Abstract
A three-dimensional (3D) cryoelectron microscopy reconstruction of the prototype Atadenovirus (OAdV [an ovine adenovirus isolate]) showing information at a 10.6-Å resolution (0.5 Fourier shell correlation) was derived by single-particle analysis. This is the first 3D structure solved for any adenovirus that is not a Mastadenovirus, allowing cross-genus comparisons between structures and the assignment of genus-specific capsid proteins. Viable OAdV mutants that lacked the genus-specific LH3 and p32k proteins in purified virions were also generated. Negatively stained 3D reconstructions of these mutants were used to identify the location of protein LH3 and infer that of p32k within the capsid. The key finding was that LH3 is a critical protein that holds the outer capsid of the virus together. In its absence, the outer viral capsid is unstable. LH3 is located in the same position among the hexon subunits as its protein IX equivalent from mastadenoviruses but sits on top of the hexon trimers, forming prominent “knobs” on the virion surface that visually distinguish OAdV from other known AdVs. Electron density was also assigned to hexon and penton subunits and to proteins IIIa and VIII. There was good correspondence between OAdV density and human AdV hexon structures, which also validated the significant differences that were observed between the penton base protein structures.
The Adenoviridae comprise a large family of non-enveloped, icosahedral, generally nonpathogenic viruses with a double-stranded DNA genome (∼26.1 to 45 kb) (9). Because of their wide host range, viruses such as human adenovirus type 5 (AdV5) have been developed as gene delivery vectors and applied in the clinic, especially for cancer, monogenic disorders, and vaccination (http://www.wiley.co.uk/genmed/clinical/). While some of these trials have reached phases II and III, there is still considerable room for improvement in vector technology. In particular, there is strong interest in developing vectors that are targeted to specific tissues for improved therapeutic effect (6, 11), and for this purpose, structural information on the virus particle and its constituent molecules is paramount.
The family Adenoviridae comprises four genera (Mastadenovirus, Aviadenovirus, Siadenovirus, and Atadenovirus). Of these, the best studied are the mastadenoviruses, which include viruses from numerous mammalian species and all known human AdVs. In contrast, members of the avi-, si-, and atadenoviruses have been isolated from birds, although the latter also occur widely in mammalian and reptilian species (9, 38). The prototype Atadenovirus, an ovine isolate (serotype 7; OAdV), is the only member for which any significant biological studies have been undertaken. OAdV uses a different (unknown) receptor from AdV5 and infects, but does not replicate in, human cells (15). It has also been developed as a gene delivery vector (2) and has regulatory approval to enter a phase I clinical trial for prostate cancer in 2008.
Representative genomes from all four genera have now been sequenced. Comparisons reveal a central core of homologous genes that encode replication and structural proteins, while genus-specific genes are located at each end (reviewed in reference 9). The structural proteins of mastadenoviruses were first characterized in detail for prototypes AdV2 and AdV5 (reviewed in references 14 and 28). The hexon protein is the main building block of the capsid and is relatively well conserved in sequence (53% identical) and length (912 versus 968 residues) (23). This suggests that the dimensions of all AdV capsids are likely to be similar. The penton base and fiber proteins that facilitate entry of AdV into a host cell form a complex at the five-fold axis of symmetry at each of the icosahedron's 12 vertices (30). In mastadenoviruses, proteins IIIa, VI, VIII, and IX are the remaining outer capsid proteins. The viral DNA is packaged internally with proteins VII and X (Mu), which are thought to constitute a nucleoprotein core (28). The role of core protein V is less certain, but it was recently shown that it is nonessential for virus viability, although the initial mutants produced were extremely thermolabile. Variants selected for greater thermostability showed mutations in core protein X, suggestive of an interaction between the two proteins (34). One copy of terminal protein is also bound to each end of the genome. Proteins IIIa, VI, VII, VIII, and X are also cleaved by the viral protease. In addition, the 52/55k protein and viral protease are minor constituents whose location in the virion is unknown (7).
Over the last decade or more, fundamental insight into the Mastadenovirus structure has been obtained through the continuous refinement of quasiatomic structures developed from three-dimensional (3D) reconstructions of AdV5 and derivatives, calculated using single-particle image reconstruction (12, 25, 26, 29-31) and the docking of higher-resolution structures of the hexon, penton base, and fiber proteins solved by X-ray crystallography (23, 24, 42, 44). Collectively, this work has culminated in a 6-Å-resolution reconstruction of AdV. Individual α-helices (>10 amino acids in length), suggested by secondary structure predictions and difference mapping, were used to assign electron densities for proteins IIIa, VIII, and IX (26). In essence, proteins IIIa, VIII, and IX cross-link hexon trimers through contacts in the plane of the capsid shell, while protein VI appears to be located within/beneath the hexon cavities (26). The location of protein IX was also identified from a low-resolution cryoelectron microscopy (cryo-EM) reconstruction of a mutant lacking the protein (12) and by tagging its C terminus with green fluorescent protein (GFP) or other epitopes (19).
In the atadenoviruses, proteins V and IX are absent and replaced by the genus-specific proteins p32k and LH3 (1, 13, 36). p32k is cleaved by the viral protease 12 residues from its amino terminus (36). LH3 was earlier thought to be a homologue of the multifunctional, nonstructural E1B 55-kDa protein of mastadenoviruses but was recently identified by peptide analysis as a structural protein (13), although additional roles for LH3 in virus replication are not excluded. Thus, the complement of capsid proteins differs between mast- and atadenoviruses. The latter are also more thermostable (16), suggesting a difference in particle structure.
Here we present a 3D reconstruction of OAdV featuring information to a 10.6-Å resolution (0.5 Fourier shell correlation [FSC] criterion) determined by single-particle analysis and cryo-EM. The importance of this study is that it provides new structural insights for the future targeted engineering of a unique class of gene delivery vectors. In addition, it is the only 3D structure solved for any AdV from a genus other than Mastadenovirus, allowing the first cross-genus comparisons between AdV structures. In addition, we have successfully generated viable mutants that individually lack the LH3 and p32k proteins. Negatively stained 3D reconstructions of these particles have been used to identify LH3 and suggest the possible position of p32k within the capsid. We have also assigned electron density to proteins IIIa and VIII.
MATERIALS AND METHODS
Construction of OAdV LH3 and p32k mutants.
