Abstract
Bovine herpesvirus type 1 (BHV-1) is an important component of the bovine respiratory disease complex (BRDC) in cattle. The ability of BHV-1 to transport anterogradely from neuronal cell bodies in trigeminal ganglia (TG) to nerve ending in the noses and corneas of infected cattle following reactivation from latency plays a significant role in the pathogenesis of BRDC and maintenance of BHV-1 in the cattle population. We have constructed a BHV-1 bacterial artificial chromosome (BAC) clone by inserting an excisable BAC plasmid sequence in the long intergenic region between the glycoprotein B (gB) and UL26 genes. A BAC-excised, reconstituted BHV-1 containing only the 34-bp loxP sequence within the gB-UL26 intergenic region was highly infectious in calves, retained wild-type virulence properties, and reactivated from latency following treatment with dexamethasone. Using a two-step Red-mediated mutagenesis system in Escherichia coli, we constructed a gE cytoplasmic tail-truncated BHV-1 and a gE-rescued BHV-1. Following primary infection, the gE cytoplasmic tail-truncated virus was efficiently transported retrogradely from the nerve endings in the nose and eye to cell bodies in the TG of calves and rabbits. However, following dexamethasone-induced reactivation from latency, the gE mutant virus was not isolated from nasal and ocular sheddings. Reverse transcriptase PCR assays detected VP5 transcription in the TG of rabbits infected with gE-rescued and gE cytoplasmic tail-truncated viruses during primary infection and after dexamethasone treatment but not during latency. Therefore, the BHV-1gE cytoplasmic tail-truncated virus reactivated in the TG; however, it had defective anterograde transport from TG to nose and eye in calves and rabbits.
Bovine herpesvirus type 1 (BHV-1) is an alphaherpesvirus that causes abortion, respiratory, and genital infections in cattle (29, 38), but usually does not cause encephalitis. BHV-5 is another bovine alphaherpesvirus, which causes fatal encephalitis in calves (7, 13). Following primary infection of respiratory and ocular epithelia, both BHV-1 and BHV-5 establish latency in trigeminal ganglia (TG) (6, 26). BHV-1 replicates in the nasal and ocular epithelia during primary infection. It is believed that capsids together with tegument then enter sensory nerve endings of the ophthalmic and maxillary branches of the trigeminal nerve located in the nasopharynx and eye. These virus particles are transported retrogradely to cell bodies in TG, where the virus establishes a lifelong latent infection (17). The episomal form of viral genomic DNA, transcripts originating from the latency-related gene, and proteins encoded by the latency-related gene can be detected in TG during latency (16, 17, 26). However, there is no evidence of virus DNA replication and productive infection during latency (15, 17). Periodic reactivation from BHV-1 latency usually results in nasal and ocular virus shedding. In this case, infectious virus particles are transported anterogradely to axon termini in the nasopharynx and eye, where they infect epithelial cells, resulting in virus replication and shedding (8, 16, 17).
Several reports (19, 20) indicate that BHV-1 glycoprotein E (gE)-deleted viruses fail to reactivate from latency and are not shed in the nose and/or eye secretions following dexamethasone-induced reactivation. Results from studies in our laboratory suggested that gE was required for anterograde transport of the virus from TG to nerve endings in the respiratory and ocular epithelia (S. I. Chowdhury, unpublished data). In this study, we investigated the significance of the BHV-1gE cytoplasmic tail in the anterograde transport of BHV-1 from TG to respiratory and ocular epithelia. We constructed a BHV-1gE cytoplasmic tail-truncated virus by using a BHV-1 bacterial artificial chromosome (BAC) clone. Two BHV-1 BAC clones have been developed recently (23, 34). However, in both cases, the BAC plasmid was inserted in place of nonessential genes, replacing in one case the thymidine kinase gene (23) and in other case the gE and gG genes (34). Therefore, it was imperative to construct a BHV-1 BAC clone in which no viral genes are deleted or affected. We have constructed a BHV-1 BAC clone by inserting an excisable BAC plasmid sequence in the long intergenic region between the gB and UL26 genes. Infectious BHV-1 was readily reconstituted after transfection of cloned BHV-1 BAC DNA (pBHV-1 BAC) into Madin-Darby bovine kidney (MDBK) cells, and the BAC sequence could be excised following cotransfection of pBHV-1 BAC DNA with a Cre recombinase-expressing plasmid DNA. The BAC-excised reconstituted vBHV-1lox containing only the 34-bp loxP sequence in the gB-UL26 intergenic region is highly infectious in calves, retained wild-type (wt) virulence properties, and reactivated from latency. Further, we used two-step Red-mediated mutagenesis to construct a gE cytoplasmic tail-truncated BHV-1 mutant. In calves, the mutant BHV-1gE cytoplasmic tail-truncated virus is significantly attenuated and has defective anterograde transport. Therefore, we believe that sequences within the BHV-1gE cytoplasmic tail are important for BHV-1 anterograde transport from TG to the nose and eye.
MATERIALS AND METHODS
Virus strain and cell line.
The BHV-1 Cooper (Colorado-1) strain, obtained from the American Type Culture Collection (Manassas, VA), was propagated and titrated in MDBK cells as previously described (11).
Plasmids and bacterial strains.
The mini-F plasmid pHA2 (1), containing a BAC plasmid and enhanced green fluorescent protein cassette flanked by loxP, was provided by N. Osterrieder at Cornell University. BHV-1gB and UL26 transfer plasmids were constructed and maintained in Escherichia coli strain XL1-Blue grown at 37°C in Luria-Bertani (LB) medium. E. coli strain DH10B was provided by N. Osterrieder and was used to maintain the mini-F plasmid. The wt BHV-1 BAC clone (pBHV-1 BAC) and the pBHV-1 BAC gEAm453 mutant clone (gE cytoplasmic tail truncated) were maintained in GeneHogs E. coli (Invitrogen). E. coli strain SW105 was provided by N. G. Copeland (NCI, Frederick, MD) and was used for Red recombination (36).
Preparation of radiolabeled mock- and virus-infected cell lysates and immunoprecipitation.
Steady-state and pulse-labeling of infected MDBK cell proteins with [35S]methionine-cysteine, pulse-chase assay, immunoprecipitation with rabbit BHV-1 gI-specific antibody (3, 37), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses were performed as described earlier (3). Endo-β-N-acetylglucosaminidase H (endo-H) and glycopeptidase F treatments of the immunoprecipitated proteins were performed as described earlier. Steady-state 32P labeling of infected MDBK cell proteins was performed for 2 to 3 h starting at 8 h postinfection as described earlier (3).
Construction of gB-BAC-UL26 insertion plasmid.
