Abstract
Hepatitis delta virus (HDV) is a pathogenic RNA virus with a plant viroid-like genome structure. HDV encodes two isoforms of delta antigen (HDAg), the small and large forms of HDAg (SHDAg and LHDAg), which are essential for HDV RNA replication and virion assembly, respectively. Replication of HDV RNA depends on host cellular transcription machinery, and the exact molecular mechanism for HDV RNA replication is still unclear. In this study, we demonstrated that both isoforms of HDAg interact with transcription factor YY1 (Yin Yang 1) in vivo and in vitro. Their interaction domains were identified as the middle region encompassing the RNA binding domain of HDAg and the middle GA/GK-rich region and the C-terminal zinc-finger region of YY1. Results of sucrose gradient centrifugation analysis indicated the cosedimentation of the majority of SHDAg and a portion of the LHDAg with YY1 and its associated acetyltransferases CBP (CREB-binding protein) and p300 as a large nuclear complex in vivo. Furthermore, exogenous expression of YY1 or CBP/p300 in HDV RNA replication system showed an enhancement of HDV RNA replication. Interestingly, the acetyltransferase activity of p300 is important for this enhancement. Moreover, SHDAg could be acetylated in vivo, and treatment with cellular deacetylase inhibitor elevated the replication of HDV RNA and acetylation of SHDAg. All together, our results reveal that HDAg interacts with cellular transcription factor YY1 and its associated acetyltransferases CBP and p300 in a large nuclear complex, which in turn modulates the replication of HDV RNA.
Hepatitis D virus (HDV) was discovered in 1977 and was originally thought to be a nuclear antigen associated with hepatitis B virus (HBV) infections (48). Subsequent studies indicated that HDV is a defective virus requiring surface antigen of helper HBV for virion assembly and infectivity (4, 49, 52). Coinfection or superinfection of HBV-infected patients with HDV often correlated with more severe acute and rapidly progressive liver diseases than in patients infected with HBV alone (17). HDV is a negative-stranded RNA virus with a 1.7-kb circular RNA genome (13, 45, 57), which is folded into an unbranched rod-like structure due to the high degree of intramolecular self-complementarity (35, 57). The genome of HDV shares several characteristics of plant viroids, except that HDV encodes proteins, and its genome is three or four times larger (53). It is suggested that HDV RNA may evolve from a viroid-like virus, which captures cellular RNAs from the human transcriptome (5, 6, 50).
Apart from the HDV RNA genome and HBV envelope, the HDV particle also contains hepatitis delta antigen (HDAg), the sole known protein encoded by the only open reading frame of HDV RNA (26, 27, 46). The HDAg occurs as two protein species, the small HDAg of 195 amino acid residues in length (SHDAg; 24 kDa) and the large HDAg of 214 amino acid residues in length (LHDAg; 27 kDa) (59). The amino acid sequence of these two HDAg species are identical except that LHDAg is 19 amino acids longer than SHDAg at its C terminus, which results from an RNA editing event during viral replication (8). However, the two isoforms of HDAg exhibit different functions in the HDV life cycle. SHDAg is essential for HDV RNA replication (11, 36), while LHDAg is crucial for virion assembly (9). Several functional domains, including an oligomerization domain, bipartite nuclear localization signal, and two arginine-rich motifs (ARMs), of HDAg have been characterized (43). HDAg also undergoes numerous posttranslational modifications, such as phosphorylation, acetylation, methylation, and isoprenylation, which play different roles in the HDV life cycle (29). Additionally, both LHDAg and SHDAg possess the ability to affect cellular RNA polymerase II transcription (42) and elongation (60), and LHDAg is capable of activation of expression of numerous reporter genes (23, 24, 58). A recent study also indicates that LHDAg activates transforming growth factor β (TGF-β) signaling pathway, which may induce liver fibrosis in HDV patients (15). These observations suggest that HDAgs participate in the regulation of cellular gene expression, and this feature may contribute to the pathogenesis of HDV infection.
Replication of HDV RNA is independent of its helper virus HBV (36) and does not involve any DNA intermediates (13). It occurs in the nucleus via a proposed double rolling-circle mechanism (43, 54). In this model, the HDV circular RNA genome serves as a template for RNA-dependent RNA transcription to produce multimeric antigenomic intermediates, and the intermediates then form the circular unit-length antigenome via self-cleavage and self-ligation. The antigenomic RNA then acts as a template for the production of genomic RNA through a similar rolling-circle mechanism. Since neither HDV nor mammalian cells encode any RNA-dependent RNA polymerases, the replication of HDV RNA may rely on a redirection of a host DNA-dependent RNA polymerase to become an RNA-dependent RNA polymerases (37, 54). The host RNA polymerase II is suspected to be responsible for HDV RNA replication, while some studies also suggest that replication of HDV RNA is carried out by two different cellular RNA polymerase activities (25, 30, 43, 54).
Several studies have shown that the HDAg associates with several cellular factors and specific nuclear bodies, such as delta-interacting protein A, RNA polymerase II, nucleolar proteins B23 and nucleolin, and PKR (6, 12, 31, 38, 60). These cellular components also contribute to the life cycle of HDV, while the underlying mechanism is still unknown. Interestingly, among these HDAg-interacting factors, RNA polymerase II, B23, and nucleolin have a common interacting partner, the multifunctional transcription factor YY1 (Yin Yang 1) (19, 33, 40, 61), and these three cellular factors also have been shown to facilitate the replication of HDV RNA (31, 38). YY1 is a ubiquitous zinc finger transcription factor that is involved in activating or repressing vast numbers of cellular and viral promoters and is capable of interaction with numbers of transcription factors, coactivators, and corepressors (22, 55). It is generally believed that YY1 modulates the transcription activity of promoters mainly through recruitment of different transcriptional regulatory partners (22, 55). Since replication of the HDV genome depends on cellular transcription machinery and is modulated by RNA polymerase II, B23, and nucleolin, it raises the possibility that HDAg interacts with these nuclear proteins and recruits YY1 and its associated transcription regulatory factors to modulate HDV replication. For example, CBP/p300 proteins are transcriptional coactivators recruited by YY1 for its trans-activation activities. They participate in several physiological processes and are able to modulate activities and functions of cellular and viral proteins (55). Recently, HDAg was found to be acetylated by p300, and acetylation of SHDAg may affect its subcellular localization and its activities to support HDV RNA replication (47). These results further strengthen the possible involvement of YY1 in HDV replication.
In this study, we demonstrated that the transcription factor YY1 interacts with both isoforms of HDAg. SHDAg is cosedimented with YY1 in a high-molecular-weight nuclear complex in sucrose gradient centrifugation analysis, and the YY1-associated acetyltransferases CBP and p300 are also included in this complex. Overexpression of YY1, CBP, or p300, but not the acetyltransferase-defective mutant of p300, can enhance the replication of HDV RNA. Furthermore, HDAg can be acetylated, and addition of trichostatin A (TSA), a potent inhibitor for cellular deacetylases, increases the acetylation level of SHDAg as well as HDV RNA replication. Taken together, these results demonstrated that YY1 and its associated acetyltransferases CBP and p300 can modulate HDV RNA replication, and they work probably through the complex formation with HDAg. Additionally, since YY1 is involved in various cellular processes (22), YY1 targeted by HDAg may contribute to the pathogenesis of HDV infection.
MATERIALS AND METHODS
Plasmids.
