/phenotypes of bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype/phenotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli strains | ||
| VCS257 | DP50 sup F[supE44 supF58 hsd53(rB mB) dapD8 lacY1 glnV44 Δ(gal-uvrB)47 tyrT58 gyrA29 tonA53Δ (thyA57)] | Stratagene |
| BL21(DE3) | F−ompT rB mB | Novagen |
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Promega |
| H1594 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB fiu::Mud1X (Ampr Strr) | 20 |
| H1875 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB cir::Mud1X fepA::Tn10 (Ampr Tetr Strr) | 20 |
| H1876 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB fiu::Mud1X cir fepA::Tn10 (Ampr Tetr Strr) | 20 |
| H5311 | H1143 derivative; araD139 Δ(argF lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB entC::Mud1X | 19; K. Hantke lab collection |
| K12 entB | entB; obtained by gene replacement with Kan (Kanr) | 27 |
| ER1100A | entF; obtained by gene replacement with Chl (Chlr) | 38 |
| ER1300H | entF; ER1100A derivative sensitive to Chl | 38 |
| S. enterica strain | ||
| LT2 | Also known as Salmonella enterica serovar Typhimurium LT2; prototroph | S. Maloy lab collection |
| Plasmids | ||
| pJAM434 | Moderately low production of MccE492 (Ampr) | 45 |
| np133 | pJAM434 mceC::Tn5 (Ampr Kanr) | 24 |
| np205 | pJAM434 mceI::Tn5 (Ampr Kanr) | 24 |
| np221 | pJAM434 mceJ::Tn5 (Ampr Kanr) | 24 |
| np220 | pJAM434 mceA::Tn5 insertion in amino acid 43 (Ampr Kanr) | This work |
| pJEM15 | Overproduces MccE492 (Ampr) | 45 |
| npB4 | pJEM15 mceC::Tn5 insertion in amino acid 60 (Ampr Kanr) | This work |
| p157 | mceB (immunity) cloned into pT7-7 (Ampr) | 25 |
| pIroB | iroB of S. enterica cloned into pACYC184 (Chlr) | This work |
| pER311 | entF cloned into pET28-a with a six-His tag at the C terminus (Ampr) | 38 |
| pER307A | Domains A-PCP-TE (between amino acids 437 and 1293) of EntF cloned into pET28-a with a six-His tag at the C terminus (Ampr) | C.T. Walsh lab collection |
| pER304A | Domains A-PCP (between amino acids 437 and 1045) of EntF cloned into pET28-a with a six-His tag at the C terminus (Ampr) | C.T. Walsh lab collection |
| pACYC-EntF | entF cloned into pACYC184 as it is in pER311 (Chlr) | This work |
| pACYC-A-PCP-TE | Domains A-PCP-TE of EntF cloned into pACYC184 as in pER307A (Chlr) | This work |
| pACYC-A-PCP | Domains A-PCP of EntF cloned into pACYC184 as in pER304A (Chlr) | This work |
| pACYC-A | Domain A (between amino acids 437 and 968) of EntF cloned into pACYC184 (Chlr) | This work |
| pACYC-EntF-E750A | entF with the E750A mutation cloned into pACYC184 (Chlr) | This work |
Plasmid construction.
pACYC184 was the vector used to make constructs compatible with pHC79, the vector in which the MccE492 system is cloned (pJAM434 and pJEM15) (45). The gene iroB from S. enterica LT2 was amplified from chromosomal DNA by using the primers IRO (5′GAGAGGATTTCATATGCGTATTCTGTTTG3′) and IroBR (5′CTCCTTATCGGGATCCGATGAGATATGGCGACACAC3′). IRO generates an NdeI restriction site, which was used to ligate this amplified DNA to pT7-7 linearized with NdeI. The ligation product was used as a template to amplify the iroB gene under the control of the T7 promoter, using the primers T7pro (5′TAATACGACTCACTATAGGG3′) and IroBR. The resulting DNA was cloned into the EcoRV site of pACYC184, and the recombinants were selected by resistance to chloramphenicol and sensitivity to tetracycline. The entF gene and its domain combinations were cloned using the respective pET-28 derivatives mentioned in Table 1 as templates, with the primers T7 (5′GAAATTAATACGACTCACTATA3′) and TER (5′GGATATAGTTCCTCCTTTCA3′). The amplified DNAs were subsequently cloned into the EcoRV site of pACYC184. The DNA corresponding to the EntF adenylation domain was cloned into the pACYC184 EcoRV site, using pER307A as the template and the primers T7 and DomAR (5′TGGCGCTTATGCCTTCAGTT3′). A construct with an E750A mutation in domain A of EntF was constructed by site-directed mutagenesis, using a QuikChange mutagenesis kit from Stratagene following the conditions suggested by the manufacturer, with an extension time of 16 min. The template used was pER311, and the primers employed were A-E750A-F (5′GGCCCGACGGCAGCGGCGGTA3′) and A-E750A-R (5′TACCGCCGCTGCCGTCGGGCC3′). The resulting plasmid was introduced into an entF strain, and mutants defective in enterochelin production were selected on chrome azurol S (CAS) plates and recloned into pACYC184 as described above for pACYC-EntF. All constructs were fully sequenced. Standard techniques for cloning, electroporation, restriction analysis, and plasmid and chromosomal DNA preparations were performed as described previously (2, 32, 39).