Plasmid pHA2 contains an 8.8-kb PacI fragment containing BAC sequence, an enhanced green fluorescent protein gene cassette, and one copy of the 34-bp loxP sequence flanking either end of the PacI fragment (1). A 336-bp intergenic region between the gB and UL 26/UL26.5/UL24 genes (indicated hereafter as UL26) (GenBank accession number AJ004801) was found to be suitable for the insertion of the 8.8-kb spanning BAC mini-F plasmid sequence. The intergenic region is bracketed by two poly(A) sites. The gB (UL27) is transcribed from left to right and its poly(A) site is located between nucleotides 58223 and 58228, whereas the UL26/UL26.5/UL24 genes are transcribed from right to left and their shared poly(A) site is located between nucleotides 58564 and 58569 on the complementary strand. For the insertion of BAC plasmid sequence within the gB-UL26 intergenic region, a BAC insertion plasmid containing a PacI insertion site within the gB-UL26 intergenic region (nucleotides 58360 to 58366) and flanked by the upstream gB gene (1 kb) and the downstream UL26 (1 kb) gene was constructed. Briefly, the gB upstream (partial) region and the intergenic region were amplified as a 1.1-kb fragment using primers designated gB-for/gB-rev (Table 1) and cloned into the EcoRI/PacI sites of pNEB193 (New England Biolabs), resulting in pKS-gB. The intergenic region and downstream UL26 (partial) region were then amplified as a 1-kb fragment using primers UL26-for/UL26-rev (Table 1) and cloned into the PacI/HindIII sites of pKS-gB. In the resulting clone, pKS-gB-UL26, the PacI site was incorporated within the intergenic region of gB and UL26 (Fig. 1A). The nucleotide sequences of these clones were verified each time before combining them into one plasmid clone by sequencing (Iowa State University). To insert the mini-F (BAC) plasmid sequence in to the intergenic region of pKS-gB-UL26, plasmid pHA2 (1) was digested with PacI and the 8.8-kb fragment was isolated and cloned into the Pac I site of pKS-gB-UL26. The resulting insertion plasmid, pgB-BAC-UL26 (pKS05-34), contains the mini-F plasmid sequence flanked by 1.1 kb of gB-specific sequence on the left and 1.0 kb of UL26 gene family sequence on the right, required for homologous recombination with BHV-1 genomic DNA.
TABLE 1.
Primers used for generating BHV-1 BAC, BHV-1gEAm453, and BHV-1gEAm453R
| Purpose and primer | Sequence (restriction endonuclease) |
|---|---|
| BAC insertiona | |
| gB-for | 5′-CGAGGAATTCGATGCGCGCGCCCGGC-3′ (EcoRI) |
| gB-rev | 5′-TTCCTTAATTAAGGGGCGCCCTGCCGTGC-3′ (PacI) |
| UL26-for | 5′-TTCCTTAATTAAGTTTGGCGCGCGGTGG-3′ (PacI) |
| UL26-rev | 5′-TACCAAGCTTGTGCGGCCTCGGCGCAC-3′ (HindIII) |
| Mutagenesisb | |
| gEAm453-for | 5′-GCAGCCCTCGCCGTTCGGGTGTGCGCGCGCCGCGCAAGCTAGAAGCGCACCTACGACATCaggatgacgacgataagtaggg-3′ |
| gEAm453-rev | 5′-CGGGCCCGAAGGGGTTGAGGATGTCGTAGGTGCGCTTCTAGCTTGCGCGGCGCGCGCACACcaaccaattaaccaattctgattag-3′ |
| gEAm453 rescue-for | 5′-GCAGCCCTCGCCGTTCGGGTGTGCGCGCGCCGCGCAAGCCAGAAGCGCACCTACGACATCaggatgacgacgataagtaggg-3′ |
| gEAm453 rescue-rev | 5′-CGGGCCCGAAGGGGTTGAGGATGTCGTAGGTGCGCTTCTGGCTTGCGCGGCGCGCGCACACcaaccaattaaccaattctgattag-3′ |
Primers used for amplification of BHV-1gB and UL26 sequences for the construction of the BAC insertion vector. The underlined sequence indicates the restriction site, and bold letters indicate modified bases to accommodated the restriction site.
Oligonucleotide sequence used for point mutation using the two-step Red-mediated mutagenesis system (32) to construct the BHV-1gEAm453 mutant and BHV-1gEAm453 rescue viruses. BHV-1 gE-specific sequences are shown in uppercase letters; the italicized and italicized-underlined sequences are complementary to each other in inverse orientation. Bold letters indicate the changed nucleotide to introduce the stop codon or to rescue the original sequence in the BHV-1gE cytoplasmic tail. Nucleotides in lowercase indicate the pEPkan-S-specific sequence (for details, see Materials and Methods and reference 32).
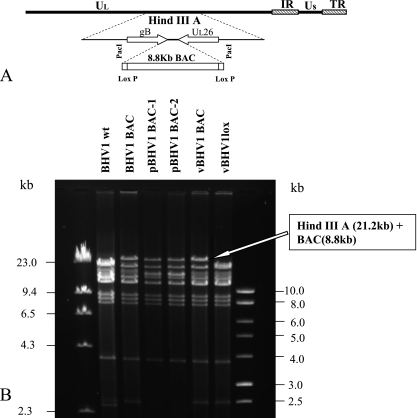
FIG. 1.
(A) Schematic showing the incorporation of a BAC plasmid into the PacI site created within the intergenic region of the gB and UL26 genes of BHV-1 genome. (B) HindIII restriction profile analysis of wt BHV-1, BHV-1 BAC recombinant virus prior to propagation in E. coli, pBHV-1 BAC clones 1 and 2 isolated from E. coli, vBHV-1 BAC (reconstituted by cotransfection of pBHV-1 BAC clone 1 into MDBK cells), and BAC-excised vBHV-1lox DNA (reconstituted by cotransfection of pBHV-1 BAC DNA and pCREin into MDBK cells).
Construction of a BHV-1 BAC clone.
To construct an infectious BHV-1 BAC clone, pgB-BAC-UL26 (pKS05-34), generated as described above, was cotransfected with full-length BHV-1 Cooper strain genomic DNA into MDBK cells by calcium phosphate precipitation as described earlier (10). Several green fluorescent protein-positive recombinant viruses were plaque purified as described earlier (2) and analyzed by sequencing two PCR fragments spanning the BAC insertion site within the gB-UL26 intergenic region and the left and right ends of the inserted BAC plasmid. Based on plaque morphology and growth characteristics, a vBHV-1 BAC recombinant was selected for further study.
To isolate the circular replicative form of vBHV-1 BAC DNA, confluent MDBK cells were infected with vBHV-1 BAC virus at a multiplicity of infection of 5. Circular replicative-form vBHV-1 BAC DNA was isolated at 2 h postinfection from virus-infected cells and electroporated into GeneHogs (Invitrogen) with 0.1-cm cuvettes at 1.5 kV, a resistance of 200 Ω, and a capacitance of 25 μF. Several pBHV-1 BAC clones were isolated in LB agar plates containing 30 μg/ml chloramphenicol and were analyzed by DNA restriction analysis. To reconstitute vBHV-1 BAC, pBHV-1 BAC clone 1 and 2 DNAs were isolated with the QIAprep miniprep kit (Qiagen, catalog no. 27104) and transfected into MDBK cells by the calcium phosphate precipitation method as described earlier (10). After analysis of the plaque morphology and growth characteristics of viruses isolated from several fluorescent plaques, one reconstituted virus was selected for further studies and designated the vBHV-1 BAC.
To excise the BAC sequence from the vBHV-1 BAC, pBHV1 BAC-1 DNA was cotransfected into MDBK cells along with DNA from a Cre expression plasmid, pCre-in (28). Several nonfluorescent BAC-excised BHV-1 viruses were plaque purified and analyzed by plaque morphology and growth characteristics. One reconstituted, BAC-excised BHV-1, containing one copy of the 34-bp loxP sequence within the gB and UL26 intergenic region (referred to hereafter as vBHV-1lox), was selected for further study. Reconstituted vBHV-1 BAC, plasmid pBHV-1 BAC clone 1 and 2, and vBHV-1lox DNAs were analyzed by HindIII restriction endonuclease digestion profiles (Fig. 1B).
Construction of BHV-1gEAm453 by two-step Red-mediated mutagenesis.