Plasmids pGEM3L and pGEM3S harboring the HDV cDNA of LHDAg and SHDAg, respectively, have been described previously (31). These two plasmids were used to generate strand-specific probes by in vitro transcription and produce LHDAg or SHDAg proteins by in vitro translation (28). Plasmids pGEX-3X-L and pGEX-3X-S are bacterial expression constructs for the glutathione S-transferase (GST)-LHDAg and GST-SHDAg fusion proteins, respectively (31). The expressing constructs of the His-tagged SHDAg polypeptides, which included the full-length SHDAg consisting of all 195 amino acids, dAg(1-195) and deletion variants consisting of the N terminus residues 1 to 88 [NdAg(1-88)], the N terminus and the middle region [NMdAg(1-143)], the middle region [MdAg(89-143)], and the C terminus plus the middle region [CdAg(89-195)] of SHDAg have been described elsewhere (31, 32). Plasmid pRSETB-YY1, a derivative of pRSETB (Invitrogen), contains the full-length cDNA of YY1 driven by the T7 promoter, which allows the production of YY1 RNA by in vitro transcription or protein by in vitro translation (44). Plasmid pcDNA3-HA-YY1 (where HA is hemagglutinin) was constructed by insertion of an excised BamHI/XbaI DNA fragment harboring YY1 cDNA from pGAL4-YY1 (44) into BamHI/XbaI-digested pcDNA3-HA. Plasmids pGST-YY1 and its serial deletion constructs pGST-YY1/1-80 (harboring residues 1 to 80 of YY1), pGST-YY1/1-154, pGST-YY1/1-198, pGST-YY1/1-295, pGST-YY1/155-198, pGST-YY1/199-295, and pGST-YY1/296-414 are bacterial expression constructs that encode various lengths of the YY1 protein fused with the C terminus of the GST protein as described previously (44). Plasmids FLAG-d-N, FLAG-d-NM, and FLAG-d-NC containing different portions of SHDAg cDNA (FLAG-d-N encompasses amino acid residues 1 to 88 of SHDAg; FLAG-d-NM encompasses amino acid residues 1 to 163 of SHDAg; and FLAG-d-NC encompasses amino acid residues 1 to 88 and 164 to 195 of SHDAg) were derived from a FLAG-tagged SHDAg expression plasmid (12).These plasmids were constructed using a QuikChange Multi Site-Directed Mutagenesis kit (Merck Co.) with the primer sets 5′-GGACCTGGGAAGAGGTGACCTCTCAGGGGAGGATTC and 5′-TCCTCCCCTGAGAGGTCACCTCTTCCCAGGTCCGGA for FLAG-d-N, 5′-TCGAGGGGAGCGCCCTGAGGGGGCGGCTTCGTCCCC and 5′-GACGAAGCCGCCCCCTCAGGGCGCTCCCCTCGATCC for FLAG-d-NM, and ′-TCCGGACCTGGGAAGAGGGGGGGCGGCTTCGTCCCC and 5′-GGGGACGAAGCCGCCCCCCCTCTTCCCAGGTCCGGA for FLAG-d-NC. Plasmid pKS/HDV1.9, which contains the entire HDV genome and about 200 additional nucleotides of HDV sequence, was used to produce HDV genomic RNA (1.9 kb) by in vitro transcription (46). Plasmids pCMV-CBP-FLAG and pSK-p300-myc (kindly provided by L.-J. Juan, Academica Sinica, Taiwan) are the expression constructs for FLAG-tagged CBP and Myc-tagged p300 in mammalian cells, respectively.
Cell lines and subcellular fractionation.
HeLa, HuH-7, and HepG2 cells were cultured as described elsewhere (62). L10 and S7, derived from HepG2 cells, have been described previously (31); they are permanent cell lines constitutively expressing LHDAg and SHDAg, respectively. SVLD3-N1 is the stable cell line permanently expressing both LHDAg and SHDAg and both HDV genomic and antigenomic RNA in HepG2 cells (14). The total cell lysates and nuclear extracts were isolated from these cells as described previously (63).
In vitro binding analysis.
GST-LHDAg, GST-SHDAg, and GST-YY1 as well as various truncation mutants of GST-YY1 were purified as previously described (51). The various deletion mutants of His-tagged SHDAg fusion proteins were also purified as described elsewhere (31, 32). For each in vitro binding analysis, 20 μl of glutathione-Sepharose 4B beads prebound with GST fusion proteins (1 to ∼5 μg) or His resins prebound with His-tagged SHDAg variants (1 μg) were incubated with HeLa nuclear extracts (200 μg) or in vitro translated proteins (5 μl) at 4°C overnight under gentle agitation. The beads were then washed four times with washing buffer (phosphate-buffered saline [PBS] containing 0.5% NP-40). Proteins bound on beads were eluted by sampling buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and processed for Western blot analysis with rabbit anti-YY1 antibodies (C-20; Santa Cruz) or autoradiography.
Immunofluorescence microscopy.
Immunofluorescence staining was performed by a method described elsewhere (30) with some modifications. Briefly, HuH-7 cells seeded on coverslips were first transfected with the expression constructs of SHDAg or HA-tagged YY1 either alone or in combination. Transfected cells were fixed with 3.7% paraformaldehyde at 48 h posttransfection. After fixation, cells were washed with PBS three times and then blocked in PBS containing 1% bovine serum albumin for 1 h at room temperature. Cells were then immunostained with rabbit anti-HDAg antiserum together with monoclonal anti-HA antibodies (Sigma Co.) for 1 h at room temperature. After being washed four times with PBS, the cells on coverslips were stained with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) and rhodamine-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) for another 1 h. After four washes in PBS, the coverslips were mounted on slides with Vectashield (Vector Laboratory, Inc., Burlingame, CA), and an immunofluorescence microscope (Leica) was used for observation.
Sucrose gradient centrifugation and in vivo coimmunoprecipitation.
The nuclear extracts of the LHDAg- or SHDAg-producing cells (L10 or S7) and their parental HepG2 cells were prepared and analyzed by sucrose gradient centrifugation as described previously (31). After centrifugation, aliquots of each fraction were analyzed by immunoblotting using human anti-HDAg antiserum or rabbit anti-YY1 antibodies. For the in vivo coimmunoprecipitation analysis of HDAg and YY1, CBP, or p300, the sucrose gradient fractions 15 to 17 obtained from L10, S7, or HepG2 cells were collected and immunoprecipitated with 20 μl of protein A-Sepharose beads, which were preconjugated with human anti-HDAg antiserum. The immunoprecipitates were washed with washing buffer (PBS containing 0.3% NP-40) and processed for immunoblotting analysis with antibodies against YY1 (C-20; Santa Cruz), CBP (C-20; Santa Cruz), and p300 (N-15; Santa Cruz).
In vitro transcription.
HDV genomic RNA (1.9 kb) was transcribed from pKS/HDV1.9, which was linearized by EcoRV digestion, with a T7 MEGAscript (Ambion) transcription kit (46). Capped RNA of SHDAg was transcribed from pGEM3S with the SP6 mMESSAGE mMACHINE (Ambion) transcription kit after linearization by EcoRI. Capped RNA of YY1 was transcribed from pRSETB-YY1 with a T7 mMESSAGE mMACHINE (Ambion) transcription kit after linearization by EcoRI.
DNA and RNA transfection.
For transient DNA transfection experiments, plasmid DNAs were transfected into HuH-7 cells by Lipofectamine 2000 transfection reagent (Invitrogen) according to the instructions provided by the supplier. For transient RNA transfection experiments, in vitro transcribed HDV genomic RNA (15 μg) and SHDAg mRNA (5 μg) were cotransfected into HuH-7 cells by DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol) transfection reagent (Invitrogen) according to the manufacturer's instructions (30). This HDV RNA transfection method has been described as the cDNA-free HDV RNA replication system (46). In some experiments, various amount of YY1 RNA (0.5 to ∼5 μg) would also be included in the transient RNA transfection procedure.
RNA preparation and Northern blotting.
The extraction of total cellular RNAs and Northern blotting analysis were preformed as described previously (31). Total cellular RNAs of HuH-7 cells were extracted by using TRI reagent (Molecular Research Center) according to the manufacturer's instructions (31). For Northern blot analysis, 20 μg of total cellular RNA was analyzed by electrophoresis on a 6% formaldehyde-1% agarose RNA gel and then transferred to a nylon membrane. The membranes were prehybridized and then hybridized with 32P-labeled strand-specific HDV RNA probes, which were generated from pGEM3L by using a SP6/T7 transcription kit (Promega) (62).
In vivo acetylation labeling and TSA treatment.