Growth conditions and MccE492 activity assay on plates.
Bacterial growth was performed as previously described (24, 25, 45), with antibiotics used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; streptomycin, 20 μg/ml; tetracycline, 50 μg/ml; and chloramphenicol, 50 μg/ml.
Activity in plates was determined by mixing an aliquot of 0.2 ml of 2 × 107 cells/ml of the sensitive E. coli strain grown in LB (with the corresponding antibiotic) with 3 ml of soft agar and overlaying the mixture onto LB plates. Three-microliter aliquots of overnight cultures were seeded onto these plates, and the presence of active MccE492 was detected by the formation of growth inhibition halos. A lawn of the sensitive E. coli strain BL21(DE3) was routinely used as the indicator strain. Depending on the size of the inhibition halo produced, in almost all cases the activity was assigned to one of the following three categories: low (+), medium (++), and high (+++). The evaluation of activity was performed after the observation of 6 to 20 inhibition halos for each case. This semiquantitative assay is comparative, and for comparison criteria, the activity produced by E. coli pJAM434 was considered low (+), that by S. enterica np133 was considered medium (++), and that by S. enterica pJAM434 was considered high (+++). There were two special cases in which the activity, although very low, was detectable by visual inspection, and the results were expressed as ±.
CAS plates were prepared as described by Schwyn and Neilands (41). When chrome azurol binds to iron, a blue color is produced in the plates. Siderophore production was detected by the appearance of a yellow halo around the colonies after 24 h of incubation.
MccE492 purification, PAGE, and immunoblotting.
MccE492 was extracted from 500 ml of supernatant of cultures of E. coli VCS257/pJEM15 or from different ent-defective hosts carrying pJEM15, using a similar procedure to one described previously (43). Cells were grown at 37°C in M9 medium supplemented with citrate and glucose (34) with shaking at 220 rpm for 20 h, and the supernatant was collected by centrifugation at 17,000 × g for 30 min. The supernatant was loaded onto a Sep-Pak C8 cartridge (Waters) previously equilibrated with 5 ml of 0.1% trifluoroacetic acid (TFA) in nanopure water. The cartridge was washed with 5 ml 30% acetonitrile (ACN)-0.1% TFA, and MccE492 was eluted with 5 ml of 40% ACN-0.1% TFA. Polyacrylamide gel electrophoresis (PAGE) was carried out under the conditions described by Schägger and von Jagow (40), and nitrocellulose membranes (Millipore) were used for immunoblot transference (2 h at 100 V and −20°C; chilled 25 mM Tris-HCl-190 mM glycine-20% methanol was used as transfer buffer). MccE492 was detected with a polyclonal antibody prepared in rabbits against the last 20 amino acids of the protein (antiserum dilution, 1:500) and with a horseradish peroxidase-linked anti-rabbit goat antibody (Pierce) (dilution, 1:20,000). The chemiluminescence reaction was performed in 100 mM Tris-HCl, pH 8.5, 1.25 mM luminol, and 0.2 mM p-coumaric acid. The reaction was started by the addition of an aliquot of 30% H2O2 (final concentration, 0.01%). The membrane was exposed to X-OMAT AR film (Kodak) for 2 to 5 min, depending on the signal obtained.
Bioinformatic tools.
The sequences of the crystallized proteins used in this work were taken from their respective Protein Data Bank (PDB) files, using the program Swiss-PDB Viewer V3.7 (http://expasy.org/spdbv). Sequence alignment was performed with the AlignX module of the VectorNT19 program (Informax), using the blosum62mt2 matrix under the standard conditions provided by the software. Three-dimensional analysis of the structures was performed using the VMD1.8.5 program (23), freely available at http://www.ks.uiuc.edu/Research/vmd/.
MALDI-TOF mass spectrometry (MALDI-TOF-MS).