Two-step Red-mediated mutagenesis was performed as described earlier (32) for point mutation and a short deletion protocol. To introduce an amber mutation at BHV-1gE amino acid residue 453 (gE open reading frame [ORF] nucleotide 1357), primers gEAm453-for (contains a C-to-T mutation at gE ORF nucleotide 1357) and gEAm453-rev (contains a G-to-A mutation at nucleotide 1357 of the noncoding complementary strand with respect to the gE ORF) were designed (Table 1). The 3′ end of the forward primer was designed to anneal to plasmid pEPkan-S, such that it overlapped with an I-SceI site located upstream of aphAI, conferring kanamycin resistance, and that the 3′ end of the reverse primer was complementary to sequences downstream of aphAI of pEPkan-S (32). PAGE-purified high-quality oligonucleotide primers were used for PCR amplification of the I-SceI aphAI cassette from pEPkan-S. In the resulting PCR product, the glutamine codon at gE residue 453 (5′-CAG-3′) is changed to a stop codon (5′-TAG-3′). Electrocompetent SW105 cells harboring pBHV1 BAC were transformed with approximately 100 ng of DpnI-digested PCR product together with 100 ng of pBAD-I-sceI (32). Following arabinose induction, several kanamycin-sensitive colonies were obtained and stored at −70°C. Positive clones were identified by PCR and sequencing and designated pBHV1 BAC gEAm453. To excise the BAC sequence, pBHV1 BAC gEAm453 DNA was cotransfected along with pCre-in into MDBK cells as described above. The BAC-excised gE cytoplasmic tail-truncated virus, designated vBHV-1gEAm453, was analyzed further by immunoblotting with BHV-1gE peptide-specific rabbit polyclonal antibody (12) (Fig. 2).
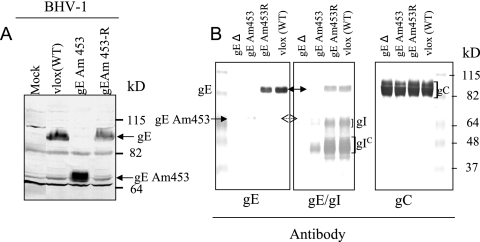
FIG. 2.
Identification and characterization of BHV-1gEAm453 virus. (A) Immunoblot analysis of cell lysates with BHV-1gE peptide-specific rabbit polyclonal antibody (12). Cells were either mock infected or infected with vBHV-1gEAm453R (rescued), vBHV-1gEAm453, and wt BHV-1 viruses. (B) Immunoblot analysis of identical Western blots containing lysates of partially purified BHV-1 gEΔ, vBHV-1gEAm453, vBHV-1gEAm453R, or wt BHV-1 virions with either anti-gE peptide rabbit antibody (as described for panel A), a cocktail of rabbit anti-gE peptide-specific and gI-specific polyclonal antibodies, or anti BHV-1 gC-specific monoclonal antibody F2 (11).
Construction of gEAm453-rescued BHV-1.
To rescue the amber mutation of BHV-1gE at amino acid residue 453, PCR products generated by gEAm453 rescue primers gEAm453 rescue-for and gEAm453 rescue-rev (Table 1) from the pEPkan-S template were used along with pBAD-I-SceI DNA as described above to transform SW105 competent cells containing pBHV1 BAC gEAm453. Two-step Red-mediated mutagenesis was performed as described above, and positive colonies identified by sequencing were selected for reconstitution of the virus by transfection into MDBK cells. A BAC-excised gEAm453-rescued virus (vBHV-1gEAm453R) was further analyzed by immunoblotting with BHV-1gE peptide-specific rabbit polyclonal antibody (Fig. 2).
Calf experiments.
Calf experiments were performed as described earlier (8). Four calves each were inoculated with reconstituted BAC-excised wt BHV-1 (vBHV-1lox) and vBHV-1gEAm453. During acute infection (1 to 20 days postinfection [dpi]) and latency (21 to 40 dpi), nasal and ocular swabs were taken at 0, 1, 5, 6, 9, 19, 23, and 36 dpi. Following reactivation, nasal/ocular swabs were taken at 40 dpi (dex 0), 41 dpi (dex 1), 42 dpi (dex 2), 44dpi (dex 4), 47 dpi (dex 7), 48 dpi (dex 8), 51 dpi (dex 11), 54 dpi (dex 14), 57 dpi (dex 17), and 64 dpi (dex 24). Nasal/ocular swabs were stored at −70°C in 1 ml of tissue culture medium supplemented with 2% penicillin and streptomycin. Sera were taken on days 0, 7, 14, 20, 40 (dex 0), 48 (dex 8), 55 dpi (dex 15), and 64 dpi (dex 24) and were stored at −20°C. Two calves infected with vBHV-1gEAm453 and vBHV-1lox were euthanized at 4 days after dexamethasone injection. All the remaining calves were euthanized at 64 dpi (dex 24), and their TG samples were collected and stored at −70°C for further analysis.
Rabbit experiments.
Twelve rabbits were infected with either the vBHV-1gEAm453 mutant or the vBHV-1gEAm453R virus, and four rabbits were infected with vBHV-1lox virus. For virus infection, rabbits were sedated with Rompun (5 mg/kg intramuscularly; Bayer Corp) and infected by intranasal and ocular instillation of 1 × 107 PFU (per each nostril or eye). At 20 dpi, six rabbits in the vBHV-1gEAm453 and vBHV-1gEAm453R groups and four rabbits in the vBHV-1lox group were injected with dexamethasone (2.8 mg/kg intramuscularly) for 5 days to reactivate the latent viruses. During acute infection (1 to 14 dpi) and latency (14 to 20 dpi), nasal and ocular swabs were collected at 0, 3, 5, 7, 10, 14, 16, 18, and 20 dpi. Following reactivation, nasal/ocular swabs were collected at 21 dpi (dex 0), 23 dpi (dex 2), 26 dpi (dex 5), 28 dpi (dex 7), and 30 dpi (dex 9). Nasal/ocular swabs were stored at −70°C as before. Three rabbits in the vBHV-1gEAm453 and vBHV-1gEAm453R groups were sacrificed at 5 dpi (primary infection), 20 dpi (latent infection), 26 dpi (dex 5), or 39 dpi (dex18). All four rabbits in the vBHV-1lox virus-infected group were sacrificed at 39 dpi. Following euthanasia, TG were collected and stored at −70°C.
Virus isolation and plaque reduction assay.
Virus isolation from nasal/ocular swabs and TG was performed as described earlier (8). Serum neutralization (SN) tests were performed by plaque reduction assay as described earlier (9).
RNA extraction and reverse transcriptase PCR (RT-PCR).
Following euthanasia, rabbit TG were collected, snap frozen on liquid nitrogen, and stored at −70°C. RNA extraction was performed as described earlier (27). Briefly, tissues were homogenized using a manual homogenizer (Power Gen 125; Fisher Scientific) in the presence of 2 ml of Trizol (Invitrogen, catalog no. 15596-026). RNAs extracted from uninfected rabbit TG and from wt BHV-1-infected MDBK cells were used as negative and positive controls, respectively. RNA concentrations were determined using a spectrophotometer (Nanodrop ND-1000).
Prior to RT-PCR, RNA was treated with 5 U of DNase I (Invitrogen, 18068-015) for 25 min at room temperature. After DNase I treatment, samples were incubated at 65°C for 10 min in the presence of 2 mM EDTA to eliminate DNase I activity. To detect viral major capsid protein (virion protein 5 [VP5]) and cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcription, a one-step RT-PCR kit (Qiagen) was used following the manufacturer's recommendations. Due to a high GC content in the herpesvirus sequence, the Q-solution included in the kit was used. Following the RT reaction (50°C for 30 min), the RT enzyme was inactivated (95°C for 15 min) prior to PCR. BHV-1 VP5-specific PCRs consisted of 40 cycles of denaturing at 94°C for 45 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s. GAPDH-specific PCRs consisted of 30 cycles of denaturing at 94°C for 1 min, annealing at 55°C for 45 s, and extension at 72°C for 1 min. For both the reactions, final extension was performed for 10 min at 72°C. As a control for DNA contamination in RNA samples, a reaction lacking the RT step was performed using the BHV-1 VP5 primers.
