Abstract
4-Hydroxy-2-alkylquinolines (HAQs), especially 3,4-dihydroxy-2-heptylquinoline (Pseudomonas quinolone signal) and its precursor, 4-hydroxy-2-heptylquinoline, are attracting much attention, mainly because of their role as signaling molecules in Pseudomonas aeruginosa. The pqsABCDE operon is centrally involved in their biosynthesis. The presence of a homologous operon in Burkholderia pseudomallei and B. thailandensis was recently reported. Thus, we have investigated the abilities of 11 Burkholderia species to produce HAQ-like molecules by liquid chromatography/mass spectrometry. We have identified 29 different HAQ derivatives produced by the only three Burkholderia species where a pqsABCDE homologue was found among available sequenced Burkholderia species genomes, including B. ambifaria, a member of the Burkholderia cepacia complex. In contrast with those of P. aeruginosa, Burkholderia HAQs typically bear a methyl group, hence their designation as 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs). We identified three families of HMAQs with a saturated or unsaturated alkyl chain at the 2′ position, in contrast with the 1′ position of P. aeruginosa, including one with an N-oxide group. Furthermore, the operon in these species contains two more genes downstream of the pqsE homologue, resulting in the hmqABCDEFG operon. While the inactivation of hmqA inhibits the production of HMAQs, the methylation of the quinoline ring requires a putative methyltransferase encoded by hmqG. Interestingly, hmqA or hmqG mutations increase the production of acyl homoserine lactones and, consequently, phenotypes under the control of quorum sensing in B. ambifaria: antifungal activity, siderophore production, and proteolytic activity. These results indicate that only HAQs bearing a methyl group (HMAQs) are involved in quorum-sensing regulation.
Bacteria produce and release an array of secondary metabolites. These molecules possess biological activities that endow the producer with increased fitness to compete with other species, infect various hosts, and colonize diverse ecological niches. 4-Hydroxy-2-alkylquinolines (HAQs), originally named “pyo compounds,” have been long known as a class of antimicrobials produced by the opportunistic bacterial pathogen Pseudomonas aeruginosa (10, 25, 58). Notably, some of these compounds inhibit the growth of Staphylococcus aureus and other gram-positive bacteria and even the development of phytoplankton (35, 36). Many HAQs also act as iron chelators (2, 49) and even immune modulants (28). HAQs have recently attracted much attention because of their role as intercellular signaling molecules in P. aeruginosa (14, 45).
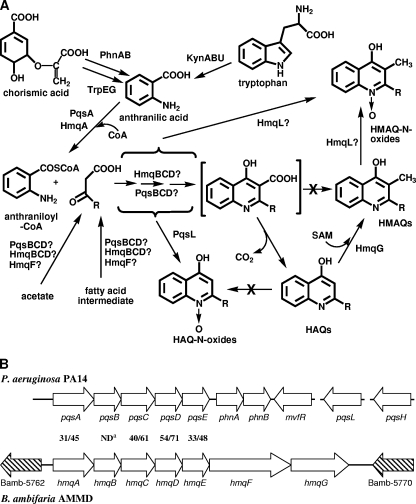
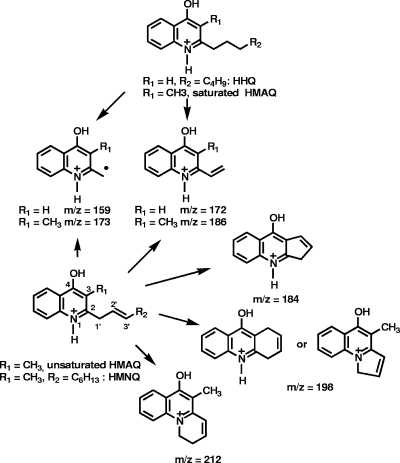
P. aeruginosa produces a large variety of HAQs belonging to various families characterized by the presence of a hydrogen or hydroxyl group at the 3 position, an N-oxide group in place of the quinoline nitrogen, and an unsaturation on the alkyl side chain (32). Among these compounds, 3,4-dihydroxy-2-heptylquinoline (Pseudomonas quinolone signal [PQS]) and 4-hydroxy-2-heptylquinoline (HHQ) act as signaling molecules and are implicated in the regulation of the expression of multiple virulence genes in this bacterium (17, 39). The pqsABCDE operon encodes the primary proteins mediating HAQ biosynthesis (Fig. 1) (14, 22). PqsA is probably an anthranilate-coenzyme A (CoA) ligase (33), while pqsB, pqsC, and pqsD encode proteins similar to β-keto-acyl-acyl carrier protein synthases (22). The last gene of the operon, pqsE, is not involved in the biosynthesis of HAQs but is instead required for the cellular response to these compounds (13, 14, 22).
FIG. 1.
(A) Biosynthetic pathway of HAQs in P. aeruginosa and HMAQs in Burkholderia. (B) Comparison of the P. aeruginosa PA14 (http://pseudomonas.com) region containing the pqsABCDE operon with the B. ambifaria AMMD region containing the hmqABCDEFG operon (http://burkholderia.com). Values represent identities/similarities between the different gene products of these operons. Searches for identity and similarity were performed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/). a, indicates that, as reported in the text, HmqB exhibits identity/similarity only with the amino-terminal half of PqsB.
The phnAB operon catalyzes the biosynthesis of anthranilic acid, the primary precursor of HAQs (14). Both pqsABCDE and phnAB are positively controlled by the LysR-type transcriptional regulator MvfR (PqsR) (5, 14, 38), whose coinducing ligands are HHQ and PQS (57, 61). The conversion of HHQ into PQS is mediated by PqsH, a putative monooxygenase not controlled by MvfR (14, 61), while the synthesis of HAQ N-oxides requires the pqsL gene (14). The production and activity of HAQs are part of a sophisticated regulatory network mediated by small intercellular signaling molecules called quorum sensing (QS) (15). QS, which relies on the activation of specific transcriptional regulators by autoinducers functioning as diffusible signals, regulates the expression of factors involved in host-microbe interactions, such as virulence factors (29, 59). In P. aeruginosa, the QS system, via the regulators LasR and RhlR, influences the production of HAQs on various levels: LasR upregulates the transcription of both mvfR and pqsH, while RhlR represses the transcription of pqsABCDE (57, 62). The production, regulation, and functions of HAQs in P. aeruginosa are thus under intense scrutiny in order to obtain a better understanding of virulence expression in this bacterium.
Formerly included in the pseudomonad family, the Burkholderia genus, of the Betaproteobacteria, comprises more than 40 species that inhabit remarkably diverse ecological niches, as they have been isolated from soil, plant rhizosphere, water, insects, fungi, hospital environments, and infected humans. Many Burkholderia species have developed beneficial interactions with their plant hosts and have considerable ecological importance: several species of Burkholderia have proven to be very efficient biocontrol and bioremediation agents (7, 53). In contrast, a number of Burkholderia species, especially members of the Burkholderia cepacia complex, are also increasingly prevalent opportunistic human pathogens that cause severe respiratory infections in people suffering from cystic fibrosis (CF) or chronic granulomatous disease (8). In addition, two other species, B. mallei and B. pseudomallei, are well-characterized primary pathogens of animals and humans. B. mallei is the etiological agent of glanders, which is mainly a horse disease but affects humans in rare cases (44). The saprophyte B. pseudomallei is the causative agent of melioidosis, a potentially fatal septicemic infection of animals and humans (6). B. thailandensis is very closely related to B. pseudomallei but is generally considered to be avirulent (4).
Not surprisingly, these versatile bacteria release a large variety of extracellular products including proteases, siderophores, toxins, antifungals, and antimicrobials, many of which are under QS control (56). While many Burkholderia strains, including B. ambifaria, carry only one QS system homologous to the P. aeruginosa Las and Rhl systems, some strains, such as B. pseudomallei and B. thailandensis, harbor five different QS transcriptional regulators and three acyl homoserine lactone (AHL) synthases (19). Evidence that the QS systems operating in Burkholderia are crucial for full virulence in various animal models has accumulated over the past few years.
With the help of a bioassay, HAQs similar to those produced by P. aeruginosa were recently detected in concentrated extracts of B. pseudomallei and B. thailandensis cultures (16). We have investigated the abilities of Burkholderia species to produce HAQ-like molecules. The present work describes the precise identification of the distinct HAQ derivatives produced by three Burkholderia species, including B. ambifaria, a member of the B. cepacia complex, and unique insights into their biosynthetic pathway. We found that in contrast with those of P. aeruginosa, Burkholderia HAQs contain predominantly an unsaturated aliphatic side chain and are typically methylated (at the 3 position), hence their designation as 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs). This methylation results from the presence of a putative methyltransferase encoded by the last gene of the hmqABCDEFG operon. Finally, we show that the synthesis of HMAQs negatively regulates the production of AHLs in B. ambifaria and consequently modulates QS-regulated phenotypes.
MATERIALS AND METHODS
Bacteria and culture conditions.