An ∼9.56-kb NaeI/XmaI fragment containing a left-hand portion of the OAdV genome was subcloned from plasmid pOAdV287100 (37) into a Bluescribe plasmid, pBS9.6 (Stratagene, San Diego, CA). The unique NcoI site within the LH3 gene was cut, made blunt ended with T4 DNA polymerase, and religated under standard conditions. The net effect of the mutation (not in the original design) was to replace the N-terminal 19 amino acids with 9 residues of different sequence (Fig. 2). The remainder of the protein was unaffected. The mutated pBS9.6 plasmid was cut with KpnI (partial) and XmaI, and the ∼9.2-kb fragment was subcloned into pOAdV-3 (17) to reconstruct the full-length potentially infectious genome. To remove the p32k reading frame, pBS9.6 was cut with SmaI/PmlI and religated. The new plasmid was digested with BseRI to release an ∼120-bp fragment from within the p32k gene. This was replaced by ligating into the BseRI sites a synthetic double-stranded oligonucleotide that had compatible ends (5′GCTGCTATGGTCTTTGAAATTTTTTCTTCCTTTT and 5′AAGGAAGAAAAAATTTCAAAGACCATAGCAGCTG on the top and bottom strands, respectively). This maintained the likely splice acceptor site on the top strand of OAdV, terminated the p32k reading frame at residue 25 on the bottom strand (codon underlined), and introduced a new PvuII site (in boldface above) to facilitate screening. Plasmid from a positive clone was digested with BsiWI to prepare an ∼3.8-kb fragment. This was subcloned into BsiWI-cut pOAdV-3 to introduce the mutation into the OAdV genome prior to virus rescue.
FIG. 2.
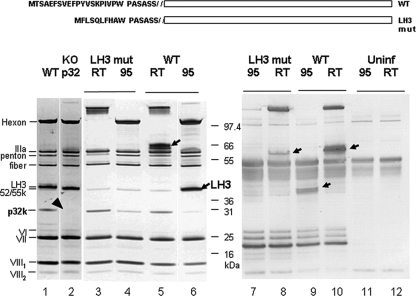
Characterization of LH3 and p32k mutants. (A) Sequence comparison of the WT and LH3 mutant (LH3 mut) in which the amino-terminal 19 amino acids were replaced with 9 residues of different sequence. (B) SDS-PAGE and Western blot analysis. Samples in lanes 1 and 2 were heated at 95°C before loading. The apparent sizes of hexon and LH3 proteins are heat dependent (13). To identify LH3, viral particles were therefore either not heated (room temperature [RT]) or were heated in loading buffer at 95°C (95) for 5 min and analyzed by SDS-PAGE. Lane 1, heated WT virus; lane 2, p32k mutant virus; lanes 3 and 4, LH3 mutant virus; lanes 5 and 6, WT virus. Western blotting was performed with polyclonal rabbit antiserum raised against WT OAdV. Lanes 7 and 8, LH3 mutant virus; lanes 9 and 10, WT virus; lanes 11 and 12, uninfected-cell lysate. Arrows and the arrowhead indicate the key LH3 and p32k proteins, respectively.
Virus rescue, purification, and characterization.
The OAdV used for these cryo-EM studies was structurally wild type (WT) but carried a 6-base insertion that disrupted the open reading frame for the nonstructural gene E4.1. pOAdV-3-based plasmids carrying the appropriately mutated gene were digested with I-SceI to release the linear full-length genomes (17). The DNA was then transfected into permissive ovine cells for virus rescue using cationic lipid CS087 (5). Viruses were propagated, purified by double-CsCl banding, and characterized as described previously (3). The particle/infectious particle ratios were 10 (wild type [WT]), 16, 155 (LH3 mutant), and 170 (p32K mutant). Genome integrity of rescued viruses was confirmed by restriction enzyme digestion and nucleotide sequencing. The phenotype was confirmed by structural protein profiles as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 12% Bis-Tris NuPAGE gels and silver staining (Silver Express; both kits from Invitrogen) according to the manufacturer's instructions.
A polyclonal rabbit antiserum against WT OAdV particles was generated by three intramuscular injections in the presence of Freund's adjuvant (IMVS Veterinary Services Division, Adelaide, South Australia).
Sample preparation and imaging.
The WT OAdV sample (in 10 mM Tris-HCl [pH 8], 8.5% [wt/vol] sucrose, 0.5% polyethylene glycol 400) was concentrated to 60 μl (1.2 × 1012 virus particles [vp]/ml) using a Vivaspin concentrator (500 μl, 30-kDa molecular mass cutoff, and polyethersulfone membrane) at 4°C and 16,200 × g. The sample was then diluted with 600 μl of 10 mM Tris (pH 7.0)-100 mM MgCl2 before concentrating it a second time by the same approach. A final dilution and concentration cycle was employed to ensure efficient buffer exchange (final volume, 20 μl; ∼3.6 × 1012 vp/ml). The washed sample was then applied to freshly glow-discharged (5 mA; vacuum, 5 × 104 Pa; 4 to 7 s in a Bal-Tec glow-discharge unit) Quantifoil grids (hole size, 1.2 μm; spacing, 1.3 μm), before flash freezing in a FEI Vitrobot system (12°C, 100% humidity, with fresh blot pads used every three grids, ensuring optimal blotting and thin ice). High concentrations and minimal glow discharging ensured that sufficient numbers of virus remained suspended in vitreous ice rather than attaching to the carbon. The loaded grids were transferred to a liquid N2-cooled Gatan 686 cryo-holder and imaged to film (Kodak SO-163) at a low electron dose (∼24 e/Å2) in a Tecnai F30 at liquid N2 temperatures (∼−176 to −180°C) set to a calibrated magnification of ×59,000. Defocus settings were in the range of ∼0.4 to 2.0 μm according to real-time Fourier transform measurements taken during imaging with Digital Micrograph and were subsequently confirmed using EMAN′s ctfit program (18).
The LH3 and p32k mutant OAdV preparations (LH3, 1.5 × 1012 vp/ml; p32k, 1.05 × 1012 vp/ml) were washed using an approach similar to that used for the WT OAdV. The LH3-modified OAdV sample was however not concentrated in the final step, while the p32k OAdV sample was (2.1 × 1012 vp/ml, final concentration). Both samples were imaged before and after concentration, ensuring that neither was damaged as a result of the washing and/or concentration steps. For data collection, both samples were then applied to glow-discharged (5 mA; vacuum, 5 × 104 Pa, 60 s) carbon-coated grids (ProSciTech) and stained with uranyl acetate (1%), before being imaged to a charge-coupled device (Gatan 4K Ultrascan, twofold binning) in a Tecnai F30 (300 keV, magnification of ×59,000, −0.06- to 0.6-μm defocus) at room temperature.
Image analysis.