For in vivo acetylation labeling experiments, approximately 107 HepG2 or SVLD3-N1 cells were plated into 150-mm culture dishes. The culture medium was changed after 16 h, and the cells were pulse labeled with 1 mCi/ml [3H]acetate for 1 h. Nuclear extracts of labeled cells were isolated and subjected to immunoprecipitation analysis with 20 μl of protein A-Sepharose beads, which were preconjugated with human anti-HDAg antiserum. The immunoprecipitates were washed with washing buffer (PBS containing 0.3% NP-40) and resolved by SDS-PAGE. The gel was then dried and detected with fluorography. For TSA treatment, approximately 2 × 106 HuH-7 cells were seeded into 100-mm culture dishes. Cells were transfected with HDV genomic RNA and HDAg mRNA 16 h after seeding. At 48 h posttransfection, transfected cells were treated with 1 μM TSA (dissolved in dimethyl sulfoxide [DMSO] at the concentration of 1 mM) for 16 h; then the culture medium was changed, and the cells were cultured for another 24 h. Nuclear extracts and total cellular RNA from the transfected cells were prepared and subjected to Western blotting and Northern blotting analysis for detection of HDAg expression by human anti-HDAg antiserum and HDV replication by strand-specific HDV RNA probes.
RESULTS
Both LHDAg and SHDAg interact with YY1.
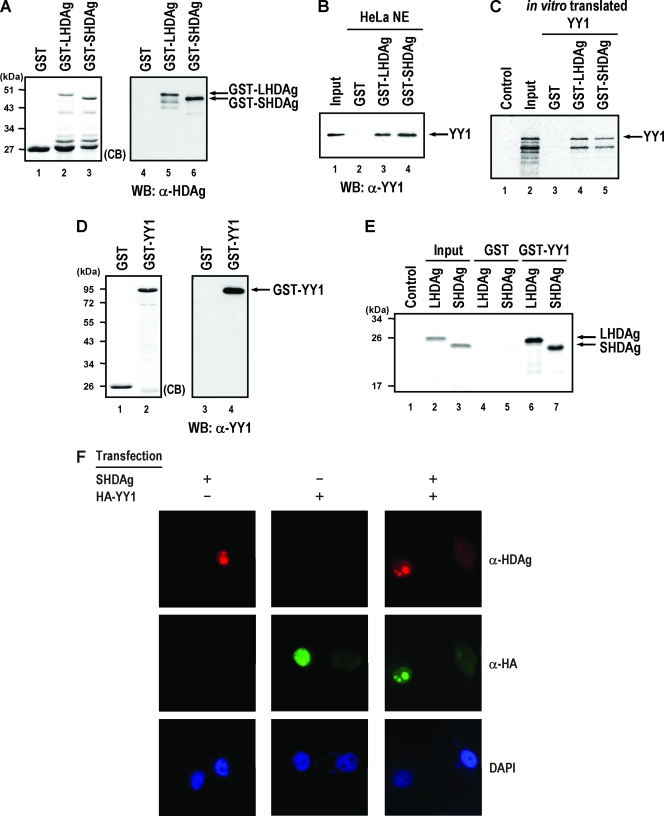
Several HDAg-interacting factors, including RNA polymerase II, B23, and nucleolin, have a common interacting partner, the multifunctional transcription factor YY1 (19, 33, 40, 61), and these three cellular factors also have been shown to facilitate the replication of HDV RNA (31, 38). These results suggest that YY1 may interact with HDAg and be involved in modulating HDV replication. To test this possibility, we first examined whether LHDAg or SHDAg interacts with YY1 by using in vitro GST pull-down analysis. To this end, the partially purified GST-LHDAg or GST-SHDAg fusion proteins (Fig. 1A, lanes 2 and 3), and nuclear extracts of HeLa cells were used for GST pull-down assay. As shown in Fig. 1B, cellular YY1 was precipitated by purified GST-LHDAg or GST-SHDAg fusion proteins (lanes 3 and 4) but not by GST (lane 2). Additionally, we performed similar GST pull-down assays as described in the legend of Fig. 1B, in which the in vitro translated 35S-labeled YY1 was used instead of HeLa nuclear extracts. As shown in Fig. 1C, in vitro translated 35S-labeled YY1 was pulled down by purified GST-LHDAg or GST-SHDAg fusion proteins (lanes 4 and 5) but not by GST (lane 3). These results indicated that YY1 interacts specifically with both isoforms of HDAg since binding between YY1 and GST was not observed. In addition, GST pull-down assays with partially purified GST-YY1 fusion protein (Fig. 1D, lane 2) and in vitro translated 35S-labeled LHDAg or SHDAg were also performed in the presence of RNase A. As shown in Fig. 1E, results demonstrated that both isoforms of HDAg were precipitated by purified GST-YY1 fusion protein (lanes 6 and 7) but not by GST alone (lanes 4 and 5). Furthermore, immunofluorescence microscopy analysis (Fig. 1F) showed that, when expressed alone, YY1 was distributed more homogeneously in the nucleoplasm while SHDAg localized in nucleoplasm and nucleoli. However, the nucleolar distribution of YY1 was greater when it was coexpressed with SHDAg. Taken together, these results indicate that HDAg interacts with YY1 in vivo and in vitro, and the association of YY1 with both isoforms of HDAg is through a direct protein-protein interaction.
FIG. 1.
Both LHDAg and SHDAg interact with YY1. (A) Purified GST, GST-LHDAg, and GST-SHDAg fusion proteins were analyzed by SDS-PAGE and stained with Coomassie brilliant blue (CB; lanes 1 to 3) or verified by immunoblotting with antibodies against HDAg (lanes 4 to 6). (B) In vitro binding assay of GST-HDAg fusion proteins and cellular YY1. The nuclear extracts (NE) of HeLa cells (200 μg) were incubated with glutathione-Sepharose 4B beads (20 μl) prebound with GST, GST-LHDAg, or GST-SHDAg (4 μg each), and the bound fractions were analyzed by SDS-PAGE followed by immunoblotting with antibody against YY1. Input, 50 μg of HeLa nuclear extracts (lane 1). (C) In vitro binding assay of GST-HDAg fusion proteins and in vitro translated 35S-labeled YY1. Glutathione-Sepharose beads (20 μl) bound with GST, GST-LHDAg, or GST-SHDAg (1 μg each) were incubated with in vitro translated 35S-labeled YY1 (10 μl). The beads were then washed, and proteins on the beads were analyzed by SDS-PAGE followed by autoradiography. Control, 2 μl of in vitro translated mixture in the absence of YY1 mRNA; input, 1 μl of in vitro translated 35S-labeled YY1. (D) Purified GST and GST-YY1 fusion proteins were analyzed by SDS-PAGE and stained with Coomassie brilliant blue (CB; lanes 1 and 2) or verified by immunoblotting with antibodies against YY1 (lanes 3 and 4). (E) In vitro binding assay of GST-YY1 and in vitro translated 35S-labeled HDAgs. Glutathione-Sepharose beads (20 μl) bound to GST or GST-YY1 (4 μg each) was incubated with the in vitro translated 35S-labeled LHDAg and SHDAg (10 μl each) in the presence of RNase A (20 μg/ml). The beads were then washed, and proteins on the beads were analyzed by SDS-PAGE followed by autoradiography. Control, 2 μl of in vitro translated mixture in the absence of HDAg mRNA; input, 1 μl of in vitro translated 35S-labeled LHDAg (lane 2) or SHDAg (lane 3). (F) Subcellular localization of SHDAg and YY1. Expression plasmids of SHDAg, HA-tagged YY1, or both were used to transfect HuH-7 cells. Transfected cells were fixed 2 days posttransfection and analyzed by immunofluorescence assay. Cells were immunostained with rabbit anti-HDAg antiserum and monoclonal anti-HA antibodies, followed by fluorescein isothiocyanate-conjugated goat anti-mouse IgG, rhodamine-conjugated goat anti-rabbit IgG, and DAPI (4′,6′-diamidino-2-phenylindole) staining, and observed by an immunofluorescence microscope. WB, Western blotting; α, anti.
Mapping the interaction regions between HDAg and YY1.
Several functional domains have been recognized in HDAgs. These include the N-terminal coiled-coil domain, the nuclear localization signal domain, and two ARMs separated by a helix-loop-helix motif; these domains are important for HDAg oligomerization, HDAg and HDV RNA nuclear transport, and HDV RNA-binding activities of HDAg, respectively (43). Furthermore, LHDAg contains the nuclear export signal and packaging signals in its C terminus (43). On the other hand, a number of functional domains have also been characterized in YY1. These include two acidic regions (amino acid residues 1 to 69 and 81 to 154) separated by a histidine cluster (amino acid residues 70 to 80), a central GA/GK-rich domain, and four C-terminal zinc finger DNA binding domains (55). The N-terminal one-third region is the trans-activation domain of YY1, whereas the central GA/GK-rich domain and C-terminal zinc finger domain are trans-repression domains. The central GA/GK-rich and C-terminal zinc finger domains are also important protein-protein interaction domains for YY1 that associate with numerous cellular factors (55). Since the functional domains of both HDAg and YY1 have been well delineated, an investigation of the interaction region required for their association may provide certain functional insights into the biological relevance of the interaction between these two proteins.