Burkholderia strains used in this study are shown in Table 1. Escherichia coli SM10 λpir (thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir) served as a donor for conjugation experiments (51). Unless otherwise stated, B. thailandensis and B. cepacia complex strains were routinely grown in tryptic soy broth (TSB) (Difco) at 37°C with shaking (240 rpm) or on TSB agar plates. B. pseudomallei strain 1026B and B. mallei strain ATCC 23344 were grown with shaking (200 rpm) in LB broth (Difco) for 36 h at 37°C. When required, chloramphenicol was used at 40 μg/ml for B. ambifaria HSJ1 and at 100 μg/ml for B. thailandensis E264.
TABLE 1.
Burkholderia strains used in this study
| Straina | Source and location | Reference or source |
|---|---|---|
| B. cepacia (gen. I) LMG1222 | Onion, United States | 37 |
| B. cepacia (gen. I) LMG18821 | CF, Australia | 37 |
| B. multivorans (gen. II) LMG17588 | Soil, United States | 37 |
| B. multivorans (gen. II) LMG16660 | CF, United Kingdom | 37 |
| B. cenocepacia (gen. III) LMG19240 | Wheat, Australia | 55 |
| B. cenocepacia (gen. III) J2315 | CF, United Kingdom | 37 |
| B. cenocepacia (gen. III) LMG18830 | CF, Australia | 37 |
| B. stabilis (gen. IV) LMG18870 | CF, Canada | 37 |
| B. vietnamiensis (gen. V) LMG22486 | Water treatment, United States | 43 |
| B. vietnamiensis (gen. V) LMG18835 | CF, United States | 37 |
| B. dolosa (gen. VI) LMG21819 | CF, United States | 9 |
| B. dolosa (gen. VI) LMG21443 | Root, Senegal | 9 |
| B. ambifaria (gen. VII) HSJ1 | CF, Canada | This study |
| B. pyrrocinia (gen. IX) LMG21824 | CF, United Kingdom | 9 |
| B. thailandensis E264 | Soil, Thailand | 4 |
| B. mallei ATCC 23344 | Mule, China | 12 |
| B. pseudomallei 1026b | Clinical isolate | 11 |
For B. cepacia complex strains, genomovar (gen.) status is indicated in parentheses.
Feeding experiments with labeled anthranilic acid were performed by supplementing cultures at an optical density at 600 nm [OD600] of 2.0 with 20 mg/liter 3,4,5,6-tetradeutered anthranilic acid. The experiment with 15N-anthranilic acid (CDN isotopes) was performed in the same way but with 50 mg/liter. Experiments with tryptophan were performed in M63 medium plus 0.1% dextrose supplemented with 10 mg/liter 2′,4′,5′,6′,7′-pentadeutered l-tryptophan (CDN isotopes).
Assays with 6-FABA.
Anthranilic acid analogue 6-fluoro-2-aminobenzoic acid (6-FABA) stock was prepared at a concentration of 300 mM in methanol. B. ambifaria cells were grown as previously described in TSB minus or plus 6-FABA. Concentrations of 0.25 mM, 0.5 mM, and 1 mM were tested. Samples were taken when the maximum production of HMAQs is normally achieved (stationary phase) and prepared for liquid chromatography (LC)/mass spectrometry (MS) analysis as explained below. Cultures were performed in triplicate.
The competing concentration of anthranilic acid used for complementation was twice that of the analogue. Samples were taken at the beginning of the exponential phase and prepared for LC/MS analysis. Cultures were performed in triplicate.
Construction of hmqA and hmqG mutants.
Briefly, a 757-bp internal fragment of hmqA and a 525-bp fragment of hmqG were amplified from B. ambifaria HSJ1 using primers hmqAKOF, with an XbaI site (Table 2), and hmqAKOR, with a KpnI site, for the hmqA gene and using primers hmqGR and hmqGF with the same restriction sites for the hmqG gene. The PCR products were digested with KpnI and XbaI and ligated into the XbaI and KpnI sites of the suicide vector pKNOCK-Cm (1). The construct was then electroporated in E. coli SM10 cells. The resulting plasmid was mobilized from E. coli SM10 into B. ambifaria HSJ1 cells by mating. Single-crossover insertion mutants were selected on TSB agar containing 20 μg/ml gentamicin and 40 μg/ml chloramphenicol. The insertion of the plasmid into the target gene was confirmed by PCR.
TABLE 2.
Primers used in this study
| Primer | Primer sequence (5′-3′)a |
|---|---|
| hmqAKOF | GCTCTAGAGCAAGTTCGACGCTACGAGACG |
| hmqAKOR | GGGGTACCCCAGGTTCCAGCCGTACTTGAC |
| hmqTKOF | GGGGTACCCCGGTGCTACGACACTGAAGA |
| hmqTKOR | GCTCTAGAGCTAGCCGTCCTCGTCCAGATA |
| hmqGR | CTAGTCTAGACTAGTTGTGGACCAGGAACATGAA |
| hmqGF | CGGGGTACCCCGTTGCTCGACGAACTCCTGT |
Restriction sites designed into the primers are underlined.
The same procedure was used for the construction of the B. thailandensis E264 hmqA mutant: a 788-bp internal fragment of hmqA was amplified from E264 using primers hmqTKOF, with a KpnI site, and hmqTKOR, with an XbaI site, and cloned into the XbaI and KpnI sites of pKNOCK-Cm. Single-crossover insertion mutants were selected on TSB agar containing 20 μg/ml gentamicin and 100 μg/ml chloramphenicol.
LC/MS analyses.
For HAQ quantification, culture samples were prepared, as described previously (14, 31), by adding methanol containing 20 mg/liter internal standard (PQS-d4 [5,6,7,8-tetradeutero-3,4-dihydroxy-2-heptylquinoline]) to culture samples for a final concentration of 50%. The mixture was centrifuged for 10 min, and the supernatant was collected for analysis. For B. pseudomallei 1026b and B. mallei GB8, cell-free supernatants were obtained by centrifugation at 10,000 × g for 20 min. Five milliliters of the supernatants was acidified by the addition of glacial acetic acid (20 μl/ml medium), and this was extracted three times with ethyl acetate (1:1, vol/vol); the extracts were combined and evaporated at 35°C under a gentle stream of nitrogen. The residue was then resuspended in 5 ml of methanol. For AHL quantification, 480 μl of culture samples was mixed with 120 μl of acetonitrile containing 50 mg/liter of internal standard (PQS-d4) for a final concentration of 10 mg/liter.
The analyses were performed with a Quattro II (Waters, Mississauga, Ontario, Canada) triple-quadrupole mass spectrometer in positive electrospray ionization mode. Collision-induced dissociation was performed with argon as the collision gas at 2 × 10−3 mTorr. The mass spectrometer was coupled to an HP 1100 (Agilent Technologies, Saint Laurent, Quebec, Canada) high-performance liquid chromatograph equipped with a 3- by 150-mm C8 Luna (Phenomenex, Torrance, CA) reverse-phase column. The flow rate was 0.4 ml/min split to 10% through a Valco Tee. A water-acetonitrile gradient with 1% acetic acid was used as the mobile phase. Analysis of all the HMAQs and HAQs was performed in full scan mode using the response factor of 4-hydroxy-3-methyl-2-nonenylquinoline (HMNQ) and using PQS-d4 as an internal standard (31). Assays of cultures were carried out in triplicate, and detailed quantification of HMAQs/HAQs was performed at the time of maximum production of HMAQs (for B. ambifaria and B. thailandensis) or at the early stationary phase (B. pseudomallei). For B. pseudomallei and B. mallei, a single analysis was made from cultures sampled at 18 and 36 h, respectively. For AHL, data acquisition was performed in full scan mode with a scanning range of 130 to 350 Da. The precise quantification of C8-HSL was performed with tandem MS (MS/MS) as described previously (13).
NMR.
HMNQ was dissolved in deuterochloroform for nuclear magnetic resonance (NMR) analysis. The NMR apparatus was a Bruker SpectroSpin model operating at 500 MHz.
Phenotype assays.
Antifungal and proteolytic activities of the culture supernatants were measured by a zone-of-inhibition assay. Proteolytic and antifungal activities were evaluated on agar plates containing 1% skim milk powder and on TSB agar plates inoculated with 100 μl of Candida albicans culture, respectively. Briefly, B. ambifaria strains were grown in TSB until an OD600 of 5.0 (end of the exponential phase) was achieved. After centrifugation and filter sterilization, supernatant samples were loaded into wells made by removing agar plugs. The plates were incubated for 24 h at 30°C. Antifungal and proteolytic activities were estimated from the area (mm2) of the inhibition zone surrounding the wells. This experiment was performed in triplicate and repeated twice with similar results.
Siderophore production was determined by use of the Chrome Azurol S (CAS) assay. On CAS agar, siderophores remove iron from the CAS dye complex, resulting in a blue-to-orange color change in zones surrounding the colonies (50). B. ambifaria strains were grown in TSB until an OD600 of 2.0 was reached and were spotted (3 μl) onto CAS agar. The plates were incubated for 24 h at 37°C. The production of siderophores was estimated by measuring the area of the halo (mm2) surrounding the colonies. After 24 h, the number of CFU present in each colony was determined by plating serial dilutions on TSB agar, and siderophore production values were corrected for the number of CFU. Eight colonies per strain were examined, and the experiment was repeated twice, with similar results.