Negatives were digitized with a Nikon super Coolscan (8000ED) using recommended settings (33). The data were then coarsened 2× (Å/pixel = 2.14). Individual particle projections were picked using the cross-correlation mode of SwarmPS (40). For this purpose, a rotationally averaged template was generated from a small subset of manually picked and band-pass-filtered (high pass, 800 Å; low pass, 2 Å) projections, using cenalignint (EMAN).
The particles selected from individual images were checked for astigmatism, drift, and low amplitude values in the power spectrum (indicative of thick ice). The final data set consisted of ∼14,000 projections. Contrast transfer function (CTF) parameters were automatically fitted for projections from each micrograph, using a manually calculated structure factor, the high-frequency amplitudes of which were correctly scaled by merging with the X-ray scattered structure factor amplitudes of GroEl (http://ncmi.bcm.tmc.edu/homes/stevel/EMAN/doc/faq.html, groel.sm) from ∼22 Å onwards using sfmerge (18). The CTF parameters were then checked and manually adjusted prior to phase flipping (ctfit, EMAN).
A reconstruction with information at 10.6 Å (0.5 FSC) (35) was generated using EMAN (18). An initial model was made by further binning projections by a factor of 2 (yielding a pixel size of 4.28 Å and a surface of 256 pixels2) and aligning against a Gaussian sphere of matching diameter. This initial model was then scaled to that of the originally binned projections (512 pixels2) and used in the subsequent reconstruction. During reconstruction, the flexible fibers of OAdV were largely masked out (460 pixel diameter = 984 Å). 3D reconstructions were calculated at increasingly finer angular sampling (10o to 1o), checking FSC plots between iterations to ensure convergence of the structure prior to increasing the sampling. As the map was refined, an increasing number of projections showing poor correspondence to the structure could be automatically excluded, optimizing resolution. Class sums were also checked for agreement with corresponding reprojections. The final 3D reconstruction was calculated using ∼12,000 projections. The handedness of the OAdV structure was validated using the AdV35F structures of Saban (26) and Fabry (12) and the AdV hexon crystal structure (Protein Data Bank [PDB] 1P30) (23).
A similar approach was used to calculate the 3D reconstructions of p32k and LH3 mutant samples, in negative stain. In these cases, the digitized images were binned fourfold (final Å/pixel ratio of 4.087). Data sets of 3,140 (LH3) and 2,800 (p32k) projections were obtained after removal of drift and astigmatic images, yielding 3D reconstructions of 32 Å and 28 Å, respectively (0.5 FSC criterion) (35). The handedness of the p32k reconstruction was determined by comparison with the WT virus.
PsiPred (http://bioinf.cs.ucl.ac.uk/psipred/) was used to generate homology searches indicating probable secondary structure. For modeling purposes, α-helices of approximate lengths were generated using Sybyl (Tripos) according to secondary structure predictions. Imaging of 3D models, the docking of subunit structures from the PDB, and manual fitting of α-helices were done in Chimera (20). The docking of PDB structures was optimized using automated fitting in Chimera (20). Docked crystal structures of AdV2 penton base (PDB 1X9P) (44) and AdV5 hexon (PDB 1P30) (23) corresponded to those used previously (26) and were used here to identify structural differences between AdV and OAdV. PDB files were converted to mrc files for import into Chimera using the program pdb2mrc (18).
RESULTS AND DISCUSSION
Overview of the 3D structure of OAdV.
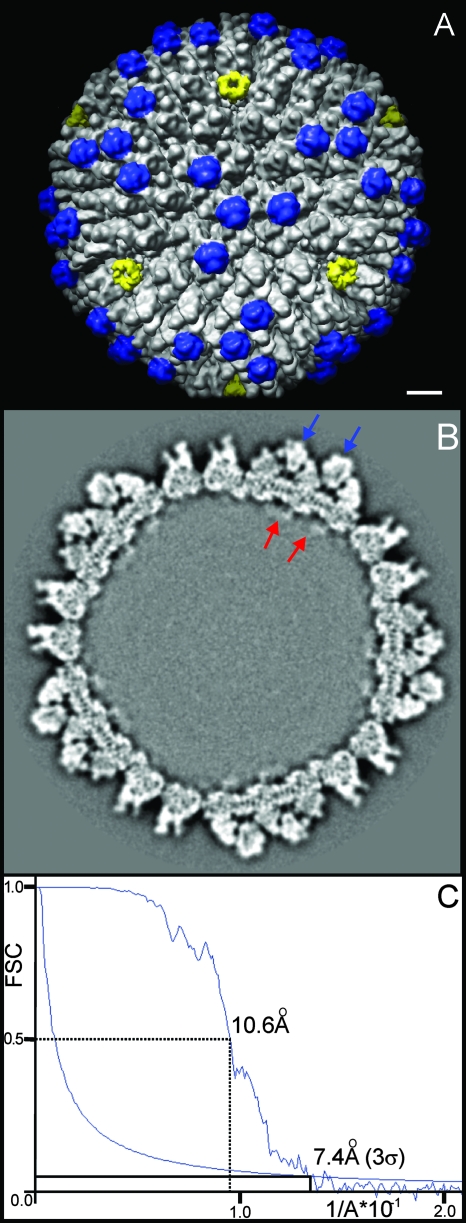
Using cryo-EM and single-particle analysis, the 3D structure of WT OAdV (Fig. 1A) was resolved at a resolution of 10.6 Å according to the FSC 0.5 criterion (Fig. 1C). OAdV is an icosahedral virus having two-, three-, and five-fold symmetry axes with a capsid diameter of ∼94 nm and approximate capsid thickness of 15 nm (Fig. 1B). The 20 facets bounded by penton complexes (Fig. 1A, yellow, at the five-fold axes), each contribute to three complete asymmetric units (see Fig. 4A) of the icosahedral structure. Thus, there are 240 copies of hexon trimer and twelve copies of the pentameric penton base complex as for AdV.
FIG. 1.
Cryo-EM structure of OAdV at 10.6-Å resolution. (A) Capsid surface oriented around one of the three-fold axes. Penton complexes are marked in yellow, indicating the approximate bounds of the icosahedral facet. A major capsid protein and key distinguishing feature of OAdV (identified below as LH3) is marked in blue. Scale bar, 10 nm. (B) Slice taken through the volume of the WT map viewed along a three-fold axis. Blue arrows indicate two LH3 densities, and red arrows are thought to correspond to p32k. Densities assigned to p32k appear to penetrate into the proposed unordered nucleoprotein core of the virus. (C) FSC plot showing peak resolutions of 10.6 Å and 7.4 Å using the 0.5 FSC and 3σ criteria, respectively.
FIG. 4.
