Abstract
The eukaryotic translation initiation factor 4E (eIF4E) (the cap-binding protein) is involved in natural resistance against several potyviruses in plants. In lettuce, the recessive resistance genes mo11 and mo12 against Lettuce mosaic virus (LMV) are alleles coding for forms of eIF4E unable, or less effective, to support virus accumulation. A recombinant LMV expressing the eIF4E of a susceptible lettuce variety from its genome was able to produce symptoms in mo11 or mo12 varieties. In order to identify the eIF4E amino acid residues necessary for viral infection, we constructed recombinant LMV expressing eIF4E with point mutations affecting various amino acids and compared the abilities of these eIF4E mutants to complement LMV infection in resistant plants. Three types of mutations were produced in order to affect different biochemical functions of eIF4E: cap binding, eIF4G binding, and putative interaction with other virus or host proteins. Several mutations severely reduced the ability of eIF4E to complement LMV accumulation in a resistant host and impeded essential eIF4E functions in yeast. However, the ability of eIF4E to bind a cap analogue or to fully interact with eIF4G appeared unlinked to LMV infection. In addition to providing a functional mutational map of a plant eIF4E, this suggests that the role of eIF4E in the LMV cycle might be distinct from its physiological function in cellular mRNA translation.
In the case of obligatory parasites, such as viruses, the absence or inadequacy of a single host factor may lead to the inability of the pathogen to multiply or to systemically invade its host (76, 79, 80). This implies that the dominant alleles of the host genes involved are associated with susceptibility and that the recessive alleles encoding nonfunctional versions of such host factors are associated with resistance. In the case of potyviruses, it has been estimated that about 40% of the known resistance genes are recessive (53).
The genus Potyvirus is one of the largest and most diverse genera of plant viruses, and its members cause severe losses of many crops, particularly vegetables (72). The flexuous potyvirus particles contain a single genomic RNA molecule of about 10,000 nucleotides encoding a polyprotein that is matured by three virus-encoded proteinases (59). The genomic RNA is polyadenylated at its 3′ end and is not capped at its 5′ end but is covalently linked to a 25-kDa virus-encoded protein named VPg (46). Among host-encoded factors required for potyvirus infection, the eukaryotic translation initiation factor 4E (eIF4E) and/or its isoform eIF(iso)4E has recently been demonstrated to play an important role. In Arabidopsis thaliana, disruption of the gene encoding eIF(iso)4E results in loss of susceptibility to at least four potyviruses, Tobacco etch virus (TEV), Turnip mosaic virus (TuMV), Lettuce mosaic virus (LMV) (16, 32), and Plum pox virus (14), and disruption of the gene encoding eIF4E compromises susceptibility to another potyvirus, Clover yellow vein virus (68). In crop species, several natural recessive resistances against potyviruses have been identified, and all were correlated with mutations in genes encoding eIF4E (for a review, see references 28 and 61). The pvr2 locus in pepper, mo1 in lettuce, and sbm1 in pea confer resistance, respectively, against Potato virus Y (64) and TEV (27), LMV (48), and Pea seed-borne mosaic virus (18). The barley rym4/5/6 resistance genes to various strains of the bymoviruses (belonging to the family Potyviridae) Barley yellow mosaic virus and Barley mild mosaic virus were also found to be mutations in the eIF4E gene (29, 73). Furthermore, the nsv locus in melon that confers resistance to a non-Potyviridae, uncapped, and nonpolyadenylated virus, Melon necrotic spot virus, also encodes eIF4E (51). Simultaneous mutations in both eIF4E and eIF(iso)4E are required to prevent Pepper veinal mottle virus infection of pepper (65). Using the yeast two-hybrid system and enzyme-linked immunosorbent assay (ELISA), it has been shown that the viral genome linked protein (VPg) of TuMV interacts with A. thaliana eIF(iso)4E (75) and that a VPg mutation that abolishes this interaction is associated with a lack of infectivity of the derived TuMV in its natural host, Brassica (33). Yeam et al. (77) recently showed that a critical substitution in the Capsicum eIF4E causing loss of interaction with TEV VPg is sufficient for resistance against TEV infection. Charron et al. (11) recently provided evidence for coevolution between pepper eIF4E and potyviral VPg. The function of eIF4E and/or eIF(iso)4E in the potyvirus cycle is still largely unknown, but the negative effect of natural mutations in these factors on the accumulation of various potyviruses in various host plants suggests that it is probably conserved (61).