RESULTS
Homologues of the pqsABCDE operon in Burkholderia species.
A search for pqsABCDE homologues in completed and uncompleted Burkholderia genome sequences (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) confirmed the presence of homologues to these genes arranged as a putative operon on chromosome II of B. pseudomallei (BPSS00481 to BPSS00485 for strain K96243) and B. thailandensis (BTH_II1935 to BTH_II1931 for strain E264), as recently reported (16). B. oklahomensis, a new species closely related to B. pseudomallei (23), was also found to contain a putative pqsABCDE operon. We noticed pqsABCDE homologues in all available sequenced B. pseudomallei, B. thailandensis, and B. oklahomensis strains. Interestingly, in silico analysis of the sequenced B. mallei genomes indicates that this species, while closely related to B. pseudomallei, does not carry this putative operon arrangement. Furthermore, we also found a corresponding putative operon on chromosome III of the biocontrol B. ambifaria strain AMMD, which belongs to the B. cepacia complex as genomovar VII (Fig. 1) (7). This region displays a GC content of 66.4%, a typical value for the B. cepacia complex. Compared with P. aeruginosa PA14 pqsABCDE gene products, B. ambifaria AMMD gene products exhibit from 31% identity for the homologue of PqsA to 54% identity for the homologue of PqsD. Interestingly, however, the second open reading frame of the B. ambifaria AMMD operon encodes a protein showing 40% identity with only the first 150 amino acids of PqsB, the region containing a domain similar to β-keto-acyl-acyl carrier protein synthases. This putative operon is well conserved among the various Burkholderia strains carrying it and demonstrates a high level of protein sequence identity. For example, between B. ambifaria AMMD and B. pseudomallei K96243, protein identities range from 59% for the PqsB homologues to 84% for the PqsD homologues. A search for pqsH confirmed the absence of homologues in these Burkholderia species.
In several other Burkholderia genomes, especially strains belonging to the Burkholderia cepacia complex and B. mallei, putative homologues of pqsA, pqsB, pqsC, and pqsD are found dispersed throughout the genomes, but none carries a homologue of pqsE, which encodes the response effector.
Apart from the homology with pqsABCDE, the genomic context surrounding this operon is quite different in these bacteria compared with P. aeruginosa. The phnAB operon, encoding anthranilate synthase, is not found near pqsABCDE homologues or elsewhere in the genomes. Furthermore, no homologue of mvfR, which codes for the transcriptional regulator of pqsABCDE and phnAB in P. aeruginosa, is present in close vicinity of the Burkholderia pqsABCDE homologues. Instead, this operon contains two additional genes at the end in all strains of Burkholderia where a putative pqsABCDE operon was identified. In silico analysis did not reveal the presence of a putative promoter upstream from these two open reading frames. These two genes are well conserved among the strains of Burkholderia carrying the pqsABCDE homologues, e.g., 72% and 75% identity, respectively, between the B. ambifaria AMMD and the B. pseudomallei K96243 homologues. Homologues to these two genes are not found in any other sequenced Burkholderia or P. aeruginosa species. In B. ambifaria and B. thailandensis, we confirmed their inclusion as part of the polycistronic operon by reverse transcription-PCR (data not shown). Accordingly, a GC-rich hairpin-forming sequence followed by several thymines, which could function as a transcriptional terminator, lies only at the 3′ end of the last gene of this operon in B. ambifaria AMMD.
Because HAQ derivatives produced by these bacteria are usually methylated (as described below) and are thus responsible for the synthesis of HMAQs, we named this operon hmqABCDEFG, with hmqF and hmqG as the two additional genes (Fig. 1).
LC/MS and LC/MS/MS identification and characterization of HMAQs produced by Burkholderia species.
We tested the following Burkholderia species for the production of HAQ derivatives: B. pseudomallei, B. thailandensis, B. mallei, and 14 other strains covering most of the species belonging to the B. cepacia complex (Table 1). HMAQs were found only in cultures of the species carrying an hmqABCDEFG operon: B. pseudomallei, B. thailandensis, and B. ambifaria.
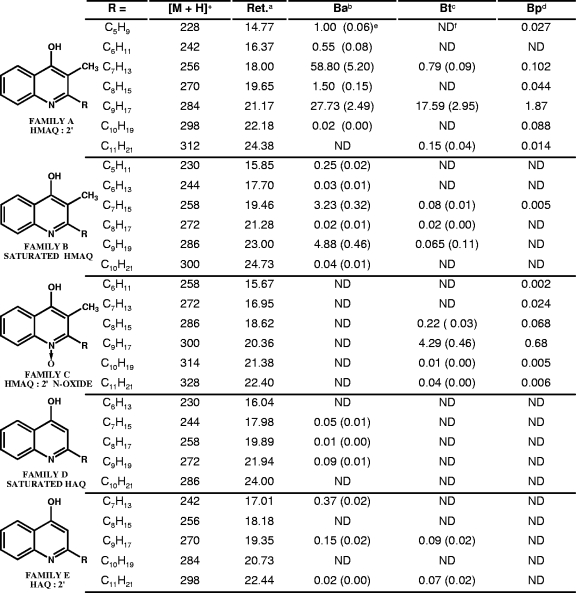
The reconstructed total-ion chromatogram (TIC) obtained from the supernatant of B. thailandensis cultivated until the maximum production of HMAQs was achieved reveals a large number of peaks dominated by one presenting a pseudomolecular ion at m/z 284. The corresponding molecule was isolated by flash chromatography and by high-performance LC, and its NMR was found to correspond to HMNQ, including the trans configuration of the double bond between the 2′ and 3′ carbons of the side chain (42). The MS/MS spectrum of HMNQ shows an intense ion at m/z 173 along with other ions of decreasing intensities at m/z 184, 186, 198, and 212. Having characterized this compound, we found two other compounds with pseudomolecular ions at m/z 256 and 312 that present the same MS/MS spectra as HMNQ. In addition to these three compounds, four additional ones are found, which present pseudomolecular ions at m/z 228, 242, 270, and 298 in B. ambifaria culture supernatants. They present the same MS/MS spectra as HMNQ and occur at regular intervals in the TIC chromatograms. All these characteristics are typical of compounds belonging to a family of molecules sharing a common structure and differing from one another by 14 Da, which corresponds to the addition of a methylene unit on a side chain (Fig. 2). B. pseudomallei also produces many molecules belonging to family A (Fig. 2).
FIG. 2.
Chemical structures, retention times, and concentrations (mg/liter) of the various congeners of the five families of HMAQs and HAQs identified in Burkholderia cultures. a, chromatographic retention time (Ret.) (minutes); b, B. ambifaria (Ba); c, B. thailandensis (Bt); d, B. pseudomallei (Bp); e, standard deviations of triplicate assays (in parentheses); f, not detected.
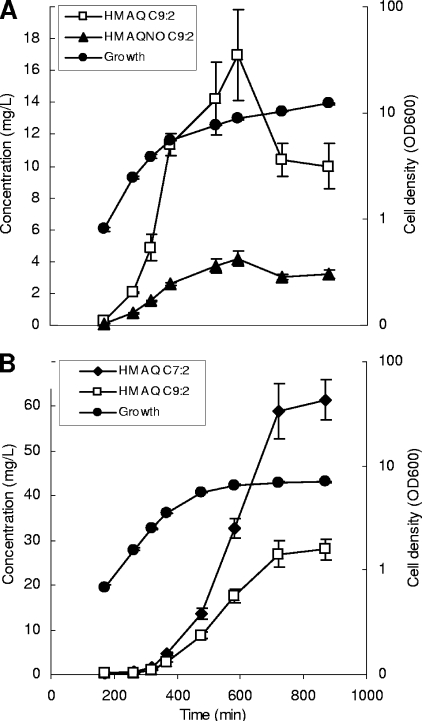
Having isolated pure HMNQ, we could determine its response factor under our analytical conditions. The concentrations of the members of this family and of all the other families of HMAQs were hence determined using this response factor. While the main HMAQ is HMNQ (m/z 284) in B. thailandensis, it is 4-hydroxy-3-methyl-2-heptenylquinoline (m/z 256) in B. ambifaria (Fig. 2). The HMAQ production kinetics for B. ambifaria and B. thailandensis performed with this response factor reveal distinct and characteristic production patterns (Fig. 3).
FIG. 3.
Production in function of growth of the major HMAQs produced by B. thailandensis (A) and B. ambifaria (B). Production was monitored until the stationary phase was reached. Error bars represent the standard deviations of data for triplicate cultures.