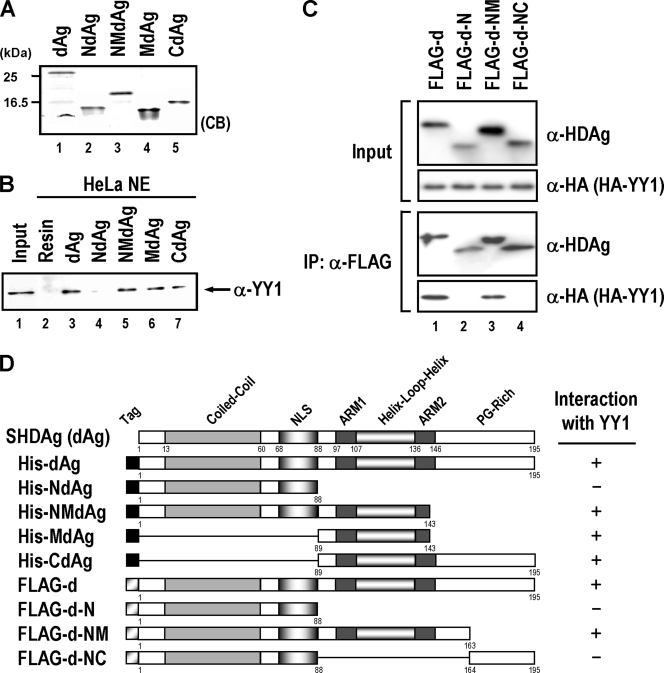
We mapped the region essential for interaction of YY1 within HDAg by using the various partially purified His-tagged HDAg fragments dAg(1-195), NdAg(1-88), NMdAg(1-143), MdAg(89-143), and CdAg(89-195) of SHDAg (Fig. 2A). These partially purified His-tagged HDAg variants and nuclear extracts of HeLa cells were used for in vitro binding analysis. As shown in Fig. 2B, except for NdAg (lane 4), all of the HDAg variants containing the middle portion fragment, including dAg, NMdAg, MdAg, and CdAg (Fig. 2B, lanes 3 and 5 to 7), could precipitate the cellular YY1. The His-tagged HDAg variants interacted with YY1 specifically, since YY1 could not be precipitate by His resins (lane 2). These results (summarized in Fig. 2D) suggest that amino acid residues 89 to 143 of HDAg are important for binding with YY1. To verify these results in vivo, we used FLAG-tagged constructs containing different domains of SHDAg for cotransfection with the HA-tagged YY1 expression construct and to analyze the interaction domain by coimmunoprecipitation. We also added RNase A in cell lysates to exclude the possible involvement of RNA in the protein-protein interaction. As shown in Fig. 2C, FLAG-tagged proteins harboring the middle portion of SHDAg (FLAG-d-NM) possess the YY1-binding activities. However, tagged proteins containing the SHDAg N-terminal portion alone (FLAG-d-N) or its N-terminal portion plus C-terminal portion (FLAG-d-NC) did not show such activities. Notably, this middle portion of HDAg encompasses the two ARMs that specifically associate with HDV RNA (10, 41) and are critical for HDV replication (39). Note also that since RNase A treatment did not hamper the interaction of these two proteins, HDV RNA may not be required for the interaction.
FIG. 2.
Mapping of the YY1-interacting region in HDAg. (A) Partial purified His-tagged SHDAg (dAg) and its various derivatives NdAg, NMdAg, MdAg, and CdAg (1 μg each) were separated by SDS-PAGE and detected by Coomassie brilliant blue (CB) staining. (B) In vitro binding assay of His-tagged SHDAg variants and cellular YY1. The nuclear extracts (NE) of HeLa cells (200 μg) were incubated with 20 μl of His-resins (lane 2) or His resins prebound with SHDAg derivatives (lanes 3 to 7) as indicated, and proteins bound on the beads were eluted and analyzed by SDS-PAGE followed by immunoblotting with anti-YY1 antibody. Input: 50 μg of HeLa nuclear extracts (lane 1). (C) Further mapping of YY1 interaction domain on SHDAg. Expression plasmids of FLAG-tagged SHDAg (FLAG-D) or its derived truncated mutants (FLAG-d-N, FLAG-d-NM, and FLAG-d-NC) were used to transfect with the expression plasmid of HA-tagged YY1 to HuH-7 cells. Transfected cells were harvested at 2 days posttransfection and then analyzed by a coimmunoprecipitation assay. For the coimmunoprecipitation, the extracts from the transfected cells were immunoprecipitated with anti-FLAG M2 affinity gel (10 μl, packed volume; Kodak) in the presence of RNase A (10 μg/ml) and incubated at 4°C for 1 h with agitation. The immunoprecipitates were washed four times with PBS buffer containing 0.3% NP-40 and processed for Western blotting (rabbit anti-HA antiserum for HA-tagged YY1 and rabbit anti-FLAG antiserum for FLAG-tagged SHDAg and its truncation mutants). (D) A schematic diagram showing the interaction region of YY1 within SHDAg. The numbers represent the positions of the amino acid residues of SHDAg and its variants. The interaction ability of SHDAg and its variants with YY1 is indicated by a plus or a minus sign. NLS, nuclear localization signal; PG-rich: proline/glycine-rich; α, anti; IP, immunoprecipitation.
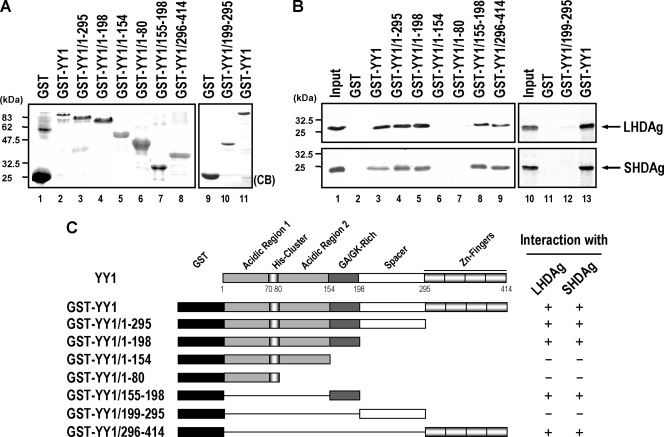
We also dissected the interaction domain of LHDAg or SHDAg within YY1. To this end, we analyzed the binding of various GST-YY1 truncated mutants (Fig. 3A) to in vitro translated 35S-labeled LHDAg or SHDAg by GST pull-down assay. As shown in Fig. 3B, both LHDAg and SHDAg interacted with a full-length GST-YY1 fusion protein (lane 3) as well as with two C-terminal deletion variants including GST-YY1/1-295 (lane 4) and GST-YY1/1-198 (lane 5). Further deletion of the C terminus of GST-YY1, as in the constructs GST-YY1/1-154 (Fig. 3B, lane 6) and GST-YY1/1-80 (lane 7), leads to a loss of the ability to associate with both isoforms of HDAg, indicating that the N-terminal 154 amino acids of YY1 were not sufficient for interacting with HDAgs. In addition, both LHDAg and SHDAg were pulled down by truncated variants GST-YY1/155-198 (Fig. 3B, lane 8) and GST-YY1/296-414 (lane 9) but not by GST-YY1/199-295 (lane 12), indicating that the fragments harboring amino acid residues 155 to 198 (central GA/GK-rich domain) and 296 to 414 (C-terminal zinc finger domain) of YY1 are important for interacting with HDAgs (summarized in Fig. 3C). It is worth noting that these HDAg-interacting regions are trans-repression domains and/or DNA binding domains of YY1 and are also responsible for interaction with other cellular factors (55).
FIG. 3.