In lettuce (Lactuca sativa), the only resistance genes currently used to protect lettuce crops worldwide are the recessive allelic genes mo11 and mo12. The mo11 gene, formerly named g, was first identified in Argentina in a Latin-type cultivar named “Gallega de Invierno” (5), while the mo12 gene, identified in Egyptian wild L. sativa lines, was named mo (66). Initially considered identical, these genes were later shown to have different specificities and to be either allelic or closely linked and therefore were renamed mo11 and mo12 (15). They have recently been cloned and sequenced in our laboratory (48). The resistance alleles mo11 and mo12, as well as the susceptibility allele mo10, were found to code for forms of the eukaryotic translation initiation factor Ls-eIF4E in lettuce. Types 0, 1, and 2 of lettuce eIF4E were named Ls-eIF4E0, Ls-eIF4E1, and Ls-eIF4E2, respectively. The type 0 sequence corresponds to that found in susceptible lettuce (Trocadéro and Salinas). The type 1 amino acid sequence (Mantilia and Floribibb) is characterized by a deletion of the triplet QGA at positions 108 to 110 and replacement by an H residue. The type 2 sequence (Salinas 88 and Vanguard 75) has an A-to-P substitution at position 70 (48).
The recessive alleles mo11 and mo12 at the mo1 locus are associated with reduced and symptomless accumulation (tolerance) or absence of accumulation (resistance) of common isolates of LMV (15, 19, 66). The result of the interaction, resistance or tolerance, depends on the virus isolate and genetic background (52, 60), but mo11 is generally associated with resistance and mo12 with tolerance (9, 20).
In order to identify which eIF4E amino acids are important for the virus cycle, we set up an experimental system by which the role of eIF4E in the virus cycle can be dissociated from its physiological function in cellular mRNA translation. It was previously observed that eIF4E from susceptible lettuce varieties (eIF4E0), but not defective eIF4E variants (eIF4E1 and eIF4E2 isolated from mo11 and mo12 varieties, respectively), is able to restore LMV susceptibility when expressed ectopically in lettuce plants carrying mo11 and mo12 (48).
We used this property to assay the effects of various amino acid mutations on the function of eIF4E in the virus cycle. The mutations were chosen based on the predicted three-dimensional (3D) structure of lettuce eIF4E, so that they would affect amino acids involved in cap recognition and eIF4G binding and surface residues close to the natural mutations associated with potyvirus resistance in lettuce mo1, pepper pvr2, and pea sbm1. The ability to complement LMV infection in mo1 plants should be affected for eIF4E mutated at amino acids required for the virus cycle. Therefore, this assay should allow an evaluation of the roles of various structural and biochemical properties of eIF4E in the potyvirus cycle.
MATERIALS AND METHODS
Plant material.
All lettuce (L. sativa) plants were grown under standard greenhouse conditions (16-h day length; 18 to 25°C) with additional light from 400-W sodium vapor pressure lamps and were maintained in insect-proof cages after inoculation. Genotypes with no mo1 resistance allele (Salinas and Trocadéro), carrying mo11 (Floribibb and Mantilia) and carrying mo12 (Salinas 88) were used in this study. Trocadéro was routinely used to propagate LMV. The pair of lettuce genotypes included in this analysis, Salinas/Salinas 88, is nearly isogenic for mo12 (25, 67).
Site-directed mutagenesis and viral constructs.
The eIF4E coding sequence isolated from susceptible lettuce (eIF4E0) was cloned into the vector pENTR/D-TOPO (Invitrogen). This recombinant plasmid was used as a template for PCR amplification to generate point mutations in the eIF4E sequence, using the QuickChange site-directed mutagenesis kit (Stratagene). The mutations introduced were W64A, F65A, W77L, R82L, E91A, Y113A, W123A, G156A, E157A, R173A, A174P, W182A, and S223L, where the first letter indicates the residue in the wild-type sequence, the number gives the position of the amino acid in the full-length lettuce eIF4E sequence, and the second letter indicates the residue present in the mutated protein.
Each of the mutant eIF4E cDNAs was then cloned independently into an LMV-derived vector (19) in such a way that the recombinant LMV carried an insertion of the eIF4E coding region in frame between the P1 and HcPro domains of LMV, with a NIaPro cleavage site resulting in the addition of 8 amino acids (PGDEVYHQ) at the C terminus of eIF4E and 5 amino acids (SDVPG) at its N terminus after in vivo processing of the eIF4E protein from HcPro (48). In the nonrecombinant LMV vector, the introduction of an artificial NIa cleavage site led to an HcPro with a slightly modified N terminus, which did not affect its biological properties (19). The LMV isolate used, LMV-0, is unable to accumulate and produce symptoms in lettuce varieties with the mo11 or mo12 gene, but insertion of the eIF4E cDNA in its genome (to obtain LMV-4E0) restores full infectivity in such varieties (48). The nonrecombinant LMV vector and LMV-4E0 were used as references throughout this work.