Samples were analyzed in a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Bruker Daltonics, Inc.) with α-cyano-4-hydroxycinnamic as the matrix (10 mg/ml in ACN-0.1% formic acid [1:1 {vol/vol}]). Data acquisition was performed in positive polarity and reflection mode, and the final spectra corresponded to 10 scans of 40 laser shots in different points selected at random. Calibration was carried out with an external standard, using a mix of peptides between 5,000 and 20,000 Da (Bruker Daltonics Inc.).
RESULTS AND DISCUSSION
Production of active MccE492 requires the synthesis of enterochelin.
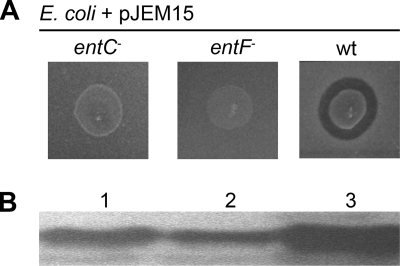
Two strains of E. coli carrying mutations in genes involved in enterochelin synthesis (entC and entF) were transformed with pJEM15, a plasmid that encodes the MccE492 system. The resulting strains did not produce antibacterial activity on a sensitive lawn (Fig. 1A). The lack of activity was not due to the absence of synthesis or secretion of MccE492 but rather to the production of an inactive form, as established by immunoblotting of purified samples from supernatant cultures of these strains (Fig. 1B). These samples did not present antibacterial activity even at protein concentrations equivalent to or higher than those in preparations obtained from the wild-type host. The mass of MccE492 isolated from the ent mutant strains was determined by MALDI-TOF-MS and corresponded to that of the unmodified form (7,887 Da), which had the N terminus cleaved at the double-glycine-type motif during export (24; M. Tello and R. Lagos, unpublished results). The lesser secreted amount of unmodified MccE492 than of the active form was consistently found in several preparations, but the cause of this effect remains to be determined. The absence of posttranslational modification has also been observed for MccE492 exported by an aroB strain, with a host mutation that affects the synthesis of enterochelin (44), but there is no report about its activity. The lack of antibacterial activity in hosts defective in enterochelin production (entA and entF mutants) has been reported for MccH47 (3), and due to similarity with MccE492 in the maturation genes and in the C-terminal region, this bacteriocin probably undergoes the same posttranslational modification (36).
FIG. 1.
Production of inactive MccE492 in hosts carrying mutations in genes involved in the synthesis of enterochelin. (A) Aliquots (3 μl) of liquid cultures of E. coli VCS257 (wt), E. coli H5311 (entC), and E. coli ER1100A (entF) carrying the MccE492 producer system pJEM15 were seeded on a lawn of sensitive cells [E. coli BL21(DE3)], and activity was assessed by the appearance of growth inhibition halos. (B) Sodium dodecyl sulfate-PAGE and immunoblotting of samples of MccE492 purified from the strains mentioned in panel A (lane 1, entC mutant; lane 2, entF mutant; lane 3, wild type).
Enterochelin is needed as a precursor of the structural variant salmochelin (6, 21), which is used for the modification of MccE492 at serine 84 (43). Salmochelins are derivatives of enterochelin that are mono-, di-, or triglucosylated on the 2,3-dihydroxybenzoyl units by the action of the C-glycosyltransferase IroB (16). The MceC maturation protein of the MccE492 system is homologous to IroB, and a mutant in mceC produces inactive MccE492 (24). Characterization in vitro of MceC activity showed that it is similar to that of IroB, although IroB has a broader substrate scope than that of MceC (33).
MceC and IroB activities are exchangeable.
To prove that IroB and MceC have the same biological activity, a complementation experiment assessing the expression of active MccE492 in an mceC mutant was carried out using S. enterica, a host that carries the iroB gene. S. enterica could not be transformed with the overproducing system pJEM15, and therefore pJAM434, a wild-type plasmid derivative that produces a moderately low activity of MccE492 due to a different orientation of the mceGHIJ gene cluster, was used (11, 24, 45). Accordingly, the MccE492 maturation gene mutants used in this set of experiments were derivatives of this plasmid. The results are presented in Table 2. np133, a mutant in mceC, did not produce any activity when this system was expressed in E. coli, whereas medium antibacterial activity was observed when it was expressed in S. enterica. Complementation by the iro gene cluster has also been observed for MccH47 and M lacking the glycosyltransferase gene equivalent (35). The high activity of the pJAM434 system expressed in S. enterica compared to that in E. coli could be explained by a gene dosage increase of the maturation genes, because S. enterica expresses IroB and IroD, the equivalents to MceC and MceD, respectively (24, 33). On the other hand, S. enterica carrying plasmids with mutations in the other maturation genes (mceI and mceJ) did not complement the production of MccE492 antibacterial activity, indicating that these genes are not replaced in this host. As a negative control, a mutant in the structural gene of MccE492 (mceA) was used. To confirm that IroB replaces MceC function, the S. enterica iroB gene was cloned as described in Materials and Methods, and the complementation assay with E. coli showed that antibacterial activity was produced (Table 2).