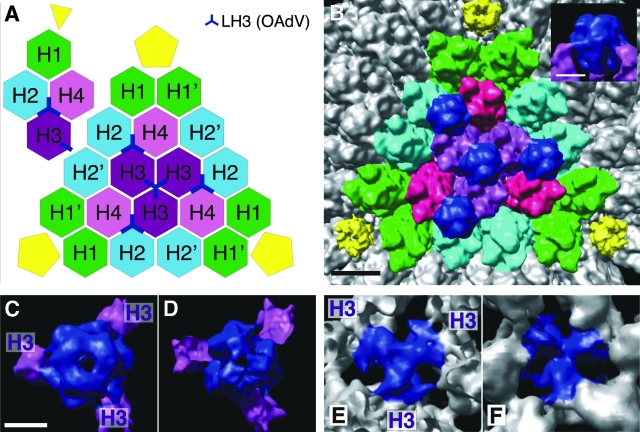
Symmetry of LH3 binding within the icosahedral facet. (A) Schematic of the hexon assignment within the icosahedral facet bounded by pentons (yellow) showing the locations of LH3 (blue) (outside view looking in). (Hexon color scheme is per Saban et al. [26].) The inset (top left) shows a single asymmetric unit of the capsid. (B) View of the icosahedral facet with hexons colored according to the scheme in panel A, indicating hexons bound by LH3 (dark blue). Scale bar, 10 nm. The inset shows that LH3 protrudes well above the hexons at the surface. (C) Top view of a single LH3 trimer as it binds to individual trimers of hexons H3 (purple). Scale bar, 5 nm. (D) Bottom view of LH3 corresponding to panel C. (E) Top view of LH3 where it binds to the bases of hexons H3. (F) Bottom view of LH3 where it binds the hexons H3, looking outwards from inside the capsid.
However, the structure of OAdV differs strikingly from those previously reported for AdV (12, 25, 26, 29-31) in that it possesses very distinctive surface protrusions (“knobs” highlighted in blue in Fig. 1A) that extend further from the surface than any protein (Fig. 1B, blue arrows) apart from the fiber (not shown). It is important to note that the central knob within the facet lies exactly on a three-fold axis (Fig. 1A). Thus, for the icosahedral symmetry of the virus to be preserved, it must be truly trimeric as opposed to a pseudo trimer. This therefore suggests that each knob is constructed from 3 identical protein monomers and that there are a total of 240 monomers on the surface of the virus (4 monomers per asymmetric unit × 60; Fig. 4A). Some density that extends beneath the viral capsid into the interior of the particle is also evident (Fig. 1B, red arrows). This is tentatively assigned as part of p32k as discussed below.
SDS-PAGE analysis of WT OAdV (Fig. 2A, lane 1) confirmed the presence of the hexon, IIIa, penton, fiber, LH3, 52/55k and p32k proteins, as well as subunits VI and VII and cleaved subunits VIII1 and VIII2, consistent with our previous reports (13, 36). Significantly, LH3 and p32k are the only two proteins that are unique to the atadenoviruses. LH3 or p32k was therefore considered most likely to comprise these surface knobs (Fig. 1A, blue).
Confirming the phenotypes of LH3 and p32k mutants.
To test this hypothesis, plasmids in which the LH3 and p32k genes were individually mutated were generated in an attempt to rescue the corresponding LH3- and p32k-deficient viruses. In the LH3 mutant, the amino-terminal 19 amino acids were (somewhat fortuitously) replaced with 9 residues of different sequence (Fig. 2), whereas in the p32k mutant the reading frame was truncated by introducing a stop codon at residue 25. Surprisingly, both mutations yielded viable viruses that grew as well as the WT, but had an LH3- or p32k-minus phenotype. This was confirmed by SDS-PAGE and Western blotting. The p32k mutant clearly lacked the corresponding protein, confirming the knockout phenotype (Fig. 2A, lane 2, arrowhead). Similarly in the LH3 mutant, LH3 (which typically migrates with apparently different sizes before and after heating, probably due to multimer formation; Fig. 2A, lanes 5 and 6), was absent from the mutant virions (lanes 3 and 4). However, infected-cell lysates analyzed by Western blotting showed that the mutant virus produced a version of the LH3 multimer but in smaller amounts than its WT counterpart (Fig. 2A, lanes 8 and 10). The mutant LH3 monomer was undetectable (compare lanes 7 and 9). We therefore conclude that amino-terminal modification of LH3 resulted in reduced levels of LH3 expression, reduced multimer formation, and, importantly, failure to assemble into virus particles (i.e., it was a phenotypic but not genetic knockout).
It is of interest that proteins LH3 and p32k are nonessential because they are Atadenovirus specific (9). Other studies of AdV mutants showed that proteins V and IX (which are specific to the mastadenoviruses) are also nonessential for viability, although virions lacking these proteins showed a greater thermolability (8, 34). Corresponding proteins in the avi- and siadenoviruses have not been identified, but their existence was predicted (13).
The surface knobs of OAdV are LH3 trimers.
To analyze the structure of the LH3 and p32k mutant viruses and to determine whether one or both of these proteins formed the surface knobs, we imaged these viruses by negative-stain electron microscopy. A 3D reconstruction for each was calculated (28-Å and 32-Å resolutions, for p32k and LH3 mutants, respectively, 0.5 FSC criterion; data not shown). The p32k particles retained an intact outer capsid, similar in diameter to the WT, although they were more prone to bending and distortion (data not shown). Nevertheless, the surface knobs identified as LH3 were clearly visible in the reconstruction (Fig. 3A, blue) indicating that p32k is not part of the surface knobs and strongly suggesting that they consist of LH3 alone. The fact that LH3 (382 residues) is more than twice the size of protein IX (140 residues) is also consistent with the extra density of the prominent surface knobs (Fig. 1A).
FIG. 3.
Structural analysis of p32k and LH3 mutants. (A) Surface view of p32k knockout virus along the three-fold axis at ∼28-Å resolution (0.5 FSC criterion; data not shown) showing the presence of the “knob-like” densities corresponding to LH3 (blue). The diameter of this particle is ∼94 nm. Scale bar, 15 nm. (B) Negative-stain images of LH3 mutant viruses. More than 95% of the viruses dissociated, as revealed by the smaller size of the remaining particles (diameter of ∼85 nm; blue arrow) and the many hexon subunits (white arrow) and fiber proteins bound to the penton complexes (yellow arrow) that are clearly seen in the background. Scale bar, 80 nm. The few intact viruses (red arrow) are larger (94 nm diameter) and have radially protruding fibers (two of which are indicated by black arrows). (C) High-magnification image of individually shed fiber/penton complexes (black arrow). What appears to be the attached penton complex and fiber head domain have been marked (red and yellow arrows, respectively). Scale bar, 18 nm.