Lettuce plants (Trocadéro) primarily inoculated by biolistics (Helios Gene Gun; Bio-Rad, Hercules, CA) were homogenized in 25 mM Na2HPO4 containing 2% diethyldithiocarbamate and used to rub-inoculate assay plants as previously described (58). Symptoms were monitored daily, and the leaves were harvested for virus titration 15 days postinoculation (p.i.), which is the time point when symptoms and virus accumulation in susceptible plants reach their maximum. The infection parameters were compared within rather than across experiments. Reverse transcription-PCR detection of the viral progeny was performed as described previously (31), and the identity of each progeny to the mutant inoculated was assessed by restriction and, in some instances, sequence analysis. Semiquantitative double-antibody sandwich ELISA was performed as described previously (19) after 10× dilution of the plant extracts, so that the relationship between the A405 and the antigen concentration was linear.
Expression of wild-type and mutant eIF4E proteins in E. coli.
Using the Gateway Technology (Invitrogen), each of the mutated eIF4Es, as well as the wild type, was transferred from pENTR/D-TOPO into the pDEST17 destination vector, to allow production of N-terminal fusions with a six-His tag. The constructs were introduced into Escherichia coli (strain BL21-AI). Cells were grown at 37°C to an A600 of 0.4 in 100 ml of LB medium containing ampicillin. eIF4E expression was induced by addition of 0.2% arabinose and continued for 3 h at 30°C. The bacterial pellet was frozen and suspended in 4 ml Hex buffer, pH 7.8 (20 mM HEPES/KOH, 1 mM dithiothreitol, 0.2 mM EDTA, 0.2% Tween 20, 15% glycerol, 0.4 M NaCl). Lysozyme (0.5 mg/ml) and phenylmethylsulfonyl fluoride (1 mM) were added, and the cell suspension was maintained for 45 min at 4°C with gentle shaking. The suspension was sonicated and centrifuged (100,000 × g; 1 h; 4°C), and the supernatant was incubated for 45 min with 100 μl of Ni-nitrilotriacetic acid Sepharose CL-6B (Qiagen) equilibrated in Hex buffer. The beads were washed extensively with Hex containing 10 mM imidazole until the A280 reached a constant value. Proteins were eluted twice with 200 μl of HEX containing 250 mM imidazole. The His-tagged protein fractions were pooled and diluted in 1.2 ml of buffer A (20 mM HEPES-KOH, pH 7.5, 1 mM dithiothreitol).
The expression and purification of wheat eIF4G will be published elsewhere (K. Browning, personal communication).
Cap-binding, eIF4G-binding, and VPg-binding assays.
To evaluate their cap-binding abilities, the Ni2+-purified eIF4Es were subjected to m7GTP-Sepharose affinity chromatography. After calibration of the protein concentration, the proteins were incubated with 100 μl of m7GTP-Sepharose 4B (Amersham Biotech) at 4°C for 45 min. The beads were extensively washed with buffer A (see above), and the proteins retained were eluted with 100 μl of 100 μM m7GDP. The fractions obtained were analyzed by Western blotting using a rabbit polyclonal antibody raised against lettuce eIF4E.
The eIF4G-binding ability of purified eIF4E was evaluated by fluorescence spectroscopy using a synthetic peptide, pep4G (KKYSRDFLLKF), derived from A. thaliana eIF4G (At3g60240; GenBank accession no. NP_567095) and including the 4E-binding motif YXXXXLΦ (36), where X represents any amino acid and Φ is a hydrophobic amino acid. The fluorescence of tryptophans is sensitive to changes in their environment upon ligand binding. This feature was also exploited to quantitatively monitor the interactions between various forms of purified eIF4E and their ligands, m7GTP and VPg0. All spectra were acquired at 25°C on a Safas Xenius spectrophotometer (Monaco). The excitation wavelength was set to 280 nm. Measurements were made in 1 ml buffer A (see above) containing 100 mM KCl and 10% glycerol. The concentration of eIF4E was set to 0.5 μM. Tryptophan fluorescence quenching at 337 nm was monitored as a function of increasing amounts (0.005 to 5 μM) of ligand (either m7GTP, pep4G, or VPg) in the mixture. For each concentration of ligand added, the fluorescence value retained was the mean value from 2 minutes of collection. Dissociation constants were deduced from data sets including at least three independent titration experiments. Bovine serum albumin (Sigma) was used to control for nonspecific binding. The dissociation constants were deduced by fitting the raw data to the simple interaction model A + B ↔ AB (40).