Most of the family B congeners, which display pseudomolecular ions at m/z 230, 244, 258, 272, 286, and 300, were found in B. ambifaria and B. thailandensis supernatants (Fig. 2). They also occur at regular intervals in the TIC chromatogram, and their MS/MS spectra all display ions at m/z 173, 186, and 200. The molecular weights of these compounds indicate that they are saturated analogues of the compounds belonging to the unsaturated HMAQ family A with a C5-to-C10 side chain. Fewer members of family B are produced and at lower concentrations in B. thailandensis than in B. ambifaria, while only minute amounts of the C7 congener are found in B. pseudomallei cultures (0.005 mg/liter).
In order to confirm that the double bond of the other members of family A is indeed between the 2′ and 3′ carbons of the side chain, as in HMNQ, and to further support the hypothesis that compounds from family B are saturated analogues of family A, the culture supernatants were hydrogenated using deuterium gas. The ions at m/z 228, 242, 256, 270, 284, 298, and 312 disappeared and new ions appeared at m/z 232, 246, 260, 274, 288, 302, and 316 at the same retention time as those of their corresponding saturated analogues. The MS/MS spectra of these deuterated compounds all presented fragment ions at m/z 173, 187, and 202. These results confirmed the hypothesis that family B congeners are linear saturated analogues of family A.
Cultures of B. thailandensis and B. pseudomallei also contain another family of congeners (family C) presenting pseudomolecular ions at m/z 272, 286, 300, 314, and 328 (Fig. 2). The molecular masses of these ions are 16 Da higher than those of the corresponding congeners of family A, indicating that they could be analogues of HMNQ containing an N-oxide and an unsaturated alkyl chain in C7, C8, C9, C10, and C11. Their MS/MS spectra present fragment ions at m/z 172, 173, 184, 186, and 198. Upon treatment of the B. thailandensis supernatant with zinc in acetic acid, conditions known to reduce N-oxides, these pseudomolecular ions disappeared, and the levels of pseudomolecular ions of family A increased, supporting the hypothesis that family C congeners are N-oxide derivatives. Interestingly, no family C congeners are produced by B. ambifaria. Since in P. aeruginosa, the production of HAQ N-oxides requires the presence of pqsL (32), we analyzed the Burkholderia genomes and found genes coding for proteins with a very good level of homology to PqsL in B. pseudomallei K96243 (BPSS0829) (52% identity and 64% similarity) and B. thailandensis E264 (BTH_II1576) (51% identity and 65% similarity) but, consistently, none in B. ambifaria.
HHQ along with its saturated C8 and C9 congeners (family D), typically produced by P. aeruginosa, are found only in B. ambifaria cultures and at very low levels (Fig. 2). However, another family of congeners presenting pseudomolecular ions at m/z 242, 270, and 298 (family E) was detected in B. thailandensis and B. ambifaria but not in B. pseudomallei (Fig. 2). These congeners are structurally related to family D but with an unsaturated C7, C9, and C11 side chain. The retention times of these compounds are slightly different from those bearing the double bond between the 1′ and the 2′ carbons of the side chain, such as those produced by P. aeruginosa, an indication that the double bond could be located further along the chain. In B. ambifaria cultures treated with deuterium, the pseudomolecular ions at m/z 242 and 270 disappear, and two new ions are observed at m/z 246 and 274 at the corresponding retention times of C7 and C9 saturated congeners. Their MS/MS spectra present ions at m/z 159 and 173. A single C9 HAQ congener containing a double bond at the 1′-2′ position was detected in B. pseudomallei but at a very low concentration (0.02 mg/liter).
Importantly, and similarly to a pqsA mutation in P. aeruginosa, knocking out hmqA in B. thailandensis and B. ambifaria completely abrogated the production of all five families of HAQ derivatives, in agreement with the proposed role of the hmq operon in the synthesis of these molecules.
Anthranilic acid is the precursor of HMAQs.
In order to verify that hmq operon-carrying bacteria derive all HMAQs from anthranilic acid, like P. aeruginosa, we fed B. thailandensis and B. ambifaria cultures with deuterium-labeled anthranilic acid. Upon LC/MS analysis of the cultures, all the ions assigned to HMAQ or HAQ congeners displayed the expected addition of 4 Da to their pseudomolecular ions, confirming the incorporation of anthranilic acid into their structures. Feeding with 15N-anthranilic acid led to an increase of 1 Da in all the pseudomolecular ions mentioned above, indicating that the nitrogen of anthranilic acid is also incorporated into all HMAQ and HAQ congeners. This 1-Da increase was also found in all the MS/MS fragment ions mentioned above.
Interestingly, we noticed that anthranilic acid accumulates in hmqA mutant cultures (at an OD600 of 6.0 [0.16 mg/liter for the wild type versus 8.13 mg/liter for the hmqA mutant of B. thailandensis]), suggesting that a source of excess anthranilate is available in these Burkholderia strains, contributing to the synthesis of HMAQs. In P. aeruginosa, three sources supply anthranilic acid to the cellular pool for HAQ synthesis: the anthranilate synthases TrpEG and PhnAB and the kynBUA-encoded kynurenine pathway, which provides anthranilic acid through the degradation of tryptophan (Fig. 1) (20). Although B. ambifaria and B. thailandensis do not seem to carry PhnAB homologues, anthranilate synthase trpEG genes are present (Bamb_0439 and Bamb_0438 for trpE and trpG, respectively, in B. ambifaria AMMD), and a complete set of kynBUA genes is found as a cluster on the Burkholderia genomes (Bamb_2625 to Bamb_2627 in B. ambifaria AMMD). To verify whether the kynurenine pathway contributes to HMAQ synthesis, we fed B. ambifaria and B. thailandensis cultures with deuterium-labeled tryptophan, and this led to the labeling of all HMAQs in both bacteria. This showed that tryptophan degradation via the kynurenine pathway provides at least part of the anthranilic acid pool used for HMAQ synthesis (data not shown).
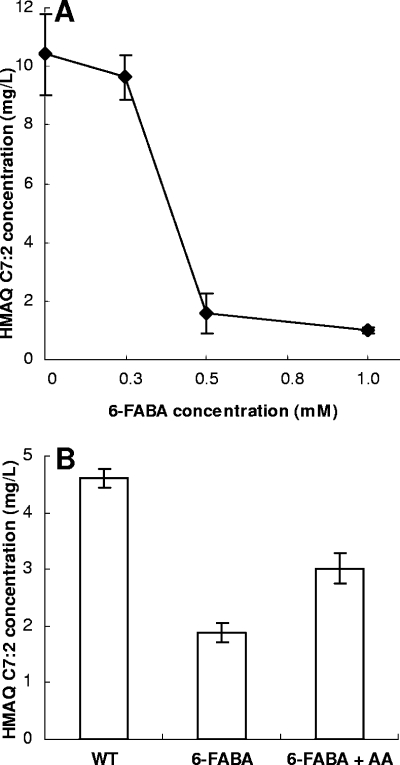
We recently reported that halogenated anthranilic acid analogues inhibit the synthesis of HAQs in P. aeruginosa and B. thailandensis, presumably by targeting the function of PqsA and/or HmqA (33). We therefore tested the effect of the anthranilic acid analogue 6-FABA on B. ambifaria cultures. The level of production of HMAQs decreased with increasing concentrations of 6-FABA added to the culture (Fig. 4A). Furthermore, the exogenous addition of anthranilic acid partially complemented this inhibition (Fig. 4B), supporting the hypothesis that HmqA is the target of 6-FABA in B. ambifaria. 6-FABA similarly inhibits B. thailandensis HMAQ production (data not shown).
FIG. 4.
Inhibition of HMAQ production by an analogue of anthranilic acid. (A) Production of HMAQ C7:2′ by B. ambifaria decreases with increasing concentrations of 6-FABA. (B) Exogenous anthranilic acid addition partially restores HMAQ C7:2′ production by B. ambifaria in the presence of 6-FABA. AA, anthranilic acid.
HmqG is required for the addition of the methyl group at the 3 position of HMAQs.
Since the hmq operon produces molecules similar to those from the pqs operon, we hypothesize that HmqA, HmqB, HmqC, HmqD, and HqmE perform functions similar to those of their homologous proteins in P. aeruginosa. However, the hmq operon presents two supplemental genes downstream of hmqE, which have no homologues in P. aeruginosa. We therefore performed BLAST analyses and conserved-domain investigations to obtain hints as to the function of the products of hmqF and hmqG, the two last genes of the Burkholderia hmq operons.
hmqF (Bamb_5768 and BTH_II1930) encodes a protein of 1,292 amino acids showing homology with several putative AMP-dependent synthetases and ligases and several putative nonribosomal peptide synthetases. Analysis of HmqF protein motifs and conserved domains in the Pfam 22.0 database (http://pfam.janelia.org/) revealed that this putative protein contains several conserved domains: an N-terminal region showing homology to the AMP-binding enzyme family (PF00501), two predicted acyl-CoA dehydrogenase domains (PF02770 and PF00441), and a predicted phosphopantetheine attachment site in the C-terminal region (PF00550).