The capsid of the LH3 mutant (Fig. 3B) was clearly far less stable than that of the WT. In fact there were very few intact virus particles (Fig. 3B, red arrow) and dissociated protein subunits littered the grid (yellow and white arrows). The intact nature of the virus marked by the red arrow is confirmed both by its larger size (∼94 nm) and the fact that fibers clearly radiate from its surface (e.g., black arrows). In contrast, the majority of particles (blue arrow) appeared to be smaller in size, with an estimated diameter of ∼85 nm. It is not possible to say with complete certainty at this stage whether the complete outer capsid was uniformly lost from these particles, but no knob structures were evident in the preliminary 3D reconstruction (data not shown). Similar types of particles were observed with WT “egg drop syndrome” virus, an avian atadenovirus (32), suggesting that capsid fragility may be a feature of this genus. These observations complement those obtained with the p32k mutant and suggest that LH3 plays an important structural role in capsid stabilization.
From Fig. 3C, the length of the OAdV fiber can be estimated. Measurement of what appears to be the penton/fiber assembly (red/black arrows), at ∼38 nm, is consistent with an OAdV fiber that has 25 repeats (at 15 Å per repeat) and a smaller head domain (yellow arrow) (22, 36). These measurements are consistent with the size of the AdV2 fiber shaft with 22 repeating units, which is estimated at ∼31 nm, including the 44 residues of ∼4 nm in length at the amino terminus, yielding a total estimated length of ∼35 nm (41).
Structure and function of LH3.
Having identified the prominent surface knobs of OAdV, the remaining major structural components of the capsid were assigned. Figure 4A schematically represents the structural organization of proteins within the facet bounded by the three pentons shown in Fig. 4B. Hexons H1, H1′, H2, H2′, H3, and H4 are the major constituents of the capsid (Fig. 4A) and have been named according to the original nomenclature (4). Hexon trimers H2, H3, and H4 belong to the so-called “group of nine” that preferentially dissociate together (21). In OAdV, which lacks protein IX, LH3 occupies the equivalent position between the hexon subunits (blue Y symbols in Fig. 4A).
The LH3 densities that protrude from the surface of the viral capsid (Fig. 4B, inset) extend down through the arrayed hexon trimers (Fig. 4C, viewed toward the center of the virus; and D, viewed outwards from inside the capsid). Figures 4E and 4F show the close structural interactions of the LH3 trimer with the bases of H3 trimers that surround the three-fold axis (highlighted in purple in Fig. 4A and B). LH3 (382 residues) is more than twice the size of protein IX (140 residues). The additional 242 residues of LH3 are therefore predicted to make up the bulk of the prominent surface protrusions. This also explains the absence of these knobs in AdV. However, GFP can be attached to the C terminus of protein IX without loss of virus viability (19), indicating that additional protein can also be accommodated on the AdV capsid structure. The three other LH3 trimers (blue symbols in Fig. 4A) lie between H2, H3, and H4 hexon trimers, which create a niche equivalent to that between the H3 trimers (4). This suggests that LH3 does indeed act as the “glue” that holds these capsid units together, in a manner similar to that reported for protein IX.
The three-pronged star shape of the blue LH3 density (Fig. 4E), as well as its close interaction with the three adjacent hexons, is highly reminiscent of the organization of the N terminus of protein IX (residues 1 to 56) in human AdV (26) and is consistent with the putative common roles of the two proteins in AdV and OAdV. The higher-resolution reconstruction of AdV also identified a set of four helix bundles, each having a coiled-coil arrangement. These coiled-coil domains were located toward the edge of the icosahedral facet (26) and interpreted as consisting of the C-termini (residues 92 to 131) of four protein IX subunits (three from the same facet and one from a neighboring facet). Saban et al. (26) also detected density linking the resolved N and C termini and demonstrated that residues 57 to 91 of protein IX could be sensibly docked into it. In our 10.6-Å model, we did not resolve such a four-helix cluster, although rod-like structures likely corresponding to α-helices were resolved in other parts of the model. This is consistent with the different sequences and predicted secondary structures of proteins IX and LH3 by Psipred analysis that indicated very little if any α-helical structure in LH3 (in contrast to the high helical content of pIX; data not shown).
Identification of protein VIII in OAdV.
In contrast, there is significant homology between OAdV and AdV pVIII precursors. As shown by sequence comparison and Psipred analysis, these proteins exhibit a high degree of similarity, especially at the amino terminus, and two protease cleavage sites are also conserved (Fig. 5). In AdV, protein VIII was assigned to two elongated regions of density (per asymmetric subunit) that were located on the inner surface of the capsid (12). This assignment was subsequently supported by other work which identified pVIII based on electron density and α-helix assignment (26). One of the protein VIII subunits interacted with hexons H2′, H3, and H4, and the other interacted with hexons H1′, H1, H2, and H4. However, the density nearer to the penton complex at the H1′, H1, H2, and H4 interface was partly obscured by an α-helix-rich structure that was assigned as protein IIIa once difference maps for proteins VIII and IIIa were calculated (26). Segmentation and alignment of these two protein VIII densities showed that they were virtually identical at ∼6-Å resolution. Having established the structural similarity of the two copies of protein VIII, Saban et al. (26) went on to show that a 24-residue helix predicted for a 12.1-kDa fragment of protein VIII could be docked into its electron density. Figure 6A shows a schematic of hexon assignment in the icosahedral facet, viewed from the inside, which is based on the structure reported by Saban and the OAdV data presented here (Fig. 6B). Thus, in OAdV, protein VIII was identified by its location on the inner surface of the capsid (Fig. 6B, red densities), the structural similarity with its AdV counterpart, and its copy number (2 per asymmetric unit, 120 copies per virion). Thus, protein VIII appears to stabilize the hexon subunit interactions within the capsid from below, in a manner similar to that described for AdV.
FIG. 5.
Comparison of OAdV and AdV protein VIII and penton base sequences using a BLAST search. Identity and homology between sequences are indicated by double and single dots, respectively. Black bars indicate significant α-helices, as predicted by Psipred. Black arrows indicate viral protease cleavage sites.
FIG. 6.