An ELISA-derived interaction assay was used (12) to evaluate the ability of Ls-eIF4E proteins to bind the whole wheat 4G protein. Plates were coated with eIF4E proteins (wild type, E91A, W94A, G156A, and E157A) diluted (5 μg/ml) in carbonate buffer (Na2CO3, 15 mM; NaHCO3, 35 mM). After overnight incubation at 4°C, the plates were washed and saturated with 10% fetal bovine serum in phosphate-buffered saline (1 h at room temperature) before incubation with wheat eIF4G (10 μg/ml in phosphate-buffered saline-Tween fetal bovine serum 0, 2%; 1 h; 4°C). Interactions were revealed with polyclonal antibodies against eIF4G (1/1,000; 2 h; 37°C). Titrations of the coated proteins were assessed using polyclonal antibodies against eIF4E (1/1,000; 2 h; 37°C). Both titration and interactions were followed by anti-rabbit antibodies conjugated to alkaline phosphatase. An ELISA-derived interaction assay was also used to demonstrate the interaction of LMV VPg with Ls-eIF4E (63). This assay was used to monitor the interaction of LMV VPg with Ls-eIF4E0 and the surface mutants.
Yeast complementation.
Saccharomyces cerevisiae strain J055 (cdc33-Δ: LEU2 Leu2 ura3 his3 trp1 ade2 [YCp33supex-h4E URA3]) contains a deletion of the chromosomal gene coding for eIF4E, and therefore, its survival depends on the presence of plasmid YCp33supex-h4E URA3 containing a copy of the human eIF4E cDNA, under the control of the glucose-repressible, galactose-dependent GAL promoter (24).
The cDNAs encoding each of the Ls-eIF4E mutant forms and the natural allelic forms (4E0, 4E1, and 4E2) were independently transferred into the yeast-E. coli shuttle Trp-selectable vector p426GPD (45) for expression in yeast under the control of the constitutive GPD Gal10 promoter and then transferred into strain J055. Following selection on medium containing galactose, complementation for eIF4E function was performed on glucose-containing media.
Molecular modeling.
All models were built through the interface of Swiss-PDB viewer 3.7. Calculations were submitted to Swiss Model Workspace, an automated protein-modeling server (http://swissmodel.expasy.org). Amino acid candidates for mutations were chosen after modeling the lettuce eIF4E using the murine eIF4E (Protein Data Bank accession no. 1ej1) as a template. During finalization of this article, the 3D structure of wheat eIF4E (Protein Data Bank accession no. 2idr) was made available (42). The calculations were repeated using the wheat structure. The returned scores were not significantly different from those obtained with the murine protein as a template. Models were evaluated by means of the Model Assessment package provided by SWISS-MODEL (3, 22, 71).
RESULTS
Three classes of amino acid mutations were introduced into eIF4E.
The amino acid sequences of eIF4E from various eukaryotes have been identified, including yeast, human, mouse, rabbit, fruit fly, and several plant species, among which were the model A. thaliana (62) and the crop species wheat (39), lettuce (48), pepper (64), tomato (69), and pea (18). The most conserved region in the eIF4E sequences lies in the central region (Fig. 1), which is directly involved in the cap-binding process, whereas the N terminus varies in length, shows little or no conservation, and is not required for cap-dependent translation in vitro (38). Results from previous mutagenesis experiments done on mammalian (43) and yeast (2) eIF4Es, together with the 3D modeling of eIF4E complexed with analogues of its natural ligands, allowed progress in understanding the mechanistic bases for eIF4E interactions with the mRNA 5′ cap, translation initiation factors, and regulatory proteins (37, 38). The cocrystal structure of eIF4E with an eIF4G-derived oligopeptide mimicking the eIF4E-binding domain provides a basis to establish the points of contact between eIF4E and its molecular partners during translation initiation. On the convex side of eIF4E, highly conserved surface-accessible residues were identified, which participate in the eIF4G recognition domain (38). In murine eIF4E, changing W93 into alanine prevents interaction with human eIF4GI in vitro and in vivo (55). This was also demonstrated for yeast eIF4E (54). We used this information to design the first class of mutations directed against amino acids predicted to be involved in the eIF4G-binding ability of the lettuce eIF4E (E91A, G156A, E157A, and W94A) (Fig. 2A). In lettuce, according to this model, W94 and G156 are predicted to contribute to the nonpolar part of this surface, while E91 and E157 contribute to its acidic part. These conserved surface features, therefore, may be important for the lettuce eIF4E interactions with eIF4G.
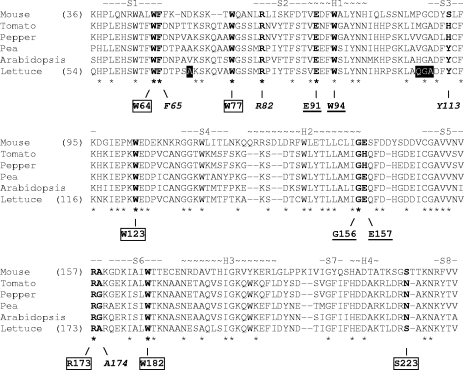
FIG. 1.