Interestingly, the same analysis for HmqG (712 amino acids) revealed the presence of a predicted methyltransferase domain (PF08242) that is part of the S-adenosyl-l-methionine (SAM)-dependent methyltransferase family. Therefore, we hypothesized that HmqG might be involved in the methylation at the 3 position of HMAQs. Accordingly, in a B. ambifaria hmqG mutant culture, the two major ions at m/z 256 and 284 (unsaturated HMAQ) disappear and are replaced by two intense ions at m/z 242 and 270 occurring at a retention time corresponding to those of C7 and C9 unsaturated HAQs of family E. This is consistent with the loss of the methyl group of the corresponding HMAQ congeners, supporting the hypothesis that HmqG acts as a methyltransferase, adding a methyl group at the 3 position of HAQs. Accordingly, the inactivation of hmqG results in the complete absence of methylation on the HMAQs normally produced (loss of families A, B, and C), with an accumulation of the unmethylated molecules (families D and E) (Table 2).
Effect of HMAQs on QS-regulated phenotypes and production of AHLs.
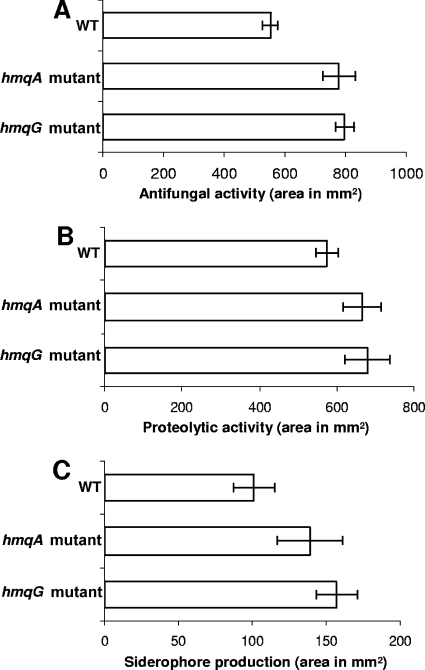
Very few QS-regulated functions have so far been identified in B. ambifaria. These functions include the production of extracellular proteases, siderophores, and an unknown antifungal compound (60). We investigated whether these QS-regulated phenotypes are affected by HMAQ production. The hmqA mutant shows increased siderophore production and proteolytic and antifungal activities compared to those of the wild type (Fig. 5). Results obtained with C. albicans were further confirmed with the same bioassay using Cryptococcus neoformans and Candida glabrata. In both bioassays, a strain deficient in HMAQ synthesis produced larger amounts of antifungal compounds (data not shown). Also, colonies of the hmqA mutant showed higher levels of protease activity on agar plates containing 1% skim milk powder and 0.2% yeast extract than did the wild type (data not shown).
FIG. 5.
Effect of hmqA and hmqG mutations on QS-regulated factors. (A) Antifungal activity against C. albicans. (B) Proteolytic activity. (C) Siderophore production. Proteolytic and antifungal activities were estimated from the areas of the clear zones surrounding wells containing supernatants of cultures. The values shown are the averages ± standard deviations for triplicate cultures. Siderophore production was estimated from the area of the halo surrounding colonies on CAS agar after 24 h at 37°C. The values shown are corrected for the number of CFU in the colony (108) and are averages ± standard deviations of data for eight colonies.
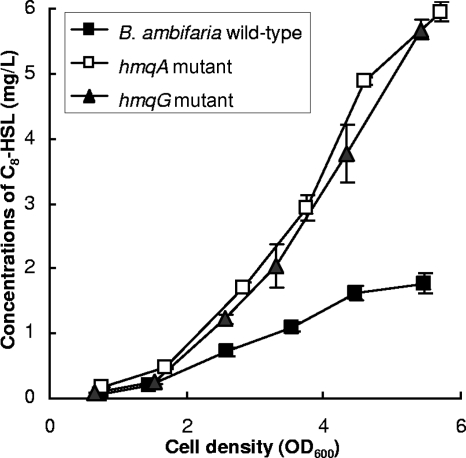
As several QS-regulated phenotypes are upregulated in an hmqA mutant background, we reasoned that the production of octanoyl homoserine lactone (C8-HSL), the predominant AHL produced by B. ambifaria (63), would be altered. Indeed, as shown in Fig. 6, the absence of HMAQs correlates with a significant increase in levels of C8-HSL production. At the end of the exponential phase, the concentration of C8-HSL detected in the supernatant of the hmqA mutant strain was twofold higher than that observed for wild-type B. ambifaria (Fig. 6).
FIG. 6.
Production in function of growth of C8-HSL by wild-type B. ambifaria and hmqA and hmqG mutants. Error bars represent the standard deviations of data for triplicate cultures.
Results obtained for phenotypic assays and the production of AHLs in the hmqG mutant, which produces only unmethylated molecules, are identical to those obtained for the hmqA mutant. These results suggest that the unmethylated HAQs are not implicated in QS regulation in B. ambifaria. Collectively, these findings indicate that only the methylated HAQs produced by B. ambifaria (HMAQs) participate in the negative regulation of C8-HSL production and consequently modulate QS-regulated phenotypes.
DISCUSSION
The homologue of pqsABCDE in Burkholderia species.
B. thailandensis and B. pseudomallei were recently reported to produce HAQs similar to those produced by P. aeruginosa (16). The importance of these versatile molecules, especially as coinducing signals in QS for the regulation of virulence in the latter bacterium, prompted us to investigate the nature and diversity of HAQ-related molecules produced by the Burkholderia genus. We searched for the presence of a pqsABCDE operon homologue in available sequenced genomic data. Like Diggle et al. (16), we confirmed the presence of homologues of the pqsABCDE genes in an operon arrangement in B. pseudomallei and in the genetically related species B. thailandensis, but we also identified another species carrying a similar operon, B. ambifaria, a member of the B. cepacia complex. Our hypothesis was that bacteria would be producers of HAQ derivatives only when carrying homologues of the pqsABCDE genes in an operon arrangement as in P. aeruginosa. Of the 11 related Burkholderia species tested, only those three produce HMAQs, supporting our hypothesis.
Moreover, we showed that the operon found in the Burkholderia strains mentioned above contains two more genes downstream of the gene homologous to pqsE that we thus named hmqF and hmqG. Apart from the homology with pqsABCDE, the genomic context surrounding hmqABCDEFG is quite different in these bacteria compared to P. aeruginosa. No anthranilate synthase reminiscent of phnAB is found nearby, and no homologue of the gene coding for the MvfR transcriptional regulator is present in the vicinity of the hmq operon. Furthermore, no putative LysR box seems to be present within the promoter of the hmqABCDEFG operon.
Chemical analysis of HMAQs.
While we actually found only traces of P. aeruginosa-type HAQs (family D), we observed that the principal products in hmqABCDEFG-carrying Burkholderia species are HAQ analogues methylated at the 3 position. After a detailed analysis of the literature, we were able to find that bacterially produced methylated HAQs with an unsaturated aliphatic side chain had been detected previously. 4-Hydroxy-3-methyl-2-heptenylquinoline from an uncharacterized Pseudomonas strain was identified (24) and was later found in various B. cepacia (unidentified genomovar) strains (48) along with HMNQ (27) and a series of congeners from family B (41). As with P. aeruginosa HAQs, HMAQs are produced as mixtures of various congeners with an alkyl chain of variable length with or without an unsaturation and/or an N-oxide function on their quinoline amino group. In order to ascertain the structures of these compounds, we first used high-performance LC to isolate the most abundant HMAQ congener produced by B. thailandensis, as its TIC chromatogram shows predominantly one major congener, compared with B. ambifaria, which produces a greater diversity of compounds. The compound isolated was determined to be HMNQ by NMR. This analytical tool confirmed that the double bond is in the trans configuration. Considering that each congener of family A with a given m/z value appears at only one retention time, and considering the regular intervals between the retention times of the various congeners of this family, we assume that all the other unsaturated HMNQ congeners also share this trans configuration.
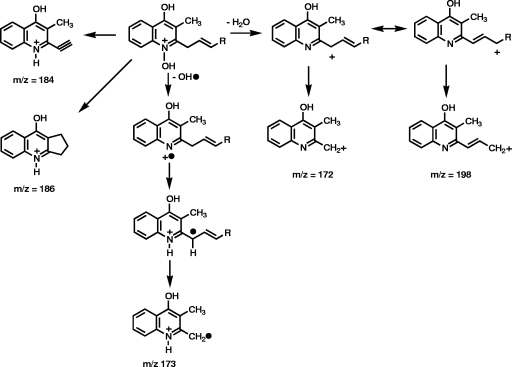
The fragmentation patterns of the pseudomolecular ions of the unsaturated HMAQs and the unsaturated HAQs (32) are quite similar, although the double bond is between the 2′ and 3′ carbons in the former and between the 1′ and 2′ carbons in the latter. The two most abundant ions of the MS/MS spectra of unsaturated HMAQ family A are at m/z 173 and 186, which correspond to the m/z 159 and 172 ions observed for unsaturated HAQs (32), with the 14-Da difference being due to the additional methyl group at the 3 position of the quinoline ring (Fig. 7). The m/z 173 fragment ion of HMNQ, or the corresponding m/z 159 fragment ion of HHQ, is a radical cation and not a cation produced by a rearrangement that would have led to the loss of the nitrogen. This is demonstrated by the fact that when 15N-anthranilic acid was fed to the bacteria, the m/z 173 ion was increased by 1 Da, thus proving that it still contains one nitrogen atom. The other daughter ion at m/z 212 in the MS/MS spectra of unsaturated HMAQs could also be structurally analogous to the m/z 198 ion observed in unsaturated HAQs. The relatively intense ion at m/z 184, however, does not have an analogous m/z 170 ion in the unsaturated HAQ MS/MS spectra and could arise from the cyclization of the 2′ carbon of the side chain with the methyl at the 3 position, followed by a loss of the residual alkyl chain with the formation of a double bond. It is also possible that the same fragmentation pathways involving the 3′ carbon of the side chain and leading to an unsaturated six-membered ring could produce that m/z 198 ion. Another possibility is that the m/z 198 and 212 ions in the MS/MS spectra of unsaturated HMAQ ions have their side chains cyclized with the quinoline nitrogen, as proposed for the HAQ MS/MS spectra.