Density assignments for proteins VIII, IIIa, and p32k. (A) Schematic showing hexon arrangement from the inside of the icosahedral facet. The probable location and orientation of OAdV protein VIII is indicated (red marker). (B) Inside view of the icosahedral facet showing positions of protein VIII (red) relative to respective hexons (lettered according to schematic in panel A) and six copies of the density tentatively assigned to p32k (orange). Scale bar, 5 nm. The inset shows increased-density rendering of three copies of p32k with α-helices tentatively docked within one copy (green). (C) The interlocking motif formed by individual copies of pIIIa is shown, where two of five copies have been marked in purple. The red density assigned to protein VIII appears to be in close contact (yellow) with pIIIa and pVIII, and the densities appear to merge. α-Helices (green) as predicted by PsiPred analysis are tentatively docked into rod-like densities of pVIII and pIIIa. Scale bar, 5 nm.
Possible identity of p32k on the inside of the capsid.
Given the location and role of LH3 protein (see above), p32k must be located elsewhere in the particle and may functionally replace protein V of AdV, although the location of protein V in that particle is unknown. Previous studies have shown an increase in capsid flexibility upon deletion of protein V in AdV (34). This was also the case for the p32k mutant, which also exhibited less capsid rigidity and was more prone to distortion (data not shown). Separate densities observed in the interior of the particle (Fig. 1B) and bound to the inside of the capsid between the bases of hexons H2, H3, and H4 and between the hexon H3 triad within the viral facet (Fig. 6B, orange) do not appear to be structurally associated with any of the other capsid proteins at the current resolution of the 3D reconstruction. Importantly, these are not stoichiometrically consistent with the density assigned to LH3 protein. We therefore conclude that the density in question is a candidate to be a part of the p32k protein. The densities are present in two copies per asymmetric unit, yielding a total of 120 per virion. Reducing the density (view not shown) facilitated the tentative docking of two α-helices of ∼12 to 16 residues in length (Fig. 6B, inset). However, there is not enough density resolved in Fig. 6B to account for the complete structure of p32K (274 residues). The C-terminal portion of the protein may be currently unresolved within the outer capsid layer. Psipred analysis suggests that the C-terminal half has abundant α-helical structure, including a single long helix of at least 24 residues (although there is no propensity for coiled-coil formation such as that which interlinks protein IX subunits in AdV (26). The amino-terminal ∼25 residues of mature p32k are very Lys/Arg-rich and could perhaps serve as an anchor point for genomic DNA in the core. Thus, we tentatively assign the densities at the interface between the capsid and the nucleoprotein core to part of p32k and suggest that this protein may form a link between the two (Fig. 1B, red arrows), with the amino terminus and its strong positive charge likely to be oriented toward the viral DNA.
Location of protein IIIa.
To assist in the identification of OAdV protein IIIa, the OAdV structure was carefully compared with the AdV structure shown by Saban et al. (26). AdV protein IIIa reportedly has a very high α-helical content (∼50%). Indeed Psipred analyses predicted that the N-terminal two-thirds of AdV IIIa contains at least 14 α-helices with a further 2 helices located in the C-terminal one-third of the protein. Saban et al. (26) resolved 12 pentameric rings on the inner surface of the capsid, below each of the 12 pentons. Each of these pentameric rings consisted of five individual clusters of at least 13 helices, resulting in each being identified as individual copies of protein IIIa. This conclusion was further supported by mass spectrometry data which concluded that 60 copies of protein IIIa were present in each virion (i.e., 12 pentamers), one subunit per asymmetric unit. Arguments were also presented to exclude proteins VI and VIII from these densities.
In OAdV, 12 pentameric ring structures were also identified below each of the 12 pentons. Consequently, we have assigned the five interlocking densities in each of these rings to five copies of pIIIa (Fig. 6C, purple). This assignment is consistent with the location of pIIIa in AdV, the reported copy number of one subunit per asymmetric unit (26), and the high degree of structural similarity predicted by Psipred for protein IIIa from AdV and OAdV (data not shown). However, because of lower resolution of the OAdV 3D reconstruction, the full complement of predicted α-helices has not been assigned. The assigned density is also insufficient to account for all of protein IIIa (568 residues before cleavage). pIIIa may also be in close contact with a subunit of protein VIII (Fig. 6C, red; apparent point of contact in yellow). Alternatively, this density may correspond to more of protein IIIa with protein VIII being obscured as in AdV (26). Difference mapping will facilitate the more precise assignment and separation of minor capsid protein densities. However, given the 10.6-Å resolution and the fact that no crystal structure has been determined for any OAdV protein, accurate difference mapping is not feasible at this stage.
Penton structure and crystal docking.
Figure 7A shows the OAdV penton complex in situ (density shaded in yellow). The first noticeable difference between AdV and OAdV is that the penton complex is not as deeply embedded in the capsid. At the resolution achieved, the complex does not appear to be held in place by protein IIIa (as is the case in AdV) or the hexons. However, it has been observed with the LH3 mutant described here, that the penton/fiber complex may be one of the more labile components of the capsid. If penton complexes disassociated from some positions, this might contribute to an apparent reduction in electron density.
FIG. 7.
OAdV penton structure. (A) View of the penton complex in situ showing the hollow cavity that runs through its central axis. Scale, bar 5 nm. (B) Penton base crystal structure (with fiber peptide in place [PDB 1X9T]; green) (43) was docked within the OAdV penton density (gray) by aligning pentagonal vertices (red arrow). The flexible Arg/Gly/Asp loop in AdV, which is absent in OAdV, emerges immediately counterclockwise to the projecting density in OAdV (yellow arrow). AdV α-helices that that do not appear to have a corresponding density in OAdV are also indicated (blue arrow). (C) Penton density and docked AdV ribbon structure viewed from below. The white arrow signifies the groove between OAdV monomer densities that is occupied by AdV sequences. (D) Docking of a single AdV penton-base monomer in the OAdV penton density showing the misalignment near the base of the complex (red arrow) and the emergence of the Arg/Gly/Asp loop in AdV, which is truncated at top right.
Homology (BLAST) searches and Psipred analyses of the OAdV and AdV penton base sequences (Fig. 5) indicate a high degree of sequence and predicted structural similarity. AdV has an ∼40-residue amino-terminal extension. Thereafter, the predicted structure of residues 40 to 287 and 390 to 571 of the AdV penton base is similar to that of OAdV through residues 3 to 249 and 268 to 452, respectively. The major difference between the two occurs in the tropism-defining residue 288-to-389 region of AdV. This consists of an α-helical domain and the Arg/Gly/Asp motif that interacts with certain cell surface integrins during virus uptake (39). The motif is missing from OAdV penton, and no other integrin-binding motifs have been identified (36).