Positions of the mutated amino acids in the sequence of lettuce eIF4E. A multiple alignment of the eIF4E amino acid sequences from mouse (Mus musculus, P63073), tomato (Lycopersicon esculentum, AAF70507), pepper (Capsicum annuum, AAS68034), pea (Pisum sativum, AAR04332), A. thaliana (O23252), and lettuce (L. sativa, AAP86602) was performed (74). Conserved amino acids are marked by asterisks below the alignment, and the structural features in the mouse eIF4E (38) are shown and labeled above the alignment (-Sx-, beta sheet; ∼Hx∼, alpha-helix). The numbers in parentheses on the left of the alignment show the numbering of the mouse and lettuce eIF4E sequences. The amino acids that have been mutated in the lettuce eIF4E are shown in boldface; mutated amino acids predicted to be involved in the eIF4G-binding ability of eIF4E are underlined, mutated amino acids supposed to affect recognition of the cap structure are boxed, and mutated amino acids located at the outer surface of eIF4E are in italics. The black boxes indicate the locations of amino acids differing in Ls-eIF4E1 (QGA in Ls-eIF4E0 deleted and replaced by H in Ls-eIF4E1) and Ls-eIF4E2 (A in Ls-eIF4E0 replaced by P in Ls-eIF4E2).
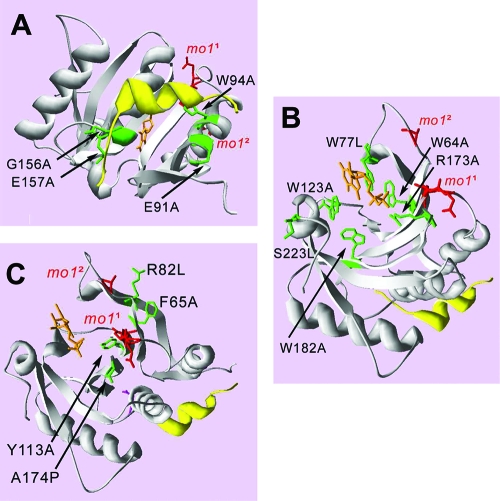
FIG. 2.
Positions of the mutated amino acids on the predicted 3D structure of lettuce eIF4E. The 3D structure of lettuce eIF4E was predicted based on its homology with murine eIF4E and is shown in a ribbon representation (48) in three different views (A to C). The positions of the cap analogue (orange nucleotide) and an eIF4G-derived peptide (yellow helix) are shown. The amino acids present in the mutant eIF4Es are displayed in green and labeled. The amino acids differing in eIF4E1 or eIF4E2 (mo11 and mo12 genotypes, respectively) compared to eIF4E0 (susceptible genotypes) are displayed in red and labeled mo11 or mo12, respectively. (A) Amino acids mutated in the domain of interaction with eIF4G. (B) Amino acids mutated in or near the cap-binding pocket. (C) Amino acids mutated at the outer surface, near the natural variations present in potyvirus resistance alleles.
The second class of mutations (W64A, W77L, W123A, R173A, W182A, and S223L) was designed to affect recognition of the cap structure (Fig. 2B). According to the 3D model of the mouse eIF4E (38), m7GDP recognition by lettuce eIF4E is mediated by π-π stacking between the guanine of the base of the cap structure and the side chains of the conserved residues W77 and W123. A van der Waals contact between the guanine N7-methyl group and the conserved W182 tightens this interaction. Other residues, like W64 and R173, are also involved in cap binding (38). Finally, it was proposed in mammals that phosphorylation of the serine corresponding to S223 in the lettuce eIF4E could have a stimulatory effect on m7GTP binding activity (38).
The third class of mutations (F65A, R82L, Y113A, and A174P) (Fig. 2C) was directed against amino acids predicted to map on the outer surface of eIF4E in the 3D vicinity of the amino acids related to potyvirus resistance in pepper, lettuce, and pea. This domain is located near the cap recognition pocket, on the face of eIF4E opposite to the eIF4G-binding site (48). Recently, the 3D structure data obtained for the wheat eIF4E confirmed that the residues known to be involved in potyvirus or bymovirus resistance are largely on the surface of eIF4E (42). In lettuce eIF4E, the only amino acid differing between eIF4E0 and eIF4E2, A70P (mo12 in Fig. 2), was predicted to be part of the loop between beta sheets 1 and 2, while the 3 amino acids Q108-G109-A110, replaced by a single histidine in eIF4E1 (mo11 in Fig. 2), are located in the neighboring loop.