FIG. 7.
Daughter ions observed in the MS/MS spectra of saturated and unsaturated HMAQs and HAQ.
The MS/MS spectra of the saturated HMAQs (family B) present two intense ions at m/z 173 and 186 that are analogous to those of their corresponding saturated HAQs (family D) but with the addition of 14 Da corresponding to the extra methyl on the quinoline ring. These two ions correspond to the quinoline moiety with the 1′ and 2′ carbons attached to it. We took advantage of this fragmentation pattern to demonstrate that the double bond of the unsaturated HMAQ congeners, as for HMNQ, is indeed between the 2′ and 3′ carbons. To do so, B. thailandensis and B. ambifaria culture supernatants were hydrogenated using deuterium, and the location of the label in the corresponding saturated compounds was investigated by MS/MS. The daughter ions of the corresponding deuterated pseudomolecular ions at m/z 260, 274, 288, and 302 were observed at m/z 173 and 187 instead of m/z 173 and 186 in the corresponding unlabeled saturated HMAQs. If the double bond had been between the 1′ and 2′ carbons, both ions would have been labeled. On the other hand, if the double bond had been located further than the 2′ position, none of these daughter ions would have been labeled. This proves that the double bond is located between the 2′ and 3′ carbons for the C7, C8, C9, and C10 members of this family, as we assumed, based on the NMR for the C9 congener HMNQ.
As in P. aeruginosa, B. thailandensis and B. pseudomallei produce another family of HMAQs with their pseudomolecular ions at 16 Da above those of the unsaturated HMAQs (family C). These molecules correspond to HMAQ N-oxides, as they disappeared when supernatants were treated with zinc in acetic acid, conditions known to reduce N-oxides. Their MS/MS spectra produced ions at m/z 172, 173, 184, 186, and 198 (Fig. 8). The m/z 173 ion is probably a radical cation related to the prominent m/z 159 ion observed in the corresponding HMAQ N-oxide through the initial loss of a hydroxyl radical (40). The m/z 172 and 298 daughter ions probably arise from the concerted loss of water, followed by chain cleavage at the positively charged carbon or at the conjugated 3′ position. We confirmed the position of the double bond on the 2′ carbon by the increase in the intensity of the unsaturated HMAQ congeners upon the reduction of the N-oxide group by zinc. The presence of these N-oxide derivatives in B. thailandensis and B. pseudomallei is consistent with the presence of a gene, which we call hmqL, that is highly homologous to the P. aeruginosa pqsL gene, which we found is required for the synthesis of HAQ N-oxides in the latter species (32). Consistently, such a pqsL homologue is absent in B. ambifaria, which does not produce the family C HMAQ N-oxides. An Arthrobacter strain from soil was previously reported to produce HMNQ N-oxide, which inhibits various gram-positive bacteria, including multiple-drug-resistant strains of Staphylococcus aureus and Staphylococcus epidermidis (30). This compound was also effective against Helicobacter pylori (52) and was found to be a potent inhibitor of mitochondrial complexes I and III (46), consistent with the well-known activities of HAQ N-oxide derivatives (26, 34, 36).
FIG. 8.
Putative structures of daughter ions observed in the MS/MS spectra of HMAQ N-oxides.
Another family of compounds that we found at low concentrations in B. thailandensis and in B. ambifaria cultures are HAQ congeners bearing a double bond at the 2′ position (family E). Although unsaturated HAQ congeners were detected previously in P. aeruginosa (32), those produced by B. thailandensis and B. ambifaria have a slightly different retention time, an indication that the position of the double bond is not the same. Upon treatment of the supernatant with deuterium, the MS/MS spectra of the deuterated pseudomolecular ions at m/z 246 and 274 present ions at m/z 159 and 173 instead of the m/z 159 and 172 normally observed with unlabeled HAQ congeners (32), while the MS/MS spectra of the deuterated HAQ congeners bearing the double bond at position 1′ show ions at m/z 160 and 174 (32). This proves that the double bond is not at the 1′ carbon and is no further than the 2′ position, as in family A (unsaturated HMAQs). In B. thailandensis, Diggle et al. (16) reported the detection of only a series of C7:1, C9:1, and C11:1 and their unsaturated HAQ congeners along with a C9 congener of 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO). In a recent report (33), we also observed ions with m/z 242, 270, and 292 ions corresponding to HAQ congeners with one unsaturation in B. thailandensis, and based on the report of Diggle et al., we initially assumed that their double bond had to be at the 1′ position. However, in view of our current results showing that unsaturated HMAQs have their double bond at the 2′ position, we performed MS/MS analysis of the product of deuteration of these unsaturated HAQ congeners. Diggle et al. assigned the position of the double bond based only upon the MS/MS spectrum of these unsaturated HAQ congeners, which could be misleading, as the position of the double bond could shift in the fragmentation mechanism. Catalytic hydrogenation with deuterium is a more reliable method and confirms that the double bond is at the 2′ position, like in all the other HMAQs, at least in the B. thailandensis strain that we are studying. The concentration of unsaturated HAQ congeners was close to our limit of detection in B. ambifaria and B. thailandensis, while none were observed in B. pseudomallei cultures.
The main difference between our results and those recently described by Diggle et al. (16) regarding the HAQ derivatives produced by B. pseudomallei and B. thailandensis is that those authors did not report the production of HMAQs. Furthermore, we did not detect any saturated HHQ congener and no saturated C9 N-oxide, and we detected only very small concentrations of the unsaturated HAQ congener that they reported for B. thailandensis. In addition, despite the sensitivity of our method, we did not find HHQ in B. cenocepacia cultures either. It is difficult to compare these last results with those of Diggle et al., as they did not quantify the compounds that they reported. We cannot exclude the possibility that B. thailandensis might produce trace amounts of saturated HAQs (family D) and HAQ N-oxide congeners; their findings could be explained by the fact that while we directly injected culture supernatant samples in our LC/MS assay without preconcentration, Diggle et al. solvent extracted and concentrated their samples 1,000 times prior to analysis (16). The fact that they did not report HMAQs is more surprising, however, since we show here that these methylated derivatives are the prominent ones. We believe that this could be because they focused only on published, known HAQ analogues described previously for P. aeruginosa (32). Moreover, since they studied their Burkholderia strains using a bioreporter designed to detect primarily PQS and HHQ, it is conceivable that it failed to detect HMAQs (21). It is very unlikely that differences in the strains investigated or culture conditions could explain these differences, as these factors have no effect in P. aeruginosa or on our tested strains. Nevertheless, it is clear that in B. ambifaria, B. thailandensis, and B. pseudomallei and under our conditions, HMAQs are more abundant, sometimes by more than 3 orders of magnitude, than unmethylated HAQs. This raises the question of the biological significance of trace amounts of HAQs in these species compared to amounts of HMAQs. In view of the very small amounts of unsaturated HAQ congeners produced by these strains in comparison with those of HMAQs, we named these operons hmqABCDEFG.
Biosynthesis of HMAQs.
Because anthranilic acid is the precursor of all HAQs (14), we tested if it is also the common precursor of HMAQs. Adding labeled anthranilic acid to a culture results in the labeling of all the HMAQs. We then found that the inactivation of hmqA in B. thailandensis and in B. ambifaria leads to the abrogation of HMAQ production, as observed for the loss of HAQs from the B. pseudomallei homologue (16) and the pqsA mutant of P. aeruginosa (14). Furthermore, 6-FABA, an analogue of anthranilic acid known to competitively inhibit PqsA (33), also inhibits HMAQ biosynthesis. The fact that this inhibition, as in P. aeruginosa, can also be reversed by an exogenous supply of anthranilic acid indicates that the genetic homology between hmqA and pqsA also translates into a common biological activity. This is in line with the reported complementation of a P. aeruginosa pqsA mutant with a B. pseudomallei homologue (16). The homologies between PqsA and various aryl-CoA ligases indicate that HmqA is likely responsible for the activation of anthranilic acid into a CoA ester (Fig. 1). Such activation of the anthranilic acyl group would be required for the closing of the second ring of the quinoline core.