Mapping of the HDAg-interacting regions in YY1. (A) Partially purified GST-YY1 and its various truncation mutants (5 μg each) were separated by SDS-PAGE and detected by Coomassie brilliant blue (CB) staining. (B) In vitro binding assay of GST-YY1 variants and in vitro translated 35S-labeled HDAgs. Glutathione-Sepharose beads (20 μl) bound with GST (lanes 2 and 11), GST-YY1 (lanes 3 and 13), or GST-YY1 variants (lanes 4 to 9 and 12) (5 μg each) were incubated with in vitro translated 35S-labeled LHDAg or SHDAg (5 μl), and proteins bound on the beads were eluted and analyzed by SDS-PAGE followed by autoradiography. Input, in vitro translated 35S-labeled LHDAg or SHDAg (5 μl for lane 1 and 1 μl for lane 10). (C) A schematic diagram showing the binding regions of HDAgs within YY1. The numbers represent the positions of the amino acid residues of YY1. The binding ability of LHDAg or SHDAg with GST-YY1 or its truncation variants is indicated by a plus or a minus sign.
YY1 forms a large complex with HDAgs in vivo.
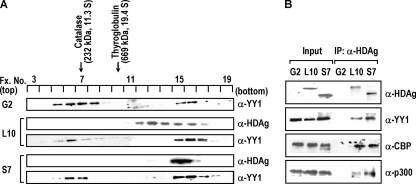
Previous studies by our laboratory had shown that both LHDAg and SHDAg were cosedimented with B23 and nucleolin in a large nuclear complex in vivo, and this HDAg-containing complex contributed to modulate HDV RNA replication (31). Since YY1 associates with these two nucleolar proteins (33, 40, 61) and since, as demonstrated above, YY1 also interacted with both isoforms of HDAg in vitro, it was interesting to investigate the status of in vivo complex formation between YY1 and HDAgs. To this end, we examined the sedimentation profiles of YY1 and HDAgs in the nuclear fractions of both types of HDAg-producing cell lines (L10 and S7) and the parental HepG2 cells by sucrose gradient centrifugation. As shown in Fig. 4A, the sedimentation profiles of YY1 were essentially similar in all of the examined cells, as two predominant peaks at fractions 5 to 8 (Fig. 4A, peak I maximum at fraction 6) and fractions 15 to 17 (peak II, maximum at fraction 16) were found. By comparing the results with two parallel sedimentation standards, catalase (fraction 7, 232 kDa) and thyroglobulin (fraction 10, 669 kDa), we found that YY1 was sedimented at a position with high molecular weight, as peak I sedimented at the position almost equal to 232 kDa and peak II sedimented at a position much larger than 669 kDa. Since cellular YY1 is 68 kDa in size when analyzed by SDS-PAGE, these results suggested that YY1 was included in large nuclear complexes by associating with other cellular components. It should be noted that the presence of either LHDAg or SHDAg somewhat altered the sedimentation pattern of YY1 since the sizes of peak II YY1-containing complexes increased slightly in L10 and S7 cell lines. The sedimentation profiles of HDAgs in both L10 and S7 cell lines were consistent with our previous results (31), in which the LHDAg sedimented broadly at fractions 12 to 17 in L10 cells and the SHDAg sedimented at fractions 15 to 17 in S7 cells (Fig. 4A). Interestingly, SHDAg showed almost cosedimentation with the peak II complex of YY1 in SHDAg-producing cells, suggesting that SHDAg associated preferentially with YY1 in the high-molecular-weight complex. However, LHDAg was more broadly distributed from fraction 12 to 17, and there was a partial overlap with the peak II complex of YY1 in the LHDAg-producing cells, indicating a partial separation of the LHDAg from YY1-associated complex. This difference of sedimentation behavior may reflect the fact that two isoforms of HDAg are associated with different cellular partners for their distinct biological functions. Additionally, since LHDAg can be farnesylated, we still cannot rule out the possibility that this modification may also influence the sedimentation behavior of LHDAg.
FIG. 4.
YY1 forms a large complex with HDAgs in vivo. (A) The nuclear extracts (500 μg) of the HDAg-producing (L10 or S7) and the parental HepG2 (G2) cells were isolated and subjected to sucrose gradient centrifugation analysis. Aliquots of each fraction were analyzed by immunoblotting using antibodies against YY1 or HDAg. Protein standards, catalase (232 kDa) and thyroglobulin (669 kDa), were run in parallel experiments, and their sedimented positions are indicated. (B) Complex formation between HDAgs, YY1, CBP, and p300. The sucrose gradient fractions 15 to 17 obtained from nuclear extracts of HepG2 (G2), L10, or S7 cells as described in panel A were pooled together and immunoprecipitated (IP) with human anti-HDAg antiserum. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with antibodies against HDAg, YY1, CBP, and p300. Input, nuclear extracts (50 μg) of G2, L10, or S7 as loading controls; α, anti.
We further verified the in vivo interaction and complex formation of either form of HDAgs with YY1. Fractions 15 to 17 of the sucrose gradients from nuclear extracts of these HDAg-producing cells were pooled together and analyzed by a coimmunoprecipitation assay with human anti-HDAg antiserum. As shown in Fig. 4B, YY1 at fractions 15 to 17 of the sucrose gradient in the HDAg-producing cells (L10 and S7) was coimmunoprecipitated with HDAg. These results indicated that YY1 was not cosedimented with HDAgs coincidentally, suggesting that YY1 interacts directly with HDAgs in vivo in the high-molecular-weight peak II complex of L10 and S7 cells. Moreover, cellular acetyltransferases CBP and p300, which are interacting partners of YY1 (2), were also coimmunoprecipitated with HDAg by anti-HDAg antiserum, suggesting that the HDAg-associated high-molecular-weight nuclear complex includes these cellular factors. Taken together, these results indicate that the components in HDAg-containing nuclear peak II complex include at least HDAg, YY1, CBP, and p300 and that this complex might participate in regulation of HDV life cycle.
YY1 enhances HDV replication.
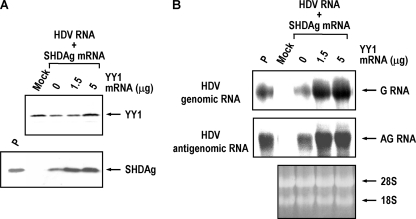
The SHDAg is required for replication of HDV RNA (11), and previous results from our laboratory showed that HDV replication was enhanced by B23 through formation of a large complex with SHDAg (31). As demonstrated above, although YY1 interacts with both LHDAg and SHDAg and was included in the HDAg-containing nuclear complexes in vivo, SHDAg cosedimented specifically with YY1 in the peak II complexes (Fig. 4A). These observations reflect the potential function of these complexes in HDV replication, and it is quite likely that YY1 might be involved in the regulation of HDV replication. To test this possibility, we employed a cDNA-free HDV RNA replication system (46) to analyze the role of YY1 in HDV replication. The HDV genomic RNA and SHDAg mRNA together with increasing amounts of YY1 mRNA were introduced into HuH-7 cells, and the expression levels of HDAg and HDV RNA in the transfected cells were then analyzed by Western blotting and Northern blotting, respectively. As shown in Fig. 5A, increasing amounts of YY1 enhanced the expression level of SHDAg in the transfected cells. Simultaneously, the expression levels of genomic and antigenomic RNA of HDV in the transfected cells also increased significantly in a YY1 dose-dependent manner (Fig. 5B) (about 3.9- to 4.9-fold increase for the genomic and about 2.2-fold for the antigenomic RNA). Hence, YY1 is able to facilitate HDV replication, apparently through the formation of complex with SHDAg and/or other cellular factors.
FIG. 5.
YY1 enhances the HDV replication. (A) YY1 enhances SHDAg expression. For each experiment, approximately 2 × 106 HuH-7 cells were cotransfected with HDV genomic RNA (15 μg), SHDAg mRNA (5 μg), and increasing amounts of YY1 mRNA (0, 1.5, or 5 μg) by DMRIE-C transfection reagent (Invitrogen). The expression levels of YY1 (2 days posttransfection) and HDAg (4 days posttransfection) were analyzed by SDS-PAGE followed by immunoblotting with antibodies against YY1 and HDAg. (B) YY1 enhances the replication of HDV RNA. The expression levels of HDV genomic (G) and antigenomic (AG) RNA in the transfected cells as described in panel A were determined by Northern blotting at 4 days posttransfection. Mock, HuH-7 cells without transfection; P, SHDAg or HDV RNAs as positive controls.