Semiquantitative PCR.
TG DNA was extracted as described earlier (8). Relative viral copy numbers in rabbit TG were determined from a standard curve generated by serially diluting purified BHV-1 viral DNA and performing PCR with major capsid protein VP5-specific forward primer 5′-TGCGGTCTGCGAGTTCATC-3′ (nucleotides 73285 to 73304) and reverse primer 5′-CGCCGCTCATGTTGTACTG-3′ (nucleotides 73517 to 73535) (GenBank accession number AJ004801). Starting with approximately 500 ng of BHV-1 viral DNA, which corresponds to a viral genomic copy number of 3.1 × 109, the DNA was serially diluted 10-fold. These serial dilutions were amplified in order to produce a standard curve of viral DNA copy number per μg of BHV-1 DNA (see below). Serial dilutions of BHV-1 DNA (standard) and DNA extracted from the TG of the infected rabbits were amplified in the same PCR run, using the same reaction conditions for each quantitation. PCR was carried out in a 50-μl volume containing 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.4 μM primers, 5% dimethyl sulfoxide, and 1.25 U Taq polymerase (BullsEye BE110203; Midwest Scientific). Briefly, following 5 min of incubation at 94°C, 35 PCR cycles were carried out. Cycles consisted of 94°C for 45 s, 55°C for 30 s, and 72°C for 45 s. Upon completion of the last cycle, the reaction mixtures were further incubated at 72°C for 10 min to ensure complete extension of the amplified products. Following PCR, 10 μl of the reaction product was analyzed by 1.6% agarose gel electrophoresis. The gel image was captured by using FOTO/Analyst PC Image version 5.00. (Fotodyne Corporation). To calculate the BHV-1 genomic copy numbers amplified from each TG samples, first the band intensity of PCR products from the dilutions of the standard BHV-1 genomic DNA was calculated using the Image J 1.37 v program (http://RSB.INFO.NIH.GOV/IJ) and plotted against approximate genome copy numbers that correlate with the specific dilution. Using this plot as a guide, the intensity of PCR products from the 2-μg TG samples were estimated and plotted in a bar graph.
RESULTS
Construction and characterization of a full-length BHV-1 BAC plasmid clone and reconstituted vBHV-1 BAC and BAC-excised vBHV-1lox viruses.
DNA restriction enzyme analysis of recombinant BHV-1 BAC (pre-E. coli passage), pBHV-1 BAC-1 and -2, vBHV-1 BAC (reconstituted from E. coli-passaged pBHV-1 BAC-1), and vBHV-1lox (BAC excised, containing one copy of loxP sequence) along with BHV-1 wt Cooper strain DNA show that the 21-kb HindIII A fragments detected both in wt BHV-1 and vBHV-1lox DNAs were approximately 30 kb (8.8 kb larger) in the digests of recombinant BHV-1 BAC, pBHV-1 BAC, and vBHV-1 BAC (Fig. 1B). Therefore, the incorporation of 8.8 kb of BAC plasmid DNA is within the HindIII A fragment. Analysis of recombinant BHV-1 BAC, pBHV-1 BAC, and vBHV-1 BAC DNA by PCR and sequencing determined that BAC-specific sequences (left and right ends) were inserted between the two poly(A) sites located within the intergenic region of the gB and UL26 genes (data not shown). Similar PCR and sequencing analysis of vBHV-1lox DNA confirmed that there is only one copy of the 34-bp loxP sequence present within the same intergenic region. Taken together, the results indicated that BAC is inserted within the gB and UL26 intergenic region in a site-specific manner and that the BAC sequence flanked by two loxP sequences is excisable by Cre-mediated loxP recombination.
Construction and characterization of gEAm453 and gEAm453-rescued (gEAm453R) viruses.
Following two-step Red-mediated mutagenesis (32), pBHV-1 BAC gEAm453 DNA was analyzed by PCR and sequencing to verify the incorporation of a stop codon at gE amino acid residue 453. The BAC-excised vBHV-1gEAm453 virus was plaque purified and analyzed by immunoblotting using a rabbit anti BHV-1gE antibody specific to gE peptide residues 381 to 396 (12) (Fig. 2). Similarly, a BHV-1 BAC gEAm453-rescued virus (vBHV-1gEAm453R) was constructed by two-step Red-mediated mutagenesis and analyzed by sequencing and immunoblotting. A BAC-excised vBHV-1gEAm453R virus was analyzed by immunoblotting with the rabbit anti BHV-1gE peptide-specific antibody (Fig. 2). The immunoblotting results shown in Fig. 2 demonstrated that the wt gE and gEAm453R have an identical molecular mass of approximately 92 kDa. The corresponding molecular mass of gEAm453 is approximately 68 kDa. Based on sequence analysis, the predicted wt gE molecular mass is 63 kDa while the corresponding molecular mass of gEAm453 is 50 kDa (13 kDa smaller). Taken together, these data indicate that the processed forms of the wt gE and gEAm453 are 29 kDa and 18 kDa larger, respectively, than predicted. This suggested that the wt gE and the gEAm453 are both processed posttranslationally (Fig. 2 and 3).
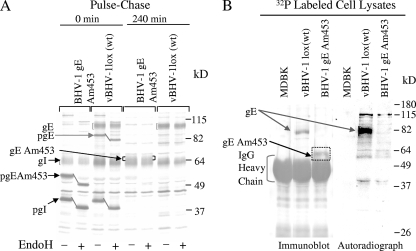
FIG. 3.
(A) Analysis of gE/gI processing and gE/gI interaction in wt BHV-1 and vBHV-1gEAm453 by immunoprecipitation with BHV-1 gI-specific antibody. Immunoprecipitated proteins were separated by SDS-PAGE (10%) and analyzed by autoradiography. (B) Analysis of gE phosphorylation in wt BHV-1 and vBHV-1gEAm453. For gE phosphorylation, MDBK cells infected with wt BHV-1 or vBHV-1gEAm453 were radiolabeled overnight in the presence of 32P. Immunoprecipitation and SDS-PAGE (10%) were performed as described for panel A. The Western blot of the gel was analyzed by immunoblotting with BHV-1gE peptide-specific polyclonal rabbit serum (12) and by autoradiography. Note that while the 92-kDa (wt) gE band is phosphorylated, the 68-kDa gEAm453 band recognized by the BHV-1gE peptide-specific antibody on the immunoblot is not phosphorylated.
Analysis of mutant gEAm453/gI incorporation in the virion envelope.
To determine gEAm453 and gI incorporation in the virion envelope, immunoblotting analysis of purified virions lysates was performed using anti-BHV-1 gE-specific antibody, a cocktail of gE- and gI-specific antibodies, and anti-gC-specific monoclonal antibody F2 (Fig. 2B). The immunoblotting results show that while the gC levels (control) were similar in the vBHV-1gEAm453 and vBHV-1gEAm453R viruses, there was a significant reduction in the amounts of gEAm453/gI incorporation in the vBHV-1gEAm453 virion envelope (Fig. 2B).
Maturation of mutant gEAm453 and gEAm453/gI interaction.