In contrast with P. aeruginosa, there is no gene responsible for the synthesis of anthranilic acid near the hmqABCDEFG operon. Nevertheless, plenty of anthranilic acid seems to be available, as this precursor accumulates in hmqA mutant cultures, as we have previously seen in a pqsA mutant of P. aeruginosa (14). While TrpEG analogues responsible for the biosynthesis of anthranilic acid in the formation of tryptophan are present, part of the anthranilic acid used for HMAQ biosynthesis also comes for tryptophan degradation, presumably through the kynurenine pathway, as shown by our labeled tryptophan experiment.
Within each HMAQ family, the relative amounts of congeners containing an odd number of carbons on their side chains are larger than those with one extra carbon, often by 2 orders of magnitude. This is analogous to the observations made with HAQs and is likely due to the successive addition of acetate groups to form an even-numbered 3-keto fatty acid intermediate, which reacts with anthranilic acid, as first hypothesized by Wells (58) and later confirmed by others (3, 47). Because of the location of the reactive keto group of the 3-keto fatty acid, this produces predominantly an odd-numbered alkyl chain in HAQs and HMAQs. The chain lengths of HMAQs also seem to vary from one Burkholderia species to another, as HMAQs in B. thailandensis and B. pseudomallei are predominantly C9 congeners, while in B. ambifaria, they are predominantly C7 congeners.
A difference between Burkholderia and P. aeruginosa is that HMAQs are predominantly unsaturated, while Pseudomonas HAQs are predominantly saturated, and in Burkholderia HMAQs and HAQs, the double bond is at the 2′ position, while in unsaturated Pseudomonas HAQs, it is at the 1′ position. This probably arises from a difference in the biosynthesis of the 3-keto fatty acid precursor. In P. aeruginosa, it is still unclear if the pqsBCD genes, which all have analogies with 3-keto fatty acid acyl carrier proteins, are responsible for the total synthesis of the 3-keto fatty acid intermediate or if they simply divert some intermediates of general fatty acid biosynthesis into the pathway leading to HAQs. In the Burkholderia strains that we studied, the pqsBCD homologues hmqBCD presumably play the same role in the biosynthesis of the 3-keto fatty acid intermediates (Fig. 1). It remains to be determined if they are responsible for the unsaturation generally encountered in HMAQs or if this occurs because of the difference in the pool of produced fatty acid intermediates between Burkholderia and P. aeruginosa. It is also possible that the hmqBCD genes are responsible for the coupling of the anthraniloyl-CoA and the 3-keto fatty acid and in the closing of the second ring of the quinoline core. An intriguing possibility is that HmqF, which is predicted to contain two acyl-CoA dehydrogenase domains and an attachment site for 3-phosphopantetheine, a functional group present in all ACP proteins, might be responsible for the 2′ unsaturation of HMAQs.
The most striking difference between HAQs and HMAQs is obviously the presence of the methyl group at the 3 position. As an hmqG mutant stops producing HMAQs and produces HAQ-like molecules instead, HmqG is likely responsible for the addition of the methyl group. Sequence analogies of the hmqG product with many SAM-dependent methyltransferases indicate that SAM is likely to be the source of the methyl group. We initially thought that this methyl group could simply be the product of the reduction of the acid group of the original 3-keto fatty acid intermediate, as this acid group must be removed to produce HHQ in P. aeruginosa. However, such an HAQ with a carboxyl function at the 3 position was never detected in P. aeruginosa supernatants. The fact that HmqG has homologies with a methyltransferase rather than a decarboxylase could be seen as a confirmation that this acid group is spontaneously lost upon the closing of the second aromatic ring or that another unknown enzyme is responsible for the loss of this acid group in these bacteria. The finding that hmqG is coexpressed with the rest of the operon substantiates that HMAQs are indeed the final products of this pathway.
The kinetics of HMAQ production show that these molecules start to accumulate in cultures near the end of the logarithmic growth phase. This is very similar to the HAQ production profile for P. aeruginosa (13, 31). In this organism, the pqsABCDE operon is controlled by MvfR, whose coinducing ligands are HAQs (61). Thus, it is conceivable that the production of HMAQs is controlled by a regulator induced by HMAQs. Accordingly, we observed that the total level of production of HAQs and HMAQs by the hmqG mutant is much lower than that observed in the wild-type strain (26.82 mg/liter total HAQs and HMAQs for the wild type versus 7.63 mg/liter for the hmqG mutant) (Table 3), suggesting a regulation of the hmqABCDEFG operon by HMAQs.
TABLE 3.
Concentrations of the major HMAQs and HAQs produced by wild-type B. ambifaria AMMD and its hmqG mutant
| HMAQ or HAQ | Mean concn (mg/liter) (SD)a
|
||
|---|---|---|---|
| [M + H]+ | B. ambifaria wild type | B. ambifaria hmqG mutant | |
| HMAQ:2′ | |||
| C7:2′ | 256 | 15.46 (0.86) | ND |
| C9:2′ | 284 | 8.26 (0.43) | ND |
| HMAQ | |||
| C7 | 258 | 0.80 (0.05) | ND |
| C9 | 286 | 1.22 (0.07) | ND |
| HHQ:2′ | |||
| C7:2′ | 242 | 0.69 (0.07) | 5.16 (0.40) |
| C9:2′ | 270 | 0.39 (0.03) | 2.31 (0.20) |
| HHQ | |||
| C7 | 244 | ND | ND |
| C9 | 272 | ND | 0.16 (0.01) |
Values in parentheses are the standard deviations of data from triplicate assays. ND, not detected.
HMAQs and QS.
In P. aeruginosa, some of the HAQs act as signaling molecules and are implicated in the regulation of the expression of multiple virulence factors, which includes products of genes controlled by AHL-mediated QS. However, in P. aeruginosa, quinolone signaling has no direct effect on the production of AHLs (13). In contrast, we show here that the synthesis of HMAQs is directly implicated in the global QS circuitry by negatively regulating the production of AHLs, at least in B. ambifaria. Furthermore, we demonstrate that the methyl group of HMAQs is crucial for the effect on C8-HSL production and QS-controlled factors, as the hmqG mutant behaves the same as the hmqA mutant. How HMAQs influence the production of AHLs is currently under investigation. One possibility is a direct regulation at the level of expression of the AHL synthase. Alternatively, the synthesis of HMAQs might modulate the availability of AHL synthesis precursors or the activity of the AHL synthase. Whether HMAQs play the same role in B. pseudomallei and B. thailandensis is still undetermined. However, available data indicate that QS in B. thailandensis is much more complex than that in B. ambifaria, as it involves multiple LuxIR homologues responsible for the production of numerous AHL signals involved in both positive and negative regulation (54).
Finally, HAQ derivatives may be more widespread than previously thought, raising the intriguing possibility of intergenus communication (18). Still, the methyl group makes the Burkholderia HAQ derivatives distinct from those of P. aeruginosa. Since these new HMAQs appear to be involved in the Burkholderia QS circuitry, and given the essential role of QS in the expression of pathogenic traits in Burkholderia (19, 56), these molecules and the enzymes involved in their biosynthesis could represent attractive targets for the development of novel therapeutics against hmqABCDEFG-carrying pathogenic Burkholderia species.