TABLE 2.
Complementation of antibacterial activity in S. enterica and E. coli/pIroB of a mutant in the mceC maturation genea
| Plasmid | Complementation in host
|
|
|---|---|---|
| E. coli | S. enterica | |
| pJAM434 (wild type) | + | +++ |
| np133 (mceC) | − | ++ |
| np205 (mceI) | − | − |
| np221 (mceJ) | − | − |
| np220 (mceA) | − | − |
| npB4 (mceC) + pACYC184 | − | ND |
| npB4 (mceC) + pIroB | + | ND |
Aliquots (3 μl) of liquid cultures of E. coli VCS257 or S. enterica LT2 carrying the indicated plasmids were seeded on a lawn of sensitive cells as described in the legend to Fig. 1, and the activity was expressed as low (+), medium (++), or high (+++) (see Materials and Methods). ND, not determined. pACYC184 was the vector used to clone iroB.
Exogenous salmochelin is imported into E. coli, mainly through the Fiu receptor, and is used as a substrate for production of active MccE492.
The experiments described above strongly suggest that MceC is required only to produce salmochelin and that once this molecule is produced, it acts as the substrate for the subsequent steps of MccE492 C-terminal modification. In order to demonstrate that MceC or its equivalent, IroB, has no other participation in MccE492 maturation but to produce salmochelin, “trans-complementation” experiments with purified salmochelin were performed. The rationale of these experiments was based on the following observations: (i) cocultures of E. coli VCS257 carrying npB4 (mceC) and S. enterica produced active MccE492 and (ii) the same result was obtained if S. enterica was replaced in the coculture by E. coli/pIroB or E. coli cells expressing cloned mceC (not shown). The simplest explanation for these results is that the salmochelin produced and secreted in the cocultures can be internalized by the host with the mutation in mceC, producing active MccE492.
The addition of purified cyclic salmochelins (monoglucosyl-enterobactin [MGE] and diglucosyl-enterobactin [DGE]) to cells carrying the mceC mutation (Table 3) produced antibacterial activity. The activity obtained with DGE was higher than the activity obtained with MGE, which could mean that the uptake of DGE in E. coli is better than that of MGE and/or that DGE is a better substrate for the modification. On the other hand, no activity was observed in the supernatant of a culture containing nonmature MccE492 that was incubated with salmochelin either purified or present in a supernatant of S. enterica or E. coli carrying iroB (not shown), discarding the possibility that just the binding of salmochelin to nonmature MccE492 is sufficient for the antibacterial activity. This result also indicates that active MccE492 is produced by an intracellular process; consequently, if the entrance of salmochelin is prevented in cells defective in both the receptors and mceC, no maturation would take place. In Salmonella, IroN is the main receptor for salmochelin uptake, and Cir and FepA play minor roles in this process (21), and in E. coli there is participation of the catecholate siderophore receptors (46). IroN is absent in nonpathogenic E. coli strains, while FepA and Cir equivalents are present in E. coli strains. A triple mutant of all catecholate receptors (fepA fiu cir) was used as an E. coli host for the MccE492 system. The mutated cells were not impaired in the production of active MccE492 when they were transformed with the wild-type system, but no antibacterial activity was observed in the system mutated in mceC (Table 3). In contrast to that obtained with the wild-type host, no activity was recovered after the addition of salmochelin to the triple catecholate receptor mutant host. To investigate further the contribution of each receptor to salmochelin uptake, combinations of mutants in fepA, fiu, and cir were used. Table 3 shows that Fiu was the main receptor for salmochelin uptake, with a low participation of FepA and Cir. These experiments do not discriminate if the uptake by Fiu is of the ferric or nonferric form of salmochelin, although it seems unlikely that the nonferric form would be imported. Until this work, there was no definitive information regarding the ability of E. coli to secrete and internalize salmochelin. In this respect, Zhu et al. (46) did not observe salmochelin iron uptake in an E. coli entC mutant, using a biological assay that detects bacterial growth under iron-depriving conditions of E. coli strains that have been supplemented with filter papers adsorbed with salmochelins S4 (the same as DGE) and S2 (linear DGE). This assay involves other steps, such as the esterase needed for iron release from ferric salmochelin, that can influence iron availability and may explain the different results found in this work. Thus, the enterochelin esterase Fes present in E. coli is unable to hydrolyze ferric DGE (28), a salmochelin used by Zhu et al. (46), and consequently, no release of iron into the cytoplasm occurs. Therefore, synthesis of MccE492 with antibacterial activity can be used as an alternative biological assay to detect the production, import, and export of salmochelin.