Immunoprecipitation and pulse-chase analysis of wt BHV-1 and vBHV-1gEAm453 virus-infected cell proteins were performed using a gI-specific rabbit polyclonal antibody (Fig. 3A). These results indicated that the cytoplasmic tail-truncated gE processing was similar to that of the wt gE protein. The abundant gE band observed after the pulse (0-min sample) was 84 kDa. The endoplasmic reticulum (ER)-processed 84-kDa wt gE and 55-kDa ER-processed cytoplasmic tail-truncated gEAm453 both were endo-H sensitive (Fig. 3A). Following a 240-min chase, both the 84-kDa (wt gE) and 55-kDa (gEAm453) bands were processed in the Golgi apparatus to mature 92- and 68-kDa bands, respectively, and they became endo-H resistant (Fig. 3A). As expected, both the wt and mutant (gEAm453) endo-H-resistant gE bands were sensitive to glycopeptidase F (data not shown).
The results also showed that immediately after pulse-labeling (0-min sample), the ER-processed, endo-H-sensitive 40-kDa prominent gI-specific bands were precipitated from both the wt and mutant virus-infected cell lysates (Fig. 3A). After 240 min of chase, the 62-kDa endo-H-resistant gI bands were immunoprecipitated with the gI-specific antibody. Taken together, these results indicated that both gEAm453 and wt gE were precipitated by the gI-specific antibody and therefore that the mutant gEAm453 formed a complex with the gI protein in the ER. This complex was then transported to the Golgi apparatus and was processed to its mature form within the Golgi apparatus.
gE phosphorylation.
Based on NetPhos 2.0 server prediction results, residues 467 to 470 (YTSL), residues 513 to 516 (YDLA), and residues 563 to 566 (YTVV) are potential tyrosine phosphorylation sites. To determine the gE phosphorylation status in wt BHV-1 and vBHV-1gEAm453, virus-infected cells were labeled with 32P. Mock- and virus-infected 32P-labeled lysates were immunoprecipitated with gI-specific polyclonal rabbit serum. The immunoprecipitated proteins were separated by SDS-PAGE, and the Western blot was analyzed using BHV-1gE peptide-specific polyclonal rabbit serum (Fig. 3B). 32P-labeled (phosphorylated) and immunoprecipitated gE-specific proteins (wt gE and gEAm453) were visualized by autoradiography. The autoradiograph of the immunoblot demonstrated that both gE forms, wt and gEAm453, were precipitated by anti-gE antibody, but only the wt gE protein was phosphorylated at detectable levels (Fig. 3B). These results also demonstrated that the 92-kDa and 68-kDa bands immunoprecipitated from the infected cell lysates of wt BHV-1 and vBHV-1gEAm453 are indeed wt- and gEAm453-specific bands.
Growth characteristics of reconstituted vBHV-1lox (wt) and vBHV-1gEAm453 viruses in MDBK cells.
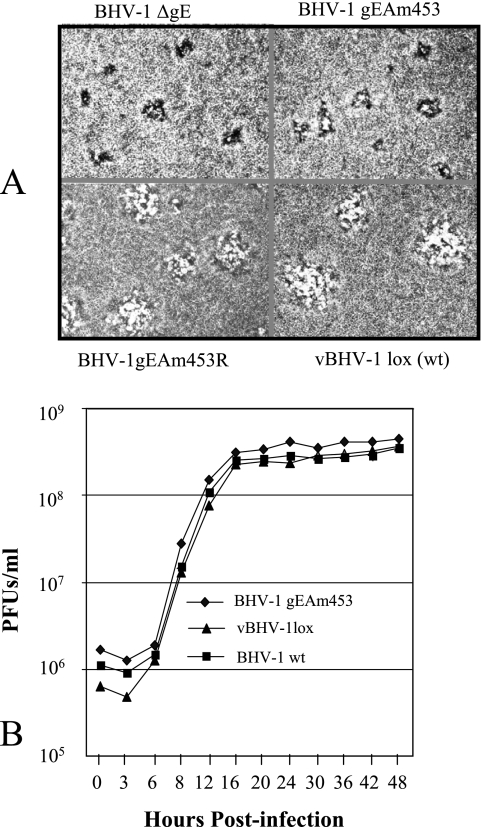
To compare the plaque morphologies of BHV-1gEAm453, BHV-1gEAm453R, and BHV-1gE-deleted viruses, MDBK cell monolayers were infected with these viruses and overlaid with a medium that contained 1.6% carboxymethyl cellulose. The infected cells were fixed at 48 h postinfection and were stained with crystal violet stain. The vBHV-1gEAm483 virus produced on average smaller plaques than those of BHV-1gEAm453R and vBHV-1lox viruses, and the plaque sizes of the BHV-1gEAm453 and BHV-1gE ORF-deleted (BHV-1gEΔ) viruses were very similar (Fig. 4A).
FIG. 4.
(A) Plaque morphology of BHV-1gEΔ, vBHV-1gEAm453, vBHV-1gEAm453R, and vBHV-1lox in MDBK cell monolayers. Viruses were inoculated onto MDBK cell monolayers, overlaid with 1.6% carboxymethyl cellulose, fixed at 48 h postinfection, and stained with 0.35% crystal violet solution. (B) One-step growth curve of BHV-1gEAm453, BHV-1lox, and wt BHV-1 viruses in MDBK cells. Confluent MDBK cells were infected at a multiplicity of infection of 5 PFU per cell with viruses. After 1 h of adsorption at 4°C, residual input viruses were removed. The cultures were washed three times with phosphate-buffered saline, and 5 ml of medium was added to each flask before further incubation (37°C). At the indicated time intervals, replicate cultures were frozen. Virus yields were determined by titration on MDBK cells. Each data point represents the average for duplicate samples obtained from separate infections.
To determine the one-step growth properties of vBHV-1lox and vBHV-1gEAm453 viruses relative to the wt BHV-1, MDBK cells were infected with the respective virus as described previously (12). Virus titration results up to 48 h postinfection (Fig. 4B) show that the growth kinetics of vBHV-1lox and vBHV-1gEAm453, relative to those of wt BHV-1, are similar.
Pathogenicity of vBHV-1lox in calves.
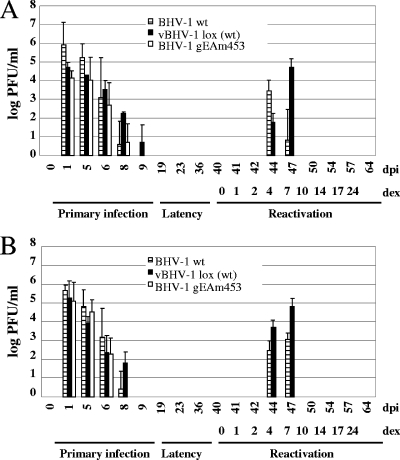
Since vBHV-1lox virus is reconstituted from BHV-1 BAC that has been maintained in E. coli, it was necessary to examine its pathogenic properties in calves. Calf experiments demonstrated that the vBHV-1lox virus is highly pathogenic for calves because it causes respiratory and ocular symptoms that are similar to those caused by wt BHV-1. From 3 dpi until 7 dpi, infected calves have conjunctivitis and excessive nasal exudates. Infected calves also showed loss of appetite and lethargy for several days. During primary infection, vBHV-1lox virus was recovered from the nasal and ocular swabs of infected calves with a higher yield than the wt BHV-1. Following reactivation initiated by dexamethasone, the vBHV-1lox virus was detectable in the ocular and nasal swabs during 4 to 7 days after dexamethasone treatment (Fig. 5). From these studies we concluded that vBHV-1lox virus maintained wt-level pathogenicity and was suitable for mutagenesis studies.
FIG. 5.
Isolation and quantitation of BHV-1, vBHV-1lox (wt), and vBHV-1gEAm453 viruses present in nasal (A) and ocular (B) swabs of calves during the acute, latent, and postreactivation stages of infection. Virus was isolated at the indicated intervals. Virus quantitation was performed by plaque assay on MDBK cells, and data represent averages and standard deviations for each group.