F65 and R82 are predicted to lie in two beta sheets (S1 and S2, respectively) (Fig. 1) near the loop between S1 and S2, with their side chains protruding on the outer surface of eIF4E (Fig. 2C). Amino acid changes in this loop, near the cap recognition pocket, are directly associated with resistance to potyviruses in pepper, lettuce, and pea (18, 48, 64) and to bymoviruses in barley (73). In lettuce and pepper eIF4Es, another class of mutations associated with potyvirus resistance are located in the loop between H1 and S3 (Fig. 1). The Y113A mutation was designed to disturb the S3 short strand in order to slightly alter the surface topology in this area. Finally, to test the involvement of the third loop forming the edge of the cap-binding pocket, S5-S6, A174P was introduced.
LMV-0 is an isolate that is unable to accumulate and produce symptoms in a mo11 and mo12 background. All the recombinant LMVs are derived from this non-resistance-breaking isolate and contain the lettuce eIF4E coding region as a translational fusion between the viral P1 and HcPro domains that is proteolytically processed in vivo to yield the free proteins (19, 48). The insertion of wild-type cDNA or each of the 14 mutant eIF4E cDNAs resulted in infectious recombinant LMV, as shown by symptom development after biolistic inoculation of the susceptible lettuce variety Trocadéro (data not shown). This was also the case in another susceptible variety, Salinas, which was consistently used as a reference in this study (Table 1). Therefore, neither the wild-type nor the mutated eIF4E inserts seemed to have strong adverse effects per se on accumulation of the carrier LMV in vivo.
TABLE 1.
Summary of the properties of the LMV-4E mutants in susceptible and resistant lettuce
| Class of mutation | eIF4E mutation | Delay in symptom emergencea
|
Reduced virus accumulationb
|
||||
|---|---|---|---|---|---|---|---|
| Salinas (mo10) | Salinas 88 (mo12) | Mantilia (mo11) | Salinas (mo12) | Salinas 88 (mo10) | Mantilia (mo11) | ||
| eIF4G binding | E91A | − | − | − | − | − | > |
| W94A | − | − | − | − | − | − | |
| G156A | − | − | − | − | * | − | |
| E157A | − | − | − | − | − | − | |
| Cap binding | W64A | − | * | * | − | *** | ** |
| W77L | − | − | ** | * | − | *** | |
| W123A | − | − | − | − | − | − | |
| R173A | − | *** (WA)c | ** | − | *** (WA) | *** | |
| W182A | − | ** (WA) | ** | − | *** (WA) | *** | |
| S223L | − | − | − | > | > | − | |
| Outer surface | F65A | − | * | *** | − | *** | − |
| R82L | − | − | − | − | > | − | |
| Y113A | − | − | − | − | − | − | |
| A174P | − | − | − | − | > | − | |
−, no difference with LMV-4E0; *, delay of more than 3 days after emergence of symptoms induced by LMV-4E0; **, more than 6 days; ***, more than 14 days.
−, no significant difference in accumulation compared to LMV-4E0; *, reduction of more than 15%; **, more than 40%; ***, more than 70%; >, accumulation increased compared to that of LMV-4E0.
WA (when applied) means that in only one experiment of three were LMV accumulation and symptoms observed in the resistant lettuce, although in susceptible plants inoculated with the same LMV inoculum, infection occurred in three experiments of three.
Mutations in the eIF4G-binding area do not affect the ability of eIF4E to restore LMV susceptibility in mo1 plants.
The recombinant viruses LMV-4E-E91A, -W94A, -E157A, and -G156A were inoculated into susceptible or resistant lettuce plants. The timing of symptom appearance and the accumulation of virus progeny 15 days p.i. were compared to those of nonrecombinant LMV and LMV-4E0. The persistence of the eIF4E insert in the replicating virus during the course of all of the experiments was confirmed by reverse transcription-PCR and expression of the 4E mutants controlled by Western blot assays in Trocadéro (data not shown).