TABLE 3.
Trans-complementation assays of an mceC mutant, using purified forms of cyclic salmochelins, and dependence on the ferric-catecholate receptorsa
| E. coli strain | Complementation with plasmid
|
|||
|---|---|---|---|---|
| pJEM15 (wild type) | npB4 (mceC)
|
|||
| No addition | +MGE | +DGE | ||
| DH5α (fepA+fiu+cir+) | +++ | − | ++ | +++ |
| H1876 (fepA fiu cir) | ++ | − | − | − |
| H1875 (fepA fiu+cir) | +++ | − | ++ | +++ |
| H1594 (fepA+fiu cir+) | ++ | − | ± | ± |
Trans-complementation was performed by supplementing the previously seeded indicated strains with 3 μl of purified salmochelin (1 mM MGE or DGE). The antibacterial activity was measured and evaluated as described in Table 2 (± indicates a detectable but very low activity).
Together, these results demonstrate that even though MccE492 maturation is an intracellular process, the salmochelin needed for the production of active MccE492 can be provided exogenously, with uptake mediated mainly by Fiu, and that in vivo, the salmochelin synthesis pathway can be separated from the process in which the covalent bond between salmochelin and MccE492 is formed. It is clear that salmochelin can parasitize the import and export of the enterochelin system, at least to a degree detectable by the MccE492 activity assay.
Production of active MccE492 requires EntF in a function different from that involved in the production of enterochelin-salmochelin.
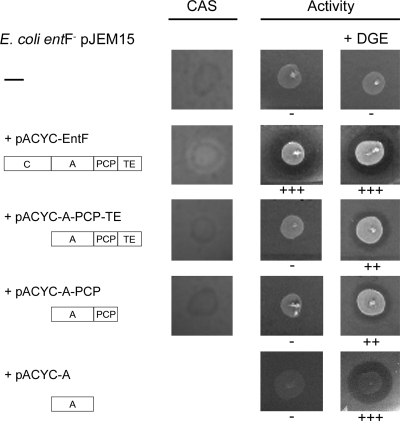
If the only cause for the production of inactive MccE492 in hosts defective in the production of enterochelin is the incapacity of these cells to produce enterochelin, the antibacterial activity should be recovered by the addition of the reaction products which are mutated in the host. Active MccE492 was produced when the entC strain E. coli H5311/pJEM15, a mutant that is unable to convert chorismate into isochorismate, a precursor for 2,3-dihydroxybenzoic acid (DHB) production, was incubated with DHB (reviewed in reference 12). A similar experiment was carried out with an entB host. Since EntB is a bifunctional enzyme that participates in the production of DHB and in the synthesis of enterochelin from DHB (18), the entB host was grown in the presence of DGE, and MccE492 activity was produced. The same result was obtained with enterochelin (not shown). Strikingly, incubation of the entF mutant (E. coli ER1100A/pJEM15) with salmochelin (or enterochelin [not shown]) did not produce the recovery of MccE492 activity (Fig. 2). This result suggests that EntF participates in MccE492 maturation not only by the production of enterochelin.
FIG. 2.
Dependence on the EntF A domain for production of active MccE492. The combinations of cloned domains of EntF indicated in the figure (left) were evaluated for the ability to complement the production of enterochelin (CAS plates) and the production of active MccE492 when supplemented with DGE, as described in Table 3. The host used was E. coli ER1300H (entF)/pJEM15. E. coli BL21(DE3) was used as the indicator strain.
The adenylation domain of EntF, but not the adenylation activity, is necessary for the production of active MccE492.