CBP and p300 also enhance HDV replication.
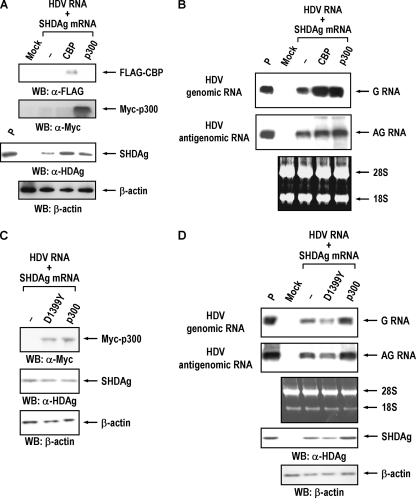
Two groups of factors, histone acetyltransferase and histone deacetylase (HDAC), known to function as coactivator and corepressor, respectively, interact with YY1 and modulate YY1-mediated transcription regulation (55). As shown in Fig. 4B, two of these YY1-associated transcriptional coactivators, acetyltransferases CBP and p300, were also included in the high-molecular-weight complex of HDAgs and YY1 in vivo. It is thus likely that YY1 might modulate HDV replication through its association with CBP or p300. To examine this possibility, we introduced CBP or p300 expression constructs into HuH-7 cells 12 h prior to RNA transfection with HDV genomic RNA and SHDAg mRNA, and the expression level of SHDAg or HDV RNA in the transfected cells was then analyzed. As shown in Fig. 6A, exogenous expression of CBP or p300 in the HDV replication system elevated the expression level of SHDAg. At the same time, the expression levels of genomic (about 3.8- to 4.0-fold increase) and, to a less extent, of antigenomic RNA (about 1.9- to 2.5-fold increase) of HDV were also enhanced by exogenous expression of CBP or p300 (Fig. 6B). Therefore, both CBP and p300 have the ability to enhance HDV replication. To further elucidate the trans-activation activities of these histone acetyltransferases on HDV replication and expression, we conducted similar experiments with an acetyltransferase-defective mutant of p300 (carrying the mutation D1399Y) (44). As shown in Fig. 6D, the D1399Y mutant of p300 inhibited HDV replication (about 43% reduction for the genomic and about 24% reduction for the antigenomic RNA) while overexpression of p300 enhanced HDV RNA replication and SHDAg expression. These results suggest that the acetyltransferase activity of p300 is important for HDV replication.
FIG. 6.
CBP and p300 modulate the HDV replication. (A) CBP and p300 enhance SHDAg expression. For each experiment, approximately 2 × 106 HuH-7 cells were transfected with 10 μg of empty vector (−) or expression construct of FLAG-tagged CBP or Myc-tagged p300 by Lipofectamine 2000 transfection reagent (Invitrogen). At 12 h posttransfection, cells were then transfected with HDV genomic RNA (15 μg) and SHDAg mRNA (5 μg) by DMRIE-C transfection reagent (Invitrogen). The expression levels of CBP, p300 (1 day posttransfection), and HDAg (4 days posttransfection) were analyzed by SDS-PAGE followed by immunoblotting with anti-FLAG tag, anti-Myc tag, or anti-HDAg antibodies. (B) CBP and p300 enhance the replication of HDV RNA. The expression levels of HDV genomic (G) and antigenomic (AG) RNA in the transfected cells as described in panel A were determined by Northern blotting at 4 days posttransfection. (C) Acetyltransferase-defective mutant of p300 inhibits HDV replication. This experiment is similar to that shown in panel A, except that 10 μg of empty vector (−) or expression construct of Myc-tagged p300 (p300) or its mutant D1399Y (D1399Y) was used for the transfection experiment. At 12 h posttransfection, cells were then transfected with HDV genomic RNA (10 μg) and SHDAg mRNA (10 μg) by DMRIE-C transfection reagent (Invitrogen). The expression levels of p300 and HDAg (1 day posttransfection) were analyzed by SDS-PAGE followed by immunoblotting with anti-Myc tag or anti-HDAg antibodies at 24 h posttransfection. (D) The expression levels of HDV RNAs and SHDAg in the transfected cells as described in panel C were determined by Northern blotting and Western blotting at 4 days posttransfection. Mock, HuH-7 cells without transfection. P, SHDAg or HDV RNAs as positive controls; WB, Western blotting; α, anti.
HDAg is acetylated in vivo, and the addition of TSA enhances HDV replication.
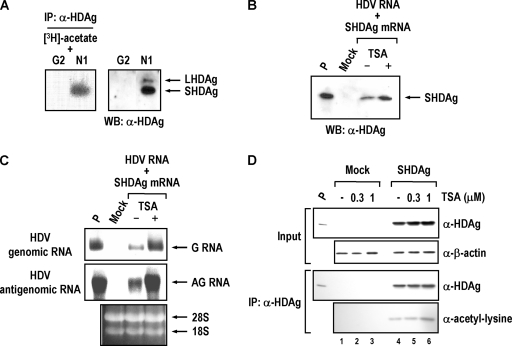
Several cellular and viral proteins were acetylated by CBP or p300, and such posttranslational modification altered their functions or activities (3, 18, 20). Since CBP and p300 interact with HDAgs and facilitate HDV replication, it raises the possibility that CBP and p300 modulate HDV replication through acetylation of HDAgs. To test this hypothesis, we performed an in vivo acetylation labeling assay to analyze the acetylation status of HDAg. The SVLD3-N1 cells (HDV genome cDNA-integrated HepG2 cell line) and its parental HepG2 cells were labeled with [3H]acetate, and then the nuclear extracts of labeled cells were subjected to immunoprecipitation analysis with human anti-HDAg antiserum. As shown in Fig. 7A, SHDAg in the nuclear extracts of the SVLD3-N1 cells was specifically labeled by [3H]acetate. No such signal could be detected in the control HepG2 cells. This result indicates that SHDAg can be acetylated in vivo.
FIG. 7.
SHDAg is acetylated, and TSA treatment enhances HDV replication. (A) SHDAg is acetylated in vivo. Approximately 107 HDV-producing SVLD3-N1 (N1) or its parental HepG2 (G2) cells were pulse labeled with 1 mCi/ml [3H]acetate for 1 h. Nuclear extracts of the labeled cells were subjected to immunoprecipitation assay with anti-HDAg antiserum and then analyzed by SDS-PAGE followed by fluorography (left panel). Nuclear extracts of untreated HepG2 and SVLD3-N1 cells were analyzed in parallel by Western blotting (WB) with antibodies against HDAg (right panel). (B) Treatment of TSA enhances SHDAg expression. For each experiment, approximately 2 × 106 HuH-7 cells were cotransfected with HDV genomic RNA (15 μg) and SHDAg mRNA (5 μg) by DMRIE-C transfection reagent (Invitrogen). At 48 h posttransfection, the transfected cells were treated with 1 μM TSA (+) or DMSO (−) for 16 h (see Materials and Methods). The expression level of SHDAg at 4 days posttransfection was analyzed by SDS-PAGE followed by immunoblotting with antibodies against HDAg. (C) Treatment of TSA enhances the replication of HDV RNA. The expression levels of HDV genomic (G) and antigenomic (AG) RNA in the transfected cells as described in panel B were determined by Northern blotting at 4 days posttransfection. (D) Treatment of TSA enhances acetylation of SHDAg. For each experiment, approximately 2 × 106 HuH-7 cells were transfected with (lanes 4 to 6) or without (lanes 1 to 3) the expression plasmids of SHDAg. At 36 h posttransfection, the transfected cells were treated, respectively, with 0.3 μM TSA, 1 μM TSA, or DMSO (−) for 12 h. Cells were then harvested, and the lysates were immunoprecipitated with anti-HDAg antiserum. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with antibodies against HDAg and acetyl-lysine (Cell Signaling technology). Mock, HuH-7 cells without transfection; P, SHDAg or HDV RNAs as positive controls; α, anti; WB, Western blotting; IP, immunoprecipitation.