Acknowledgments
This study was supported by a Canadian Institutes of Health Research operating grant to E.D. and an NSERC research grant to F.L., L.V. and V.D. were recipients of a postdoctoral fellowship and of a Ph.D. scholarship, respectively, from the Fondation Armand-Frappier.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26824-828. [DOI] [PubMed] [Google Scholar]
- 2.Bredenbruch, F., R. Geffers, M. Nimtz, J. Buer, and S. Haussler. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 81318-1329. [DOI] [PubMed] [Google Scholar]
- 3.Bredenbruch, F., M. Nimtz, V. Wray, M. Morr, R. Müller, and S. Häussler. 2005. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 1873630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48317-320. [DOI] [PubMed] [Google Scholar]
- 5.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 9814613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 511481-1490. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 412797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornforth, J. W., and A. T. James. 1956. Structure of a naturally occurring antagonist of dihydrostreptomycin. Biochem. J. 63124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 1792116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeShazer, D., D. M. Waag, D. L. Fritz, and D. E. Woods. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30253-269. [DOI] [PubMed] [Google Scholar]
- 13.Déziel, E., S. Gopalan, A. P. Tampakaki, F. Lépine, K. E. Padfield, M. Saucier, G. Xiao, and L. G. Rahme. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55998-1014. [DOI] [PubMed] [Google Scholar]
- 14.Déziel, E., F. Lépine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 1011339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diggle, S. P., P. Cornelis, P. Williams, and M. Camara. 2006. 4-Quinolone signalling in Pseudomonas aeruginosa: old molecules, new perspectives. Int. J. Med. Microbiol. 29683-91. [DOI] [PubMed] [Google Scholar]
- 16.Diggle, S. P., P. Lumjiaktase, F. Dipilato, K. Winzer, M. Kunakorn, D. A. Barrett, S. R. Chhabra, M. Camara, and P. Williams. 2006. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13701-710. [DOI] [PubMed] [Google Scholar]
- 17.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Camara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 5029-43. [DOI] [PubMed] [Google Scholar]
- 18.Eberl, L. 2006. From a local dialect to a common language. Chem. Biol. 13803-804. [DOI] [PubMed] [Google Scholar]
- 19.Eberl, L. 2006. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 296103-110. [DOI] [PubMed] [Google Scholar]
- 20.Farrow, J. M., III, and E. C. Pesci. 2007. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J. Bacteriol. 1893425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher, M. P., S. P. Diggle, S. A. Crusz, S. R. Chhabra, M. Camara, and P. Williams. 2007. A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ. Microbiol. 92683-2693. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 1846472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glass, M. B., A. G. Steigerwalt, J. G. Jordan, P. P. Wilkins, and J. E. Gee. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 562171-2176. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto, M., and K. Hattori. 1967. 2-(2-Heptenyl)-3-methyl-4-quinolinol from a Pseudomonas. Chem. Pharm. Bull. (Tokyo) 15718-720. [DOI] [PubMed] [Google Scholar]
- 25.Hays, E. E., I. C. Wells, P. A. Katzman, C. K. Cain, F. A. Jacobs, S. A. Thayer, E. A. Doisy, W. L. Gaby, E. C. Roberts, R. D. Muir, C. J. Carroll, L. R. Jones, and N. J. Wade. 1945. Antibiotic substances produced by Pseudomonas aeruginosa. J. Biol. Chem. 159725-750. [Google Scholar]
- 26.Hoffman, L. R., E. Deziel, D. A. D'Argenio, F. Lepine, J. Emerson, S. McNamara, R. L. Gibson, B. W. Ramsey, and S. I. Miller. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10319890-19895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homma, Y., Z. Sato, F. Hirayama, K. Komo, H. Shirahama, and T. Suzui. 1989. Production of antibiotics by Pseudomonas cepacia as an agent for biological control of soilborne plant pathogens. Soil Biol. Biochem. 21723-728. [Google Scholar]
- 28.Hooi, D. S., B. W. Bycroft, S. R. Chhabra, P. Williams, and D. I. Pritchard. 2004. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 726463-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7459-471. [DOI] [PubMed] [Google Scholar]
- 30.Kamigiri, K., T. Tokunaga, M. Shibazaki, B. Setiawan, R. M. Rantiatmodjo, M. Morioka, and K. Suzuki. 1996. YM-30059, a novel quinolone antibiotic produced by Arthrobacter sp. J. Antibiot. (Tokyo) 49823-825. [DOI] [PubMed] [Google Scholar]
- 31.Lépine, F., E. Déziel, S. Milot, and L. G. Rahme. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 162236-41. [DOI] [PubMed] [Google Scholar]
- 32.Lépine, F., S. Milot, E. Déziel, J. He, and L. G. Rahme. 2004. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J. Am. Soc. Mass Spectrom. 15862-869. [DOI] [PubMed] [Google Scholar]
- 33.Lesic, B., F. Lépine, E. Déziel, J. Zhang, Q. Zhang, K. Padfield, M. H. Castonguay, S. Milot, S. Stachel, A. A. Tzika, R. G. Tompkins, and L. G. Rahme. 2007. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 31229-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lightbown, J. W., and F. L. Jackson. 1956. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonist of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 63130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long, R. A., A. Qureshi, D. J. Faulkner, and F. Azam. 2003. 2-n-Pentyl-4-quinolinol produced by a marine Alteromonas sp. and its potential ecological and biogeochemical roles. Appl. Environ. Microbiol. 69568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machan, Z. A., G. W. Taylor, T. L. Pitt, P. J. Cole, and R. Wilson. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30615-623. [DOI] [PubMed] [Google Scholar]
- 37.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 23027-34. [DOI] [PubMed] [Google Scholar]
- 39.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1822702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao, X.-S., R. E. March, and C. D. Metcalfe. 2003. A tandem mass spectrometric study of the N-oxides, quinoline N-oxide, carbadox, and olaquindox, carried out at high mass accuracy using electrospray ionization. Int. J. Mass Spectrom. 230123-133. [Google Scholar]
- 41.Moon, S.-S., P. M. Kang, K. S. Park, and C. H. Kim. 1996. Plant growth promoting and fungicidal 4-quinolinones from Pseudomonas cepacia. Phytochemistry 42365-368. [Google Scholar]
- 42.Moon, S.-S., P. M. Kang, K. S. Yoon, S.-J. Yun, and B. B. Park. 1995. Synthesis of plant growth promoting and fungicidal 4-quinolinones metabolites of Pseudomonas cepacia. Bull. Korean Chem. Soc. 161128-1130. [Google Scholar]
- 43.Nelson, M. J., S. O. Montgomery, W. R. Mahaffey, and P. H. Pritchard. 1987. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl. Environ. Microbiol. 53949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nierman, W. C., D. DeShazer, H. S. Kim, H. Tettelin, K. E. Nelson, T. Feldblyum, R. L. Ulrich, C. M. Ronning, L. M. Brinkac, S. C. Daugherty, T. D. Davidsen, R. T. Deboy, G. Dimitrov, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, H. Khouri, J. F. Kolonay, R. Madupu, Y. Mohammoud, W. C. Nelson, D. Radune, C. M. Romero, S. Sarria, J. Selengut, C. Shamblin, S. A. Sullivan, O. White, Y. Yu, N. Zafar, L. Zhou, and C. M. Fraser. 2004. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA 10114246-14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 9611229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reil, E., G. Hofle, W. Draber, and W. Oettmeier. 1997. Quinolones and their N-oxides as inhibitors of mitochondrial complexes I and III. Biochim. Biophys. Acta 1318291-298. [DOI] [PubMed] [Google Scholar]
- 47.Ritter, C., and M. Luckner. 1971. Biosynthesis of 2-n-alkyl-4-hydroxyquinoline derivatives (pseudane) in Pseudomonas aeruginosa. Eur. J. Biochem. 18391-400. [DOI] [PubMed] [Google Scholar]
- 48.Roitman, J. N., N. E. Mahoney, W. J. Janissiewicz, and M. Benson. 1990. A new chlorinated phenylpyrrole antibiotic produced by the antifungal bacterium Pseudomonas cepacia. J. Agric. Food Chem. 38538-541. [Google Scholar]
- 49.Royt, P. W., R. V. Honeychuck, V. Ravich, P. Ponnaluri, L. K. Pannell, J. S. Buyer, V. Chandhoke, W. M. Stalick, L. C. DeSesso, S. Donohue, R. Ghei, J. D. Relyea, and R. Ruiz. 2001. 4-hydroxy-2-nonylquinoline: a novel iron chelator isolated from a bacterial cell membrane. Bioorg. Chem. 29387-397. [DOI] [PubMed] [Google Scholar]
- 50.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 16047-56. [DOI] [PubMed] [Google Scholar]
- 51.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 52.Taniguchi, M., J. Kazami, and K. Tanaka. May 2000. Preparation of quinolones as anti-Helicobacter pylori. Japan patent JP2000143632 A 20000526.
- 53.Trân Van, V., O. Berge, S. N. Ke, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield component in low fertility sulphate acid soils of Vietnam. Plant Soil 218273-284. [Google Scholar]
- 54.Ulrich, R. L., H. B. Hines, N. Parthasarathy, and J. A. Jeddeloh. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J. Bacteriol. 1864350-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 15491-96. [DOI] [PubMed] [Google Scholar]
- 56.Vial, L., M. C. Groleau, V. Dekimpe, and E. Déziel. 2007. Burkholderia diversity and versatility: an inventory of the extracellular products. J. Microbiol. Biotechnol. 171407-1429. [PubMed] [Google Scholar]
- 57.Wade, D. S., M. W. Calfee, E. R. Rocha, E. A. Ling, E. Engstrom, J. P. Coleman, and E. C. Pesci. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1874372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells, I. C. 1952. Antibiotic substances produced by Pseudomonas aeruginosa. Synthesis of Pyo Ib. Pyo Ic, and Pyo III. J. Biol. Chem. 196331-340. [PubMed] [Google Scholar]
- 59.Williams, P., K. Winzer, W. C. Chan, and M. Camara. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3621119-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wopperer, J., S. T. Cardona, B. Huber, C. A. Jacobi, M. A. Valvano, and L. Eberl. 2006. A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl. Environ. Microbiol. 721579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao, G., E. Déziel, J. He, F. Lépine, B. Lesic, M. H. Castonguay, S. Milot, A. P. Tampakaki, S. E. Stachel, and L. G. Rahme. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 621689-1699. [DOI] [PubMed] [Google Scholar]
- 62.Xiao, G., J. He, and L. G. Rahme. 2006. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 1521679-1686. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, H., F. Yao, D. P. Roberts, and T. G. Lessie. 2003. AHL-deficient mutants of Burkholderia ambifaria BC-F have decreased antifungal activity. Curr. Microbiol. 47174-179. [DOI] [PubMed] [Google Scholar]