The nonribosomal peptide synthetase EntF is a modular protein of 142 kDa with four domains, namely, an N-terminal elongation/condensation domain (C), an adenylation domain (A), a peptidyl carrier protein (PCP), and the C-terminal thioesterase domain (TE), all of which are needed for enterochelin production (reviewed in reference 12). The activities of some of these domains (individually and in combination) have been tested in vitro (15, 38), and due to the diversity in function, it is unlikely that all the domains are involved in this stage of MccE492 maturation. To circumscribe the domain(s) involved, combinations of these domains were cloned and coexpressed with the MccE492 system in an entF mutant background, and the putative effect on MccE492 maturation was assessed by the addition of salmochelin to the culture. As expected, none of the constructs carrying a domain deletion complemented the production of enterochelin, as determined on CAS plates, and none of them produced antibacterial activity in the absence of salmochelin (Fig. 2). The control with the whole entF gene fully complemented both phenotypes (MccE492 activity and enterochelin production). The complementation assays were positive with the combinations A-PCP-TE, A-PCP, and A, demonstrating that only the EntF A domain is required for an MccE492 maturation stage that is ahead of salmochelin production. To investigate if the participation of the EntF A domain is related to its adenylation activity, a mutant without this activity was designed and constructed, and its abilities to synthesize enterochelin and to produce active MccE492 were tested.
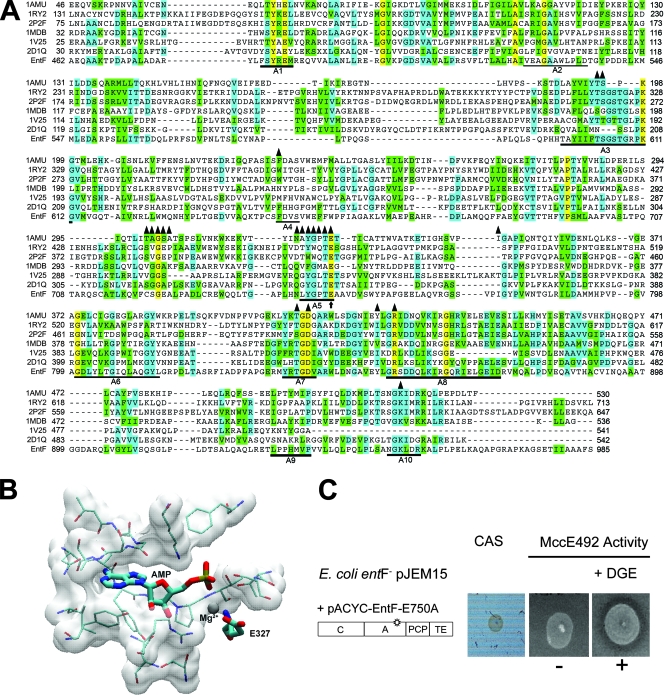
A comparative study among proteins containing an adenylation domain and known crystal structure homologues to EntF was performed. The adenylation domain of EntF belongs to the superfamily of “adenylate-forming enzymes” and is conserved in all its members (9). Although the chemistry of aminoacyl-adenylate formation is analogous to the ribosomal pathway in activating its substrate, there is no structural homology with the aminoacyl tRNA synthetases (aaRSs) (9). Only two proteins of the nonribosomal peptide synthetase group with the A domain have been crystallized, including structure 1AMU, a phenylalanine-activating subunit of gramicidin synthetase 1 that participates in gramicidin S synthesis (10); and structure 1MBD, which corresponds to DhbE, a protein that activates DHB and participates in the synthesis of the catecholate siderophore bacillibactin (30). Figure 3A shows an alignment of a region of the adenylation domains of six crystallized proteins of this superfamily where it is possible to detect residues that are 100% conserved. The selection of a residue for site-directed mutagenesis was focused on those close to or at the active site, so the residues that were 5 Å or less from this site, using the structure of 1AMU crystallized with AMP-Mg as a reference, were identified (Fig. 3A). In spite of their unrelated structures, the comparison of the well-characterized active sites of aaRSs with crystallized A domains in the presence of their substrates showed strong similarities, with chemically equivalent residues for ATP binding and for the stabilization of the negatively charged transition state (1, 8). The conserved residue E750 in the EntF A domain (29) was selected for mutagenesis because in the 1AMU crystal structure the corresponding residue (E327) is in contact with Mg2+ (Fig. 3B). This could be chemically equivalent to a key glutamic acid residue in aaRSs (10). The change E750A was introduced in EntF, and as predicted, no synthesis of enterochelin was observed (Fig. 3C). However, this mutant was able to produce active MccE492 in the presence of DGE, indicating that the adenylation activity of this protein is not essential for MccE492 maturation. Trans-complementation was observed only in the presence of DGE.
FIG. 3.