Pathogenicity of vBHV-1gEAm453 in calves.
In contrast to calves infected with the vBHV-1lox virus, vBHV-1gEAm453 virus-infected calves showed only mild conjunctivitis, and they did not display severe clinical symptoms during primary infection. These results suggested that the vBHV-1gEAm453 virus was attenuated in calves.
During acute infection, virus was detected in nasal and ocular swabs for 6 dpi with vBHV-1gEAm453. However, in this case, nasal and ocular virus shedding was 2 to 3 days shorter than that in calves infected with vBHV-1lox. Following reactivation from latency, the vBHV-1lox virus was recovered from nasal (Fig. 5A) or ocular (Fig. 5B) swabs as early as 4 days after dexamethasone treatment, and virus shedding lasted up to 7 days postreactivation. In contrast, the vBHV-1gEAm453 virus was not detectable up to 24 days after dexamethasone injection (Fig. 5A and B).
TG explants of two calves infected with vBHV-1gEAm453 and vBHV-1lox were examined for infectious virus at 4 days postreactivation. Infectious virus was recovered from one calf infected with vBHV-1gEAm453 and from both the calves infected with vBHV-1lox.
In vivo properties of vBHV-1gEAm453 and vBHV-1gEAm453R in rabbits.
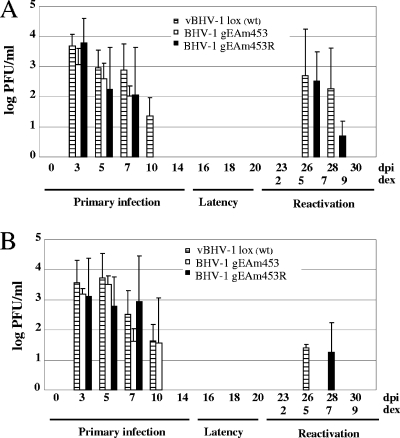
Rabbit experiments were performed to determine (i) relative nasal and ocular virus shedding during primary infection and during reactivation in rabbits infected with vBHV-1lox, vBHV-1gEAm453, and vBHV-1gEAm453R viruses and (ii) relative VP5-specific transcription and DNA copy numbers in TG of rabbits infected with vBHV-1gEAm453 and vBHV-1gEAm453R during primary infection, latency, and reactivation from latency. vBHV-1gEAm453 virus did not reactivate from latency in calves, whereas the vBHV-1lox and vBHV-1gEAm453R viruses reactivated from latency in rabbits and the virus was shed in the nasal and ocular secretions (Fig. 6).
FIG. 6.
Nasal (A) and ocular (B) swabs of rabbits infected with vBHV-1lox (wt), vBHV-1gEAm453, and vBHV-1gEAm453R viruses. Virus quantitation was performed by plaque assay on MDBK cells, and data represent averages and standard deviations for each group.
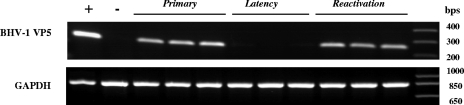
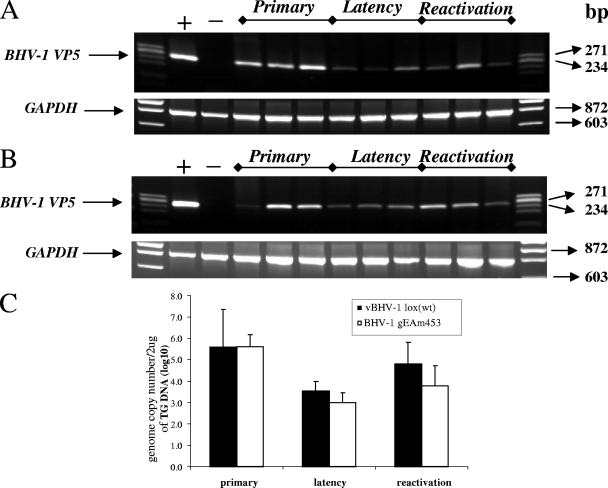
Both during primary infection and at 5 days postreactivation, VP5-specifc transcription was detected in TG of rabbits infected with vBHV-1gEAm453 (Fig. 7) and vBHV1gEAm453R (data not shown). As expected, during latency VP5 transcription was not detected in TG of rabbits infected with either virus strain. Additionally, semiquantitative PCR results (Fig. 8) indicated that similar numbers of genomic copies were present in the TG of rabbits infected with either virus strain during primary infection (approximately 105 copies/μg total DNA) and latency (approximately 103.5 for the rescued virus and 103 for the gE mutant). At 5 days postreactivation, 104.8 and 103.8 genomic copy numbers were detected for the rescued and mutant viruses, respectively (Fig. 8C). Following reactivation from latency, VP5 transcription was detected in TG of rabbits infected with gEAm453 rescued and gEAm453 mutant BHV-1 viruses. In contrast, following reactivation from latency only the gE rescued virus was detected in nasal or ocular swabs. Taken together, the results indicated that both the rescued and the gE mutant viruses were transported retrogradely from nerve endings in the nose or eye to cell bodies in the TG. However, following reactivation from latency, the gE cytoplasmic tail-truncated virus had defective anterograde transport, because we were unable to detect virus in ocular or nasal swabs.
FIG. 7.
RT-PCR amplification of BHV-1 VP5 (251-bp) specific mRNA sequence from TG of rabbits infected with vBHV-1gEAm453 virus. A 251-bp VP5 transcript-specific sequence was amplified using forward primer 5′-TGCGGTCTGCGAGTTCATC-3′ and reverse primer 5′-CGCCGCTCATGTTGTACTG-3′ (GenBank accession number AY261359) from DNase I treated RNA. To verify lack of DNA contamination, a reaction without RT using the BHV-1 VP5 primers was performed, and no product was detected (not shown). As a control, the GAPDH (841-bp) sequence was amplified using forward primer 5′-TGTTCCAGTATGATTCCACCC-3′ and reverse primer 5′-TCCACCACCCTGTTGCTGTA-3′ from each respective sample. The positive control is infected MDBK cellular RNA, and the negative control is uninfected rabbit TG RNA.
FIG. 8.
(A and B) PCR amplification of BHV-1 VP5 (251-bp) specific sequence from the DNA extracted from the TG of rabbits infected with vBHV-1gEAm453 (A) and vBHV-1gEAm453R (B). As a control, the GAPDH (841-bp) sequence was amplified using forward primer 5′-TGTTCCAGTATGATTCCACCC-3′ and reverse primer 5′ TCCACCACCCTGTTGCTGTA-3′ from each respective TG DNA. (C) Determination of BHV-1 viral genome copy number by semiquantitative PCR. A standard curve was generated using serial dilution of known amounts of purified wt BHV-1 genome. Two micrograms of total rabbit TG DNA was used as a template for the PCR. The intensity of each PCR product was determined for calculating genomic copy numbers as described in Materials and Methods. Error bars indicate standard deviations.
SN titers in calves infected with BHV-1gEAm453 and vBHV-1lox.