In all host genotypes, 7 to 10 days after inoculation with these constructs, a faint vein clearing became evident in emerging leaves, followed by mosaic symptoms 2 to 3 days later (data not shown). The timing and severity of the symptoms induced by these four mutants were similar to those of LMV-4E0 (Fig. 3). Indeed, the expression from its genome of Ls-eIF4E0 rendered LMV-4E0 able to overcome the resistance associated with mo11 and mo12 (induction of symptoms and virus accumulation), contrary to the wild-type nonresistant breaking LMV isolate from which it derives. Accumulation of the recombinant viruses was estimated by ELISA and expressed as a percentage of the average value measured for LMV in Salinas. In the susceptible genotypes Salinas and Trocadéro, all four mutants accumulated to similar levels (Fig. 4A and data not shown). In the mo1 varieties Mantilia, Salinas 88 (Fig. 4A and Table 1), and Floribibb (data not shown), there was no significant difference in accumulation, except in the case of LMV-4E-G156A, whose accumulation in Salinas 88 was 65% of the titer measured for LMV-4E0. Table 1 summarizes these results and shows that the four eIF4E mutations designed to affect eIF4G binding did not strongly affect the biological properties of the derived recombinant viruses. E91A, W94A, E157A, and G156A were therefore still able to restore full LMV susceptibility in lettuce genotypes containing mo11 and mo12, suggesting that in lettuce, these mutations do not interfere with the function of eIF4E in the LMV cycle. To confirm that the mutations introduced had the expected biochemical effect, the abilities of the mutant eIF4Es to bind eIF4G were assayed. All eIF4G factors, including plant factors, display an eIF4E-binding motif, YXXXXLΦ, where X represents any amino acid and Φ is a hydrophobic residue. In mammals, this motif interacts with the dorsal surface of eIF4E (37, 38). Oligopeptides containing this eIF4E-binding motif bind eIF4E (36). As no full-length lettuce eIF4G sequence is currently available, an oligopeptide, pep4G, was synthesized according to the Arabidopsis eIF4G sequence and was tested for its ability to bind lettuce eIF4E (40). In mammalian eIF4E, the homologue of the lettuce W94 is in close contact with the peptide (37, 38). Therefore, we hypothesized that binding of eIF4G or pep4G might affect the W94 fluorescence of lettuce eIF4E. We first tested this hypothesis using lettuce eIF4E produced as a histidine fusion in E. coli and purified by affinity chromatography. When increasing concentrations of pep4G were added to purified eIF4E0, a fluorescence decrease proportional to the amount of complex formed was observed, leading to a saturation plateau, while the fluorescence signal did not show significant changes upon addition of an unrelated protein, bovine serum albumin (data not shown). This indicated that pep4G affected the fluorescence of lettuce eIF4E in a specific manner, as expected if the close environment of W94 was changed by the interaction with this peptide. A dissociation constant of 0.11 ± 0.01 μM was deduced from the data collected with eIF4E0 (Table 2). A similar measurement of pep4G binding to a preformed eIF4E-m7GTP binary complex resulted in tighter binding of pep4G, with a Kd of 0.06 μM (data not shown); this was in accordance with data previously reported in the mouse (49) and confirmed that pep4G functionally mimicked the eIF4E-binding domain of eIF4G. The dissociation constants of the interactions between the mutant eIF4Es and pep4G showed that pep4G binding was affected quantitatively, but not abolished, for these three eIF4E mutants (Table 2).
FIG. 3.
Timing of symptom emergence for each mutant virus and LMV-4E0. Plants from the varieties Salinas 88 (mo12; hatched bars) and Mantilia (mo11; solid bars) were inoculated with LMV-4E carrying the different mutations. The average number of days for symptom emergence is given for each mutant, with the standard deviation, for at least three independent experiments, each involving at least three plants. The mutants are ordered in three classes as described in the text. The arrows indicate that in some experiments, symptoms never appeared, not allowing the calculation of a standard deviation; however, the number of days for symptom emergence observed in a single experiment where they did appear is given. The asterisks indicate that the data were significantly different from the LMV-4E0 reference (P < 0.01, according to a Student test; this could not be calculated for the bars with arrows). WT refers to the nonrecombinant LMV, which never expressed symptoms in Salinas 88 and Mantilia during the time courses of all experiments.
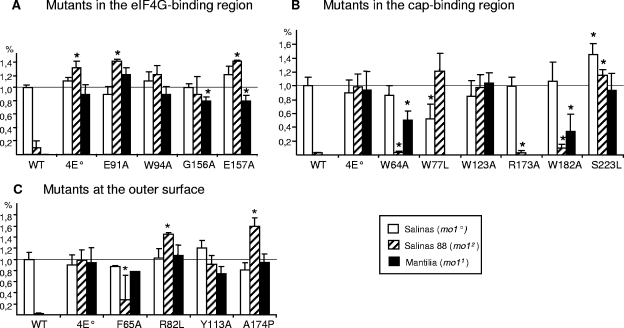
FIG. 4.