Design of the E750A mutation in the EntF A domain and its effect on MccE492 maturation. (A) Alignment of the relevant region of the EntF A domain with the sequences of different crystallized adenylation domains obtained from the following PDB files: 1AMU, 1RY2, 2P2F, 1MDB, 1V25, and 2D1Q. Residues in yellow are 100% conserved. Residues in green and blue are conserved substitutions according to Vector NTI nomenclature (green, conserved substitutions with respect to the consensus; blue, identical residues with respect to the consensus). Triangles mark the residues located 5 Å or less from the active site in the 1AMU crystal structure. The arrow marks the residue selected for mutagenesis. (B) Structure of the active site of the 1AMU protein with bound AMP-Mg. Mg2+ and AMP are shown in the structure. The selected residue (E327) is equivalent to residue E750 in EntF. (C) The EntF E750A mutant was evaluated for the ability to produce enterochelin (CAS plate) and active MccE492 in a sensitive lawn supplemented with DGE. The host used was E. coli ER1300H (entF)/pJEM15, and E. coli BL21(DE3) was used as the indicator strain.
Nolan et al. (33) demonstrated that in vitro modification of MccE492 requires only salmochelin and MceIJ for the addition of this molecule to serine 84; however, it was not reported whether the modified MccE492 produced in vitro was active. There are two possibilities for the role in vivo of EntF in MccE492 maturation, namely, that it is somehow related to the modification process or, more unlikely, that it is involved in a completely different, as yet unknown process that requires the EntF A domain for the production of active MccE492. The latter would be discarded easily if it was demonstrated that modified MccE492 produced in vitro in the absence of EntF is active. Regarding the former possibility, the EntF A domain could be used as a scaffolding protein required in vivo to complex MceIJ, and probably MceD and MceC, in such a way that the effective concentration of all these components plus salmochelin is augmented or, alternatively, as a chaperone for the presentation of the C-terminal amino acid of MccE492 for modification. In this respect, and to explain the results mentioned above (33), the requirement of EntF in vitro would be circumvented by the high concentrations in the assay of all the elements participating in MccE492 modification or because under the assay conditions MccE492 is partially unfolded and the C-terminal amino acid is available for modification. Most of the MceIJ characterization was carried out with a C10 model peptide that seems to be optimal for the enzymatic reaction. In any case, the EntF A domain seems to be bifunctional, with the participation of its adenylating activity for enterochelin synthesis and as a scaffolding/chaperone protein for MccE492 modification.
Concluding remarks and perspectives.
The deficiency in the production of active MccE492 when there is impairment of bacteriocin modification indicates that this step is essential for the production of MccE492 with antibacterial activity. The synthesis of active MccE492 can be divided into the following four independent steps: first, the synthesis of the unmodified peptide; second, the synthesis of salmochelin-like molecules; third, the covalent linkage process of these molecules to the C-terminal amino acid; and fourth, the processing and export of this bacteriocin. The MccE492 system appears to have coevolved with enterochelin metabolism to a point at which the end product is used not only as a substrate for further modification and subsequent addition to the bacteriocin but also in the utilization of EntF, with another activity different from that used for enterochelin synthesis. The most probable function of MccE492 modification is recognition by the catecholate siderophore (discussed in reference 35), a strategy that has been included in the so-called Trojan horse antibiotics (31) because they exploit the Fe3+-siderophore uptake system as a means of entering cells.
Acknowledgments
We are grateful to Michael Handford for critically reading the manuscript. We are indebted to M. A. Fischbach and C. T. Walsh for the generous gift of salmochelin (MGE and DGE), the Ent mutant strains, and plasmids and for communicating their results on MccE492 posttranslational modification prior to publication. The help of José J. Arbildúa, Claudio Aguilar, and Roberto Kolter in different aspects of this work is acknowledged. We also thank Roselyn Orellana for technical assistance.
Experiments in the Mass Spectrometry Unit at University of Chile were funded by a MECESUP UCH-0115 project. G.M. and M.T. received predoctoral CONICYT fellowships. This work was supported by grant 1061128 from the Fondo Nacional de Desarrollo Científico y Tecnológico.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Ador, L., S. Jaeger, R. Geslain, F. Martin, J. Cavarelli, and G. Eriani. 2004. Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase. Biochemistry 437028-7037. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. Greene Publishing Associates and John Wiley & Sons, New York, NY.