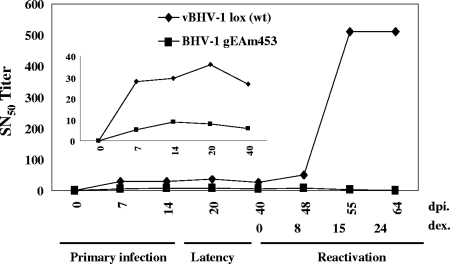
The immune response against the mutant virus was compared with that against the vBHV-1lox (wt) and gEAm453R (rescue) viruses by determining SN titers. After primary infection, calves infected with vBHV-1gEAm453 generated an average SN titer of 10 (Fig. 9), whereas in the cases of vBHV-1lox virus-infected calves (Fig. 9) and BHV-1gEAm453R virus-infected calves (data not shown), the relative SN titer was 30 (threefold higher). A secondary antibody response following reactivation from latency is a sensitive indicator of virus reactivation and virus replication in the nasal epithelium or cornea (22). As expected following reactivation, the SN titer in calves infected with vBHV-1lox spiked to 500. Conversely, in calves latently infected with the vBHV-1gEAm453 mutant, there was no change in SN titer, which correlated with our inability to detect infectious virus in nasal or ocular swabs.
FIG. 9.
SN antibody titers. Sera were collected from calves infected with vBHV-1lox and vBHV-1gEAm453 viruses. BHV-1 neutralizing titers were measured at intervals as indicated during acute primary infection, during latency, and after reactivation induced by dexamethasone. SN was performed with a constant amount of virus (100 PFU) and serial dilutions of serum. The inset illustrates the antibody titers during acute infection more clearly.
DISCUSSION
Previous studies reported that following dexamethasone-induced reactivation, gE-deleted BHV-1 was not shed in the noses and eyes of latently infected animals (19, 20). Our results from a recent study confirmed this finding and further suggested that gE-deleted BHV-1 reactivates from latency but is not transported anterogradely from TG to nerve endings in the nose or cornea (S. I. Chowdhury, unpublished data). In this study, we constructed an infectious BHV-1 BAC clone and tested its pathogenicity in calves relative to wt BHV-1. In addition, we have constructed a gE cytoplasmic tail-truncated BHV-1 mutant and examined its pathogenic potential in calves and rabbits. The salient features of our investigations are as follows. (i) The intergenic region between the gB and UL26 genes was suitable for incorporating BAC DNA into the BHV-1 genome, and E. coli-passaged BAC-excised virus retained wt-level pathogenicity in calves. (ii) Truncation of the entire cytoplasmic domain of gE led to defective gE incorporation into the envelope of BHV-1 virions. (iii) Based on VP5 (major capsid protein) transcription and relative genomic copies in the TG, both the gE-rescued and gE cytoplasmic tail-truncated BHV-1 reactivated in the TG. However, vBHV-1gEAm453 virus was not transported anterogradely from TG to nerve endings in the nose or cornea, and only the gE-rescued virus was shed in the nose or eye after reactivation. (iv) Based on SN titers, the primary immune response directed against the gE cytoplasmic tail-truncated virus was relatively weak compared to that against the gE-rescued virus.
Like the wt gE, cytoplasmic tail-truncated gE (gEAm453) is processed in the Golgi apparatus and then forms a complex with the gI glycoprotein. Therefore, consistent with previous reports on other alphaherpesviruses (4, 25, 35), the BHV-1gE cytoplasmic tail is not important for complex formation with gI, gE/gI processing, or maturation within the Golgi apparatus. Unlike the wt gE, gEAm453 was not heavily phosphorylated, and it was not incorporated into enveloped virions. In several herpesviruses, including herpes simplex virus (HSV), pseudorabies virus (PRV), and varicella-zoster virus, the YXXL motifs located within the gE cytoplasmic tail are important for tyrosine kinase phosphorylation and gE endocytosis from the cell surface (24, 30, 31). There are four tyrosine motifs within the gE cytoplasmic tail sequence, two YXXL motifs (residues 457 to 460 and/or 467 to 470), and two YXXΦ motifs (residues 513 to 516 and/or 563 to 566). Based on NetPhos 2.0 server prediction results, residues 467 to 470 (YTSL), residues 513 to 516 (YDLA), and residues 563 to 566 (YTVV) are potential tyrosine phosphorylation sites. In addition, there are two serine residues within the gE acidic domain, S493 and S499, that could be potential phosphorylation sites. Therefore, lack of phosphorylation due to the deletion of the three potential tyrosine phosphorylation sites and several serine residues was expected.
Deletion of the cytoplasmic domain of BHV-1 also resulted in defective gE incorporation into virion particles. Similar results have been found with BHV-5 (4) and PRV (30, 31). Specifically, in the case of PRV, incorporation of gE in the envelope required the YXXL motif within the gE cytoplasmic tail (30, 31). However, HSV type 1 mutant viruses without the entire gE cytoplasmic tail, including the YXXL motif, incorporated the mutant gE protein into the virion envelope (14, 18). Therefore, signals for gE incorporation must be substantially different between HSV type 1 and BHV-1, BHV-5, or PRV.
In vitro, the vBHV-1gEAm453 virus produced small plaques compared with the wt or the rescued virus, and the plaque size was similar to that produced by the gE-deleted BHV-1. This is in contrast to the case for BHV-5, where a gE cytoplasmic tail-truncated virus produced larger plaques than a gE-deleted virus (4). It is possible that the difference in plaque sizes observed between the BHV-1 and BHV-5 gE cytoplasmic tail-truncated viruses may reflect the fact that in the case of BHV-5, the gE ectodomain may partially contribute to the cell-to-cell spread of the virus, whereas in BHV-1 it does not.
Following primary infection, SN titers in animals infected with gE cytoplasmic tail mutant virus were fourfold lower than the corresponding titers in animals infected with gE-rescued virus. Our recent results showed that SN titers in animals infected with gE-deleted virus were approximately two fold lower than those for gE cytoplasmic tail mutant virus. As noted above, like the gE-deleted BHV-1, the BHV-1gE cytoplasmic tail mutant virus forms smaller plaques due to a cell-to-cell spread defect, which are very similar to the plaques formed by the gE-deleted BHV-1. Therefore, it is possible that lower neutralizing titers for both the gE-deleted and gE cytoplasmic tail mutant viruses are a result of defective cell-to-cell spread resulting in fewer infected cells. Apparently this lower spread did not affect retrograde transmission to neurons and establishment of latency, as shown by semiquantitative PCR. This was not surprising, because gE is not required for the retrograde axonal transport (19, 20).
The lack of a secondary immune response following reactivation of gE cytoplasmic tail mutant virus indicates the lack a of boost from virus replication in the nose and cornea. The RT-PCR results for VP5 transcription and semiquantitative PCR for DNA copies in TG of rabbits during primary infection, during latency, and following reactivation from latency suggested that both viruses reactivated within TG neurons, yet the mutant virus was not isolated from the nose and eye swabs. Taking these results together, we hypothesize that the gE cytoplasmic tail was necessary for the anterograde transport of infectious virus from the TG cell bodies to nerve endings in the nose or eye.
Previously it was suggested that during axonal transport, tegumented nucleocapsids and envelope proteins are transported separately and that virus envelopment takes place at or near the synapse (33). Recently, Antinone and Smith (5) reported that PRV nascent virions are transported as fully enveloped virus or within transport vesicles anterogradely within the neurons. Consistent with this notion, defective gE incorporation in the virion envelope is perhaps responsible for the defective anterograde transport of the virus. Alternatively, cytoplasmic tail-truncated gE lacking endocytosis/recycling/apical motifs probably concentrate else where on the cell surface rather than in the trans-Golgi and/or synaptic membrane, thereby indirectly rendering it nonfunctional and resulting in defective anterograde vesicular transport of nascent virions to axon termini. The latter situation is in direct contrast to our recent reports on gE cytoplasmic tail-deleted BHV-5 anterograde transport within the olfactory receptor neuron. In the case of BHV-5, the gE cytoplasmic tail sequence was not required for anterograde neuronal transport from olfactory receptor neurons to the bulb (4). It is possible that in the case of BHV-5, another viral protein(s) might have complemented for gE cytoplasmic tail-mediated anterograde transport or that glycine-rich sequences within the ectodomain of BHV-5 gE could be the axonal targeting (apical) motif. Consistent with the latter case, a BHV-5 gE mutant lacking the glycine-rich sequences showed defective anterograde transport (3). In the context of a neuron, the apical surface would be the axon terminal membrane, whereas the basolateral surface would be the cell bodies and dendritic membranes. It is known that in the absence of an apical motif, sequences/motifs within the cytoplasmic tails of membrane proteins may target the membrane proteins to axon termini (21). Since the BHV-1gE ectodomain lacks the glycine-rich sequences (3, 12), we hypothesize that in the absence of a putative glycine-rich apical motif, motifs within the BHV-1gE cytoplasmic tail may serve as a default apical motif.