The AdV penton complex structure (PDB 1X9T) (44), solved in the presence of fiber peptide) was docked into the OAdV density (Fig. 7B). The prominent protein loop corresponding to AdV residues 142 to 169, which is structurally conserved (Psipred analysis) (Fig. 5) in the OAdV penton base residues 106 to 131 but is four residues shorter, was aligned with each vertex in the overall pentagonal-shaped density of OAdV (Fig. 7B), as was also done recently in a study of CAdV-2 (27). It is immediately apparent that five α-helices around the central axis of the AdV penton base complex (blue arrow) do not fit into any corresponding density in OAdV (Fig. 7B). This suggests that a larger cavity may run through the center of the OAdV penton complex. Furthermore, with this alignment the flexible loop containing the AdV Arg/Gly/Asp motif (which is truncated in the ribbon structure) protrudes from the outer surface of the penton complex immediately adjacent to the prominent external protrusion of OAdV density (Fig. 7B, yellow arrow) that does not coincide with AdV sequence. The identity and role of this region in OAdV penton base are not yet known. While this region is a candidate to contain sequences that may influence OAdV tropism, there is currently no evidence for the involvement of a secondary receptor in OAdV uptake. Indeed, changing only the cell binding domain on the fiber protein was sufficient to alter OAdV tropism (43).
Figure 7C shows a view of the docked AdV penton structure from below. In this view, the larger cavity running through the center of the OAdV penton complex is clearly apparent. Density spikes protruding from the sides of each monomer (adjacent to white arrow in Fig. 7C) are also apparent, but the role and identity of the OAdV sequences involved remain to be determined. These appear to have no counterpart in the AdV or CAdV complexes (26, 27). In addition, there is a groove between the OAdV monomers at the base of the complex which is occupied by AdV sequences in the composite image (Fig. 7C, white arrow). The side view (Fig. 7D) further illustrates the significant misalignment between the AdV and OAdV structures (Fig. 7D, red arrow), notwithstanding their apparent similarity according to Psipred predictions. Despite this, 7 of 10 key penton base residues and most of the critical residues at the amino terminus of the OAdV fiber protein involved in penton-fiber interaction are conserved in OAdV, confirming their importance (44). In addition, the presence of fibers shed with what appears to be the penton complex bound at one end (Fig. 3C, black and red arrows, respectively) suggests a tight association between the two proteins.
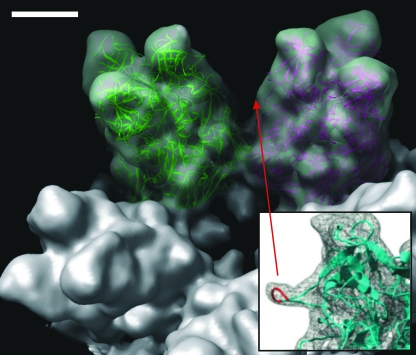
Hexon docking.
Homology searches of AdV and OAdV hexon sequences reveal gross similarity between the molecules, although differences in the known hypervariable regions (23) that separate them as distinct serotypes, are expected. Figure 8A shows hexon coordinates (PDB 1P30) docked into adjacent OAdV H1 trimers. Saban et al. highlighted a protein loop protruding from the side of each hexon monomer (Fig. 8, inset) (26) that corresponds to six residues in the HVR4 region (251 to 256 of AdV). Due to their flexibility and disorder, these were not reflected in the crystal coordinates of the hexon structure (23). Hence, Saban et al. modeled the HVR4 loop into the hexon base coordinates (Fig. 8, inset, red) using the CODA (10) protein loop prediction server (26). However, at a 10.6-Å resolution this density is not apparent in OAdV hexon. Nevertheless, the excellent fit between the AdV hexon ribbon structure and the OAdV electron density confirms the accuracy of the OAdV cryo-EM structure determined here and validates the differences that have been described for other proteins.
FIG. 8.
Docking of the AdV hexon crystal structure into OAdV. Two copies of AdV hexon crystal structure (PDB 1P30) (23) docked within adjacent H1 hexons of OAdV. Density corresponding to the HVR4 region (26) (inset) is not apparent at this resolution. Density corresponding to the penton complex has been removed. Scale bar, 5 nm.
Conclusion.
In conclusion, this study provides the first 3D structure of an adenovirus that is not a Mastadenovirus. It is evident that Atadenovirus particles (and derived gene delivery vectors) have structural features that are significantly different from those of the mastadenoviruses. The generation of two mutants allowed the identification of the LH3 protein and the tentative location of p32k protein within the capsid and showed that each was nonessential for virus assembly. Future cryo-EM of the LH3 mutant should provide greater structural detail of the inner capsid layer and permit difference mapping with the cryo-EM structure of WT virus determined here. Greater detail will also be obtained by sampling these data at a finer Å/pixel ratio and by the elucidation of OAdV protein crystal structures to facilitate the accurate difference mapping of minor capsid protein densities.
Acknowledgments
We thank Zheng-Zhou Xu for assistance with construction of the LH3 mutant.
We thank the Australian Research Council (DP0452362) and SRC for financial support.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Both, G. 2002. Identification of a unique family of F-box proteins in atadenoviruses. Virology 304425-433. [DOI] [PubMed] [Google Scholar]
- 2.Both, G. 2004. Ovine atadenovirus: a review of its biology, biosafety profile and application as a gene delivery vector. Immunol. Cell Biol. 82189-195. [DOI] [PubMed] [Google Scholar]
- 3.Both, G., F. Cameron, A. Collins, L. Lockett, and J. Shaw. 2007. Production and release testing of ovine atadenovirus vectors. Methods Mol. Med. 13069-90. [DOI] [PubMed] [Google Scholar]
- 4.Burnett, R. M. 1985. The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185125-143. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, F. H., M. J. Moghaddam, V. J. Bender, R. G. Whittaker, M. Mott, and T. J. Lockett. 1999. A transfection compound series based on a versatile Tris linkage. Biochim. Biophys. Acta 141737-50. [DOI] [PubMed] [Google Scholar]
- 6.Campos, S., and M. Barry. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr. Gene Ther. 7189-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelius, D., A. F. R. Huhmer, C. H. Shieh, E. Lehmberg, J. A. Traina, T. K. Slattery, and E. Pungor. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J. Proteome Res. 1501-513. [DOI] [PubMed] [Google Scholar]
- 8.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, A., M. Benko, and B. Harrach. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 842895-2908. [DOI] [PubMed] [Google Scholar]
- 10.Deane, C. M., and T. L. Blundell. 2001. CODA: a combined algorithm for predicting the structurally variable regions of protein models. Protein Sci. 10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas, J. 2007. Adenoviral vectors for gene therapy. Mol. Biotechnol. 3671-80. [DOI] [PubMed] [Google Scholar]
- 12.Fabry, C. M. S., M. Rosa-Calatrava, J. F. Conway, C. Zubieta, S. Cusack, R. W. H. Ruigrok, and G. Schoehn. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 241645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman, J. J., T. P. Wallis, D. A. Whelan, J. Shaw, and G. W. Both. 2005. LH3, a “homologue” of the mastadenoviral E1B 55-kDa protein is a structural protein of atadenoviruses. Virology 342159-166. [DOI] [PubMed] [Google Scholar]
- 14.Horwitz, M. S. 1990. Adenoviridae and their replication, p. 1679-1721. In B. N. Fields and D. M. Knipe (ed.), Virology, 2nd ed. Raven Press, New York, NY.