- 3.Azpiroz, M. F., and M. Laviña. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 481235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler, A. J., R. M. Tsolis, A. W. M. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183207-213. [DOI] [PubMed] [Google Scholar]
- 5.Bieler, S., L. Estrada, R. Lagos, M. Baeza, J. Castilla, and C. Soto. 2005. Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 28026880-26885. [DOI] [PubMed] [Google Scholar]
- 6.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Süssmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17471-481. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84365-380. [DOI] [PubMed] [Google Scholar]
- 8.Cavarelli, J., G. Eriani, B. Rees, M. Ruff, M. Boeglin, A. Mitschler, F. Martin, J. Gangloff, J.-C. Thierry, and D. Moras. 1994. The active site of aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 13327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti, E., N. P. Franks, and P. Brick. 1996. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzyme. Structure 4287-298. [DOI] [PubMed] [Google Scholar]
- 10.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 164174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corsini, G., M. Baeza, O. Monasterio, and R. Lagos. 2002. The expression of genes involved in microcin maturation regulates the production of active microcin E492. Biochimie 84539-544. [DOI] [PubMed] [Google Scholar]
- 12.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo, V. 1984. Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch. Microbiol. 13972-75. [DOI] [PubMed] [Google Scholar]
- 14.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands [PAI I(536) to PAI IV(536)] of uropathogenic Escherichia coli strain 536. Infect. Immun. 706365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehmann, D. E., C. A. Shaw-Reid, H. C. Losey, and C. T. Walsh. 2000. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc. Natl. Acad. Sci. USA 972509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2006. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2132-138. [DOI] [PubMed] [Google Scholar]
- 18.Gehring, A. M., K. A. Bradley, and C. T. Walsh. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 368495-8503. [DOI] [PubMed] [Google Scholar]
- 19.Hantke, K. 1983. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet. 191301-306. [DOI] [PubMed] [Google Scholar]
- 20.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 675-8. [DOI] [PubMed] [Google Scholar]
- 21.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 1003677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz, C., M. R. Bono, L. F. Barros, and R. Lagos. 2002. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc. Natl. Acad. Sci. USA 992696-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graphics 1433-38. [DOI] [PubMed] [Google Scholar]
- 24.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization, and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42229-243. [DOI] [PubMed] [Google Scholar]
- 25.Lagos, R., J. E. Villanueva, and O. Monasterio. 1999. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 181212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagos, R., M. Wilkens, C. Vergara, X. Cecchi, and O. Monasterio. 1993. Microcin E492 forms ion channels in phospholipid bilayer membranes. FEBS Lett. 321145-148. [DOI] [PubMed] [Google Scholar]
- 27.Lai, J. R., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2006. A protein interaction surface in nonribosomal peptide synthesis mapped by combinatorial mutagenesis and selection. Proc. Natl. Acad. Sci. USA 1035314-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 12711075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 972651-2673. [DOI] [PubMed] [Google Scholar]
- 30.May, J. J., N. Kessler, M. A. Marahiel, and M. T. Stubbs. 2002. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 9912120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Nolan, E. M., M. A. Fischbach, A. Koglin, and C. T. Walsh. 2007. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J. Am. Chem. Soc. 12914336-14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orellana, C., and R. Lagos. 1996. The activity of microcin E492 from Klebsiella pneumoniae is regulated by a microcin-antagonist. FEMS Microbiol. Lett. 136297-303. [DOI] [PubMed] [Google Scholar]
- 35.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 1492557-2570. [DOI] [PubMed] [Google Scholar]
- 36.Poey, M. E., M. F. Azpiroz, and M. Laviña. 2006. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 501411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pons, A.-M., N. Zorn, D. Vignon, F. Delalande, A. Van Dorsselaer, and G. Cottenceau. 2002. Microcin E492 is an unmodified peptide related in structure to colicin V. Antimicrob. Agents Chemother. 46229-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roche, E. D., and C. T. Walsh. 2003. Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis. Biochemistry 421334-1344. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 41.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 16047-56. [DOI] [PubMed] [Google Scholar]
- 42.Strahsburger, E., M. Baeza, O. Monasterio, and R. Lagos. 2005. Cooperative uptake of microcin E492 by receptors FepA, Fiu, and Cir and inhibition by the siderophore enterochelin and its dimeric and trimeric hydrolysis products. Antimicrob. Agents Chemother. 493083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, X., D. Destoumieux-Garzón, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J.-C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 27928233-28242. [DOI] [PubMed] [Google Scholar]
- 44.Vassiliadis, G., J. Peduzzi, S. Zirah, X. Thomas, S. Rebuffat, and D. Destoumieux-Garzón. 2007. Insight into siderophore-carrying peptide biosynthesis: enterobactin is a precursor for microcin E492 posttranslational modification. Antimicrob. Agents Chemother. 513546-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkens, M., J. E. Villanueva, J. Cofré, J. Chnaiderman, and R. Lagos. 1997. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J. Bacteriol. 1794789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, M., M. Valdebenito, G. Winkelmann, and K. Hantke. 2005. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 1512363-2372. [DOI] [PubMed] [Google Scholar]