Acknowledgments
We acknowledge Lynn Enquist for providing the gI-specific rabbit polyclonal antibody.
This work was supported by USDA grants 00-02103 and 2004-35204-14657 to S. I. Chowdhury.
Footnotes
Published ahead of print on 14 May 2008.
Contribution 08-273-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 746964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Mubarak, A., and S. I. Chowdhury. 2004. In the absence of glycoprotein I (gI), gE determines bovine herpesvirus type 5 neuroinvasiveness and neurovirulence. J. Neurovirol. 10233-243. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mubarak, A., Y. Zhou, and S. I. Chowdhury. 2004. A glycine-rich BHV-5 gE-specific epitope within the ectodomain is important for BHV-5 neurovirulence. J. Virol. 784806-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Mubarak, A., J. Simon, C. Coats, J. D. Okemba, M. D. Burton, and S. I. Chowdhury. 2007. Glycoprotein E (gE) specified by bovine herpesvirus type 5 (BHV-5) enables trans-neuronal virus spread and neurovirulence without being a structural component of enveloped virions. Virology 365398-409. [DOI] [PubMed] [Google Scholar]
- 5.Antinone, S. E., and G. A. Smith. 2006. Two modes of herpesvirus trafficking in neurons: membrane acquisition directs motion. J. Virol. 8011235-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashbaugh, S. E., K. E. Thompson, E. B. Belknap, P. C. Schulteiss, S. I. Chowdhury, and J. K. Collins. 1997. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn. Investig. 9387-394. [DOI] [PubMed] [Google Scholar]
- 7.Belknap, E. B., J. K. Collins, V. K. Ayers, and P. C. Schulteiss. 1994. Experimental infection of neonatal calves with neurovirulent bovine herpesvirus type 1.3. Vet. Pathol. 31358-365. [DOI] [PubMed] [Google Scholar]
- 8.Butchi, N. B., C. Jones, S. Perez, A. Doster, and S. I. Chowdhury. 2007. Role of envelope protein Us9 in the anterograde transport of bovine herpesvirus-1 (BHV-1) following reactivation in the trigeminal ganglia. J. Neurovirol. 13384-388. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury, S. I., H. Ludwig, and H. J. Buhk. 1988. Molecular biological characterization of equine herpesvirus 1 (EHV-1) isolated from ruminant hosts. Virus Res. 11127-139. [DOI] [PubMed] [Google Scholar]
- 10.Chowdhury, S. I., and W. Batterson. 1994. Transinhibition of herpes simplex virus replication by an inducible cell-resident gene encoding a dysfunctional VP19c capsid protein. Virus Res. 3367-87. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury, S. I. 1995. Molecular basis of antigenic variation between the glycoproteins C of respiratory bovine herpesvirus 1 (BHV-1) and neurovirulent BHV-5. Virology 213558-568. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury, S. I., B. J. Lee, A. Ozkul, and M. L. Weiss. 2000. Bovine herpesvirus 5 glycoprotein E is important for the neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J. Virol. 742094-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Offay, J. M., R. E. Mock, and R. W. Fulton. 1993. Isolation and characterization of encephalitic bovine herpesvirus type 1 isolates from cattle in North America. Am. J. Vet. Res. 54534-539. [PubMed] [Google Scholar]
- 14.Farnsworth, A., and D. C. Johnson. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junction to promote cell-cell spread. J. Virol. 803167-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 728133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 5181-133. [DOI] [PubMed] [Google Scholar]
- 17.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 1679-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaashoek, M. J., F. A. M. Rijesewijk, R. C. Ruuls, G. M. Keil, E. Thiry, P. P. Pastoret, and J. T. van Oirschot. 1998. Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in gC, gG, gI, gE or in both the gI and gE gene. Vaccine 16802-809. [DOI] [PubMed] [Google Scholar]
- 20.Kaashoek, M. J., F. A. C. van Engelenburg, A. Moerman, A. L. J. Gielkens, F. A. M. Rijsewijk, and J. T. Oirschot. 1996. Virulence and immunogenicity in calves of thymidine kinase- and glycoprotein E-negative bovine herpesvirus 1 mutants. Vet. Microbiol. 48143-153. [DOI] [PubMed] [Google Scholar]
- 21.Keller. P., and K. Simons. 1997. Post-Golgi biosynthetic trafficking. J. Cell Sci. 1103001-3009. [DOI] [PubMed] [Google Scholar]
- 22.Lovato, L., M. Inman, G. Henderson, A. Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 strain that contains a mutation in the latency-related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 774848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahony, T. J., F. M. Mccarthy, J. L. Gravel, L. West, and P. L. Young. 2002. Construction and manipulation of an infectious clone of the bovine herpesvirus 1 genome maintained as a bacterial artificial chromosome. J. Virol. 766660-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson, J. K., and C. Grose. 1997. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J. Virol. 714042-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizvi, S. M., and M. Raghavan. 2001. An N-terminal domain of herpes simplex virus type 1 gE is capable of forming stable complexes with gI. J. Virol. 7511897-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock, D. L., W. A. Hagemoser, F. A. Osorio, and D. E. Reed. 1986. Detection of bovine herpesvirus type 1 RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J. Gen. Virol. 672515-2520. [DOI] [PubMed] [Google Scholar]
- 27.Schang, L. M., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 716786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 974873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis and control. Adv. Virus Res. 45191-223. [DOI] [PubMed] [Google Scholar]
- 30.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 724571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tirabassi, R. S., R. A. Townley, M. G. Eldridge, and L. W. Enquist. 1997. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence of envelope incorporation, virulence, and neurotropism domains. J. Virol. 716455-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 33.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. cell Biol. 154741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapp, S., N. Osterrieder, G. M. Keil, and M. Beer. 2003. Mutagenesis of a bovine herpesvirus type 1 genome cloned as an infectious bacterial artificial chromosome: analysis of glycoprotein E and G double deletion mutants. J. Gen. Virol. 84301-306. [DOI] [PubMed] [Google Scholar]
- 35.Tyborowska, J., K. Bienkowska-Szewczyk, M. Rychlowski, J. T. Van Oirschot, and F. A. M. Rijsewijk. 2000. The extracellular part of glycoprotein E of bovine herpesvirus 1 is sufficient for complex formation with glycoprotein I but not for cell-to-cell spread. Arch. Virol. 145333-351. [DOI] [PubMed] [Google Scholar]
- 36.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitbeck, J. C., A. C. Knapp, L. W. Enquist, W. C. Lawrence, and L. J. Bello. 1996. Synthesis, processing, and oligomerization of the bovine herpesvirus 1 gE and gI membrane proteins. J. Virol. 707878-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyler, R., M. Engels, and M. Schwyzer. 1989. Infectious bovine rhinotracheitis/vulvo-vaginitis (BHV-1), p. 1-72. In G. Wittman (ed.), Herpesvirus disease of cattle, horses and pigs. Kluwer Acadademic Publishers, Hingham, MA.