We next tested whether the acetylation status of SHDAg would have any effect on HDV replication. To test this possibility, the HDV genomic RNA and SHDAg mRNA were introduced into HuH-7 cells, and TSA, a potent inhibitor of HDAC, was added to maintain the protein acetylation status in the cells. As shown in Fig. 7B and C, treatment with TSA dramatically elevated both SHDAg gene expression and HDV replication (about 4.8-fold increase for genomic and about 3-fold increase for antigenomic RNA) in the transfected cells. Furthermore, TSA treatment also increased the acetylation level of SHDAg (Fig. 7D) (about 1.5- to 3.8-fold increase). These results indicate that SHDAg is an acetylated protein and that its acetylation status affects HDV replication, which is consistent with the result reported by Mu et al. (47).
DISCUSSION
In this study, we have demonstrated that both LHDAg and SHDAg associate with the transcription factor YY1. YY1 is a multifunctional protein involved in modulating transcription activities of various promoters and was found to interact with several key transcriptional regulatory proteins (22, 55). Our experiments also indicated that HDAg interacts with YY1 via the central GA/GK-rich and C-terminal zinc-finger domains of YY1 (Fig. 3). These two regions are the major protein-protein interaction domains of YY1. Many cellular factors, such as TATA box-binding protein, CBP, p300, TFIIB, Sp1, HDAC1 to HDAC3, and B23 (22, 55), interact with YY1 through these two domains. Under appropriate conditions, three factors together, including YY1, TFIIB, and RNA polymerase II, can direct transcription on template DNA carrying the YY1 Inr element in vitro, suggesting that YY1 binds to the core promoter and recruits RNA polymerase II to the initiation complex (56). Since YY1 interacts with HDAg and both factors possess RNA polymerase II-binding activity, the recruitment of YY1 to HDV RNA template by SHDAg may facilitate the recruitment of RNA polymerase II for HDV replication and transcription.
It has been proposed that the regulation of particular promoter activity by YY1 mainly depends on its associated factors, especially interaction with coactivators or corepressors (22, 55). Coimmunoprecipitation analysis on the sucrose gradient fractions of HDAg-producing cells suggested that YY1 and YY1-interacting acetyltransferases CBP and p300 associate and form large nuclear complexes with both LHDAg and SHDAg in vivo (Fig. 4). Remarkably, our previous study showed that the nucleolar protein B23 and nucleolin also associated and cosedimented with both isoforms of HDAg in the same fractions (31). It is noteworthy that the interaction domains of B23 and nucleolin on HDAg are different from those of YY1 (31, 38) (Fig. 2). These results suggest that these large nuclear HDAg-containing complexes we observed in the sucrose gradient centrifugation analysis might contain HDAg, YY1, CBP, p300, B23, and nucleolin. Notably, the sedimentation profiles of YY1 in either control HepG2 or HDAg-producing cells are similar to those of B23, and YY1 and B23 seem to cosediment with SHDAg rather than with LHDAg (Fig. 4A) (31). These observations reflect the possible functional relevance between YY1 and B23 and, more importantly, their involvement in HDV replication since SHDAg is essential for HDV RNA replication (11, 36). In agreement with this hypothesis, exogenous expression of YY1 or B23 in an HDV RNA replication system enhanced HDV RNA replication (Fig. 5) (31). Moreover, introducing expression constructs of CBP, p300, or nucleolin in the HDV RNA replication system also enhanced HDV RNA replication (Fig. 6) (38). CBP/p300 and p300/CBP-associated factor affect activities of various cellular or viral factors by acetylation via their intrinsic acetyltransferase activity (7, 18, 20). Notably, several viral proteins also form complex with CBP/p300 and p300/CBP-associated factor, such as human immunodeficiency virus type 1 Tat, adenovirus E1A, and papillomavirus E2 proteins (7). These reports and findings suggest that through physical interactions between SHDAg and YY1, CBP, p300, B23, and nucleolin, these proteins and possibly other unknown cellular factors may form a high-molecular-weight nuclear complex, which functions as an activating complex to enhance the HDV RNA replication.
Various posttranslational modifications occur on HDAg, and these modifications affect the function of HDAg (29). Acetylation of SHDAg has been demonstrated to modulate the replication of HDV RNA. Thus, in addition to serving as transcriptional coactivators in the large nuclear SHDAg-containing complex, CBP/p300, through recruitment by SHDAg and YY1, may modulate functions of SHDAg on HDV replication through acetylation. This hypothesis is further supported by the experimental results that SHDAg could be acetylated and that the addition of TSA promotes not only the acetylation of SHDAg but also the enhancement of HDV replication (Fig. 7C and D). These findings are consistent with the results of an earlier study (47). Mu and colleagues had determined that SHDAg could interact with p300, and the lysine 72 of SHDAg was acetylated in vitro by p300. Our results further showed that both LHDAg and SHDAg associated with CBP or p300 in the large nuclear complex in vivo (Fig. 4B) and that the exogenous expression of p300 or CBP in the HDV RNA replication system enhanced HDV RNA replication (Fig. 6). These findings suggested that CBP may be also involved in HDAg acetylation. Mu and colleagues also demonstrated that LHDAg was acetylated in vivo. We did not detect acetylated LHDAg in SVLD3-N1 cells. This may be because the level of LHDAg expression may be too low to observe acetylation (Fig. 7A). Taken together, these findings provided valuable information to explain the molecular mechanisms involved in SHDAg acetylation and the possible role of CBP and p300 in the process of HDV replication. However, we could not exclude the possibility that, in addition to the acetylation of HDAg, overexpression of CBP and p300 or treatment with TSA may also activate some cellular factors contributing to the enhancement of HDV RNA replication. Along these lines, CBP/p300 also functions as a scaffold, recruiting other cellular factors in a transcription complex to activate the transcription of cellular genes (44). In this study, since the acetyltransferase-defective mutant of p300 (D1399Y) lost its ability to enhance HDV replication (Fig. 6D), the acetyltransferase activity of p300 may play a more important role than its possible scaffold function in promoting HDV replication.
Several studies indicate that HDAg has the ability to modulate host gene expression. Both LHDAg and SHDAg possess trans-suppression activity on RNA polymerase II-dependent transcription (42), and LHDAg enhances activity of reporter containing ATF, AP1, or serum response element binding sites (23, 58). Moreover, LHDAg and HBV X protein synergistically activate the serum response element-dependent pathway by activating the transcriptional abilities of Elk1 and serum response factor, respectively (24). Recently, LHDAg was shown to activate the TGF-β signaling pathway and enhance the expression level of TGF-β-induced plasminogen activator inhibitor 1, which may induce liver fibrosis in HDV patients (15). Since YY1, CBP, and p300 are potent cell growth regulators and are involved in various biological processes, such as development, differentiation, cell proliferation, cell cycle regulation, apoptosis, and tumor biology (1, 16, 21, 22, 34), interaction with YY1, CBP, and p300 may contribute to the trans-acting activities of HDAg.
Acknowledgments
We thank L.-J. Juan for the kind provision of plasmids pCMV-CBP-FLAG and pSK-p300-myc.