- 15.Khatri, A., Z. Z. Xu, and G. W. Both. 1997. Gene expression by atypical recombinant ovine adenovirus vectors during abortive infection of human and animal cells in vitro. Virology 239226-237. [DOI] [PubMed] [Google Scholar]
- 16.Löser, P., G. Cichon, G. Jennings, G. Both, and C. Hofmann. 1999. Ovine adenovirus vectors promote efficient gene delivery in vivo. Gene Ther. Mol. Biol. 433-43. [Google Scholar]
- 17.Löser, P., C. Hofmann, G. W. Both, W. Uckert, and M. Hillgenberg. 2003. Construction, rescue, and characterization of vectors derived from ovine atadenovirus. J. Virol. 7711941-11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semi-automated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 19.Marsh, M. P., S. K. Campos, M. L. Baker, C. Y. Chen, W. Chiu, and M. A. Barry. 2006. Cryoelectron microscopy of protein IX-modified adenoviruses suggests a new position for the C terminus of protein IX. J. Virol. 8011881-11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Computational Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 21.Prage, L., U. Pettersson, S. Hoglund, K. Lonberg-Holm, and L. Philipson. 1970. Structural proteins of adenoviruses. IV. Sequential degradation of the adenovirus type 2 virion. Virology 42341-358. [DOI] [PubMed] [Google Scholar]
- 22.Ruigrok, R. W. H., A. Barge, C. Albiges-Rizo, and S. Dayan. 1990. Structure of adenovirus fibre. II. Morphology of single fibres. J. Mol. Biol. 215589-596. [DOI] [PubMed] [Google Scholar]
- 23.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 779553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rux, J. J., D. Pascolini, and R. M. Burnett. 1999. Large-scale purification and crystallization of adenovirus hexon, p. 259-275. In W. S. M. Wold (ed.), Adenovirus methods and protocols, vol. 21. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 25.Saban, S. D., R. R. Nepomuceno, L. D. Gritton, G. R. Nemerow, and P. L. Stewart. 2005. CryoEM structure at 9 angstrom resolution of an adenovirus vector targeted to hematopoietic cells. J. Mol. Biol. 349526-537. [DOI] [PubMed] [Google Scholar]
- 26.Saban, S. D., M. Silvestry, G. R. Nemerow, and P. L. Stewart. 2006. Visualization of α-helices in a 6-Å resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 8012049-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoehn, G., M. El Bakkouri, C. M. S. Fabry, O. Billet, L. F. Estrozi, L. Le, D. T. Curiel, A. V. Kajava, R. W. H. Ruigrok, and E. J. Kremer. 2008. Three-dimensional structure of canine adenovirus serotype 2 capsid. J. Virol. 823192-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 29.Stewart, P., S. Fuller, and R. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 122589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart, P. L., R. M. Burnett, M. Cyrklaff, and S. D. Fuller. 1991. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67145-154. [DOI] [PubMed] [Google Scholar]
- 31.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. M. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 161189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd, D., and M. S. McNulty. 1978. Biochemical studies on a virus associated with Egg Drop Syndrome 1976. J. Gen. Virol. 4063-75. [Google Scholar]
- 33.Typke, D., R. A. Nordmeyer, A. Jones, J. Lee, A. Avila-Sakar, K. H. Downing, and R. M. Glaeser. 2005. High-throughput film-densitometry: an efficient approach to generate large data sets. J. Struct. Biol. 14917-29. [DOI] [PubMed] [Google Scholar]
- 34.Ugai, H., A. V. Borovjagin, L. P. Le, M. Wang, and D. T. Curiel. 2007. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J. Mol. Biol. 3661142-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel, M., and M. Schatz. 2005. Fourier shell correlation threshold criteria. J. Struct. Biol. 151250-262. [DOI] [PubMed] [Google Scholar]
- 36.Vrati, S., D. E. Brookes, P. Strike, A. Khatri, D. B. Boyle, and G. W. Both. 1996. Unique genome arrangement of an ovine adenovirus: identification of new proteins and proteinase cleavage sites. Virology 220186-199. [DOI] [PubMed] [Google Scholar]
- 37.Vrati, S., E. S. MacAvoy, Z. Z. Xu, C. Smole, D. B. Boyle, and G. W. Both. 1996. Construction and transfection of ovine adenovirus genomic clones to rescue modified viruses. Virology 220200-203. [DOI] [PubMed] [Google Scholar]
- 38.Wellehan, J. F. X., A. J. Johnson, B. Harrach, M. Benkö, A. P. Pessier, C. M. Johnson, M. M. Garner, A. Childress, and E. R. Jacobson. 2004. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 7813366-13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrin-alpha-v-beta-3 and integrin-alpha-v-beta-5 promote adenovirus internalization but not virus attachment. Cell 73309-319. [DOI] [PubMed] [Google Scholar]
- 40.Woolford, D., G. Ericksson, R. Rothnagel, D. Muller, M. J. Landsberg, R. S. Pantelic, A. McDowall, B. Pailthorpe, P. R. Young, B. Hankamer, and J. Banks. 2007. Swarm (PS): rapid, semi-automated single particle selection software. J. Struct. Biol. 157174-188. [DOI] [PubMed] [Google Scholar]
- 41.Wu, E., L. Pache, D. J. Von Seggern, T.-M. Mullen, Y. Mikyas, P. L. Stewart, and G. R. Nemerow. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 777225-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 angstrom resolution. Structure 21259-1270. [DOI] [PubMed] [Google Scholar]
- 43.Xu, Z. Z., and G. W. Both. 1998. Altered tropism of an ovine adenovirus carrying the fiber protein cell binding domain of human adenovirus type 5. Virology 248156-163. [DOI] [PubMed] [Google Scholar]
- 44.Zubieta, C., G. Schoehn, J. Chroboczek, and S. Cusack. 2005. The structure of the human adenovirus 2 penton. Mol. Cell 17121-135. [DOI] [PubMed] [Google Scholar]