This work was supported by grants NHRI-GT-EX89B502L, NHRI-GT-EX89B502Ls, NHRI-EX90-9002BL, NHRI-EX91-9002BL, and NHRI-EX96-9501BI from the National Health Research Institute; in part by grants NSC95-2320-B-010-049 from the National Science Council; and by “Aim for the Top University Plan” from the Ministry of Education of the Republic of China to Y.-H. Wu Lee.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Affar, E. B., F. Gay, Y. Shi, H. Liu, M. Huarte, S. Wu, T. Collins, and E. Li. 2006. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 263565-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 2721709-1717. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and E. A. Miska. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 571184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonino, F., B. Hoyer, J. W. Shih, M. Rizzetto, R. H. Purcell, and J. L. Gerin. 1984. Delta hepatitis agent: structural and antigenic properties of the delta-associated particle. Infect. Immun. 431000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branch, A. D., B. J. Benenfeld, B. M. Baroudy, F. V. Wells, J. L. Gerin, and H. D. Robertson. 1989. An ultraviolet-sensitive RNA structural element in a viroid-like domain of the hepatitis delta virus. Science 243649-652. [DOI] [PubMed] [Google Scholar]
- 6.Brazas, R., and D. Ganem. 1996. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science 27490-94. [DOI] [PubMed] [Google Scholar]
- 7.Caron, C., E. Col, and S. Khochbin. 2003. The viral control of cellular acetylation signaling. Bioessays 2558-65. [DOI] [PubMed] [Google Scholar]
- 8.Casey, J. L. 2006. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 30767-89. [DOI] [PubMed] [Google Scholar]
- 9.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 888490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao, M., S. Y. Hsieh, and J. Taylor. 1991. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J. Virol. 654057-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 645066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. W., Y. G. Tsay, H. L. Wu, C. H. Lee, D. S. Chen, and P. J. Chen. 2002. The double-stranded RNA-activated kinase, PKR, can phosphorylate hepatitis D virus small delta antigen at functional serine and threonine residues. J. Biol. Chem. 27733058-33067. [DOI] [PubMed] [Google Scholar]
- 13.Chen, P. J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 838774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, P. J., M. Y. Kuo, M. L. Chen, S. J. Tu, M. N. Chiu, H. L. Wu, H. C. Hsu, and D. S. Chen. 1990. Continuous expression and replication of the hepatitis delta virus genome in Hep G2 hepatoblastoma cells transfected with cloned viral DNA. Proc. Natl. Acad. Sci. USA 875253-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi, S. H., S. H. Jeong, and S. B. Hwang. 2007. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology 132343-357. [DOI] [PubMed] [Google Scholar]
- 16.Donohoe, M. E., X. Zhang, L. McGinnis, J. Biggers, E. Li, and Y. Shi. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 197237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farci, P. 2003. Delta hepatitis: an update. J. Hepatol. 39(Suppl. 1)S212-S219. [DOI] [PubMed] [Google Scholar]
- 18.Fu, M., C. Wang, X. Zhang, and R. G. Pestell. 2004. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem. Pharmacol. 681199-1208. [DOI] [PubMed] [Google Scholar]
- 19.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 173723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 36315-23. [DOI] [PubMed] [Google Scholar]
- 21.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 141553-1577. [PubMed] [Google Scholar]
- 22.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 251125-1142. [DOI] [PubMed] [Google Scholar]
- 23.Goto, T., N. Kato, S. K. Ono-Nita, H. Yoshida, M. Otsuka, Y. Shiratori, and M. Omata. 2000. Large isoform of hepatitis delta antigen activates serum response factor-associated transcription. J. Biol. Chem. 27537311-37316. [DOI] [PubMed] [Google Scholar]
- 24.Goto, T., N. Kato, H. Yoshida, M. Otsuka, M. Moriyama, Y. Shiratori, K. Koike, M. Matsumura, and M. Omata. 2003. Synergistic activation of the serum response element-dependent pathway by hepatitis B virus X protein and large-isoform hepatitis delta antigen. J. Infect. Dis. 187820-828. [DOI] [PubMed] [Google Scholar]
- 25.Greco-Stewart, V. S., P. Miron, A. Abrahem, and M. Pelchat. 2007. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology 35768-78. [DOI] [PubMed] [Google Scholar]
- 26.Gudima, S., S. Y. Wu, C. M. Chiang, G. Moraleda, and J. Taylor. 2000. Origin of hepatitis delta virus mRNA. J. Virol. 747204-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh, S. Y., M. Chao, L. Coates, and J. Taylor. 1990. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol. 643192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, H. M., K. N. Shih, and S. J. Lo. 1996. Disulfide bond formation of the in vitro translated large antigen of hepatitis D virus. J. Virol. Methods 6039-46. [DOI] [PubMed] [Google Scholar]
- 29.Huang, W. H., C. W. Chen, H. L. Wu, and P. J. Chen. 2006. Post-translational modification of delta antigen of hepatitis D virus. Curr. Top. Microbiol. Immunol. 30791-112. [DOI] [PubMed] [Google Scholar]
- 30.Huang, W. H., Y. S. Chen, and P. J. Chen. 2008. Nucleolar targeting of hepatitis delta antigen abolishes its ability to initiate viral antigenomic RNA replication. J. Virol. 82692-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, W. H., B. Y. Yung, W. J. Syu, and Y. H. Wu Lee. 2001. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 27625166-25175. [DOI] [PubMed] [Google Scholar]
- 32.Huang, Z. S., and H. N. Wu. 1998. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 27326455-26461. [DOI] [PubMed] [Google Scholar]
- 33.Inouye, C. J., and E. Seto. 1994. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J. Biol. Chem. 2696506-6510. [PubMed] [Google Scholar]
- 34.Kalkhoven, E. 2004. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 681145-1155. [DOI] [PubMed] [Google Scholar]
- 35.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323558-560. [DOI] [PubMed] [Google Scholar]
- 36.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 631945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, M. M. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 797951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C. H., S. C. Chang, C. J. Chen, and M. F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 2737650-7656. [DOI] [PubMed] [Google Scholar]
- 39.Lee, C. Z., J. H. Lin, M. Chao, K. McKnight, and M. M. Lai. 1993. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 672221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y. M., and S. C. Lee. 1994. Transcriptional activation of the alpha-1 acid glycoprotein gene by YY1 is mediated by its functional interaction with a negative transcription factor. DNA Cell Biol. 131029-1036. [DOI] [PubMed] [Google Scholar]
- 41.Lin, J. H., M. F. Chang, S. C. Baker, S. Govindarajan, and M. M. Lai. 1990. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J. Virol. 644051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo, K., G. T. Sheu, and M. M. Lai. 1998. Inhibition of Cellular RNA polymerase II transcription by delta antigen of hepatitis delta virus. Virology 247178-188. [DOI] [PubMed] [Google Scholar]
- 43.Macnaughton, T. B., and M. M. Lai. 2006. HDV RNA replication: ancient relic or primer? Curr. Top. Microbiol. Immunol. 30725-45. [DOI] [PubMed] [Google Scholar]
- 44.Mai, R. T., T. S. Yeh, C. F. Kao, S. K. Sun, H. H. Huang, and Y. H. Wu Lee. 2006. Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene 25448-462. [DOI] [PubMed] [Google Scholar]
- 45.Makino, S., M. F. Chang, C. K. Shieh, T. Kamahora, D. M. Vannier, S. Govindarajan, and M. M. Lai. 1987. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature 329343-346. [DOI] [PubMed] [Google Scholar]
- 46.Modahl, L. E., and M. M. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 725449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mu, J. J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 31960-70. [DOI] [PubMed] [Google Scholar]
- 48.Rizzetto, M., M. G. Canese, S. Arico, O. Crivelli, C. Trepo, F. Bonino, and G. Verme. 1977. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 18997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. delta Agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 776124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salehi-Ashtiani, K., A. Luptak, A. Litovchick, and J. W. Szostak. 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 3131788-1792. [DOI] [PubMed] [Google Scholar]
- 51.Shih, C. M., S. J. Lo, T. Miyamura, S. Y. Chen, and Y. H. Wu Lee. 1993. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J. Virol. 675823-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor, J. M. 1999. Replication of human hepatitis delta virus: influence of studies on subviral plant pathogens. Adv. Virus Res. 5445-60. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, J. M. 2006. Structure and replication of hepatitis delta virus RNA. Curr. Top. Microbiol. Immunol. 3071-23. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236197-208. [DOI] [PubMed] [Google Scholar]
- 56.Usheva, A., and T. Shenk. 1994. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell 761115-1121. [DOI] [PubMed] [Google Scholar]
- 57.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 323508-514. [DOI] [PubMed] [Google Scholar]
- 58.Wei, Y., and D. Ganem. 1998. Activation of heterologous gene expression by the large isoform of hepatitis delta antigen. J. Virol. 722089-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293124-127. [DOI] [PubMed] [Google Scholar]
- 61.Yang, T. H., W. H. Tsai, Y. M. Lee, H. Y. Lei, M. Y. Lai, D. S. Chen, N. H. Yeh, and S. C. Lee. 1994. Purification and characterization of nucleolin and its identification as a transcription repressor. Mol. Cell. Biol. 146068-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh, T. S., S. J. Lo, P. J. Chen, and Y. H. Wu Lee. 1996. Casein kinase II and protein kinase C modulate hepatitis delta virus RNA replication but not empty viral particle assembly. J. Virol. 706190-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You, L. R., C. M. Chen, T. S. Yeh, T. Y. Tsai, R. T. Mai, C. H. Lin, and Y. H. Wu Lee. 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 732841-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]