Abilities of the eIF4E mutants to restore LMV accumulation in a mo1 background. Plants from the varieties Salinas (susceptible; white bars), Salinas 88 (mo12; hatched bars), and Mantilia (mo11; solid bars) were assayed. The accumulation of the recombinant virus was determined by ELISA and expressed as a percentage of the average value measured for LMV in the variety Salinas. The samples (at least three plants for each virus) were collected from developmentally equivalent noninoculated leaves (level 3 from the top of the plant) at 15 days p.i. in at least three independent experiments. The “WT” and “4E0” labels refer to nonrecombinant LMV and LMV-4E0, respectively. (A) Mutants in the eIF4G-binding region. (B) Mutants in the cap-binding region. (C) Mutants at the outer surface of eIF4E. The asterisks indicate that the accumulation of the LMV recombinant virus was significantly different from that of LMV in Salinas (P < 0.01 according to a Student test). The error bars indicate standard deviations.
TABLE 2.
Dissociation constants of the different forms of eIF4E with their ligands
| eIF4E form |
Kd (106 mol/liter)
|
||
|---|---|---|---|
| m7GTP | VPg | pep4G | |
| eIF4E0 | 0.25 ± 0.01 | 0.27 ± 0.03 | 0.11 ± 0.01 |
| eIF4G-binding mutants | |||
| E91A | NDa | ND | 0.23 ± 0.01 |
| G156A | ND | ND | 0.30 ± 0.01 |
| E157A | ND | ND | 0.43 ± 0.02 |
| Outer-surface mutants | |||
| R82L | 0.41 ± 0.03 | 1.28 ± 0.07 | ND |
| F65A | 0.75 ± 0.05 | 2.41 ± 0.14 | ND |
| A174P | 1.56 ± 0.17 | 14.1 ± 2.3 | ND |
ND, not determined.
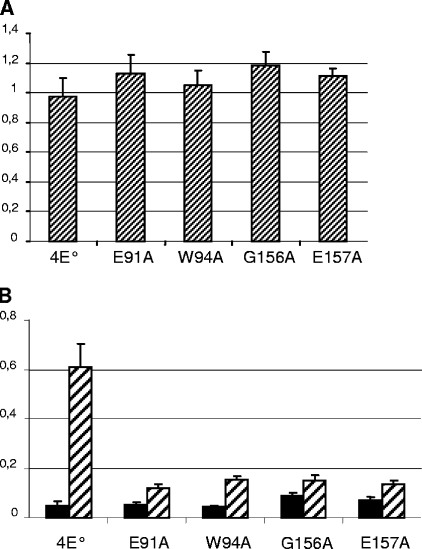
To confirm these results, and to test eIF4G binding of the mutant W94A, we developed an ELISA based on interaction of Ls-eIF4E with wheat eIF4G produced in E. coli. Figure 5 is representative of three independent experiments. Plate wells were precoated with purified eIF4E and incubated with purified wheat eIF4G. Figure 5A shows that the amounts of coated protein were equivalent for all proteins. Figure 5B shows that the wild-type Ls-eIF4E interacted with wheat eIF4G, while the optical-density signal was drastically reduced in the case of the interactions between eIF4E mutated forms and eIF4G. Therefore, wheat eIF4G binding of the four Ls-eIF4E mutants was strongly affected compared to wild-type Ls-eIF4E but not fully abolished.
FIG. 5.
Biochemical assessment of the effects of the mutations introduced in eIF4E on eIF4G binding. The results of an ELISA representative of three independent experiments are shown. Wells were precoated with 1 μg of eIF4E protein and incubated with either 2 μg of wheat eIF4G (hatched bars) or no protein (black bars). (A) Titration of the coated protein was carried out using polyclonal antibodies against eIF4E. (B) Binding complexes were detected using polyclonal antibodies against eIF4G. The absorbance values at 405 nm of four replicates and standard deviations are given.
Binding to lettuce eIF4G could not be assayed due to the unavailability of this protein in a purified form. However, our results with an oligopeptide and wheat eIF4G suggest that the mutations introduced had a debilitating effect on eIF4G binding, which contrasts with their lack of effect in the virus cycle. Therefore, either eIF4G binding is not required for the function of eIF4E in the virus cycle or the weak interaction still detectable in the mutants is sufficient to achieve this function. Both alternatives show that binding of eIF4G to eIF4E is not a limiting step in the virus cycle.
The abilities of eIF4E to bind a cap analogue and to restore LMV susceptibility in mo1 plants can be uncoupled.
In order to check the effect of the “cap-binding” mutations on the abilities of the corresponding eIF4Es to restore LMV susceptibility in resistant lettuce, the six recombinants, LMV-4E-W64A, -W77L, -W123A, -W182A, -R173A, and -S223L, were inoculated into susceptible mo11 and mo12 lettuce plants, and the timing of symptom emergence, virus accumulation, and the stability of the inserted sequence were monitored as described above. In both mo11 and mo12 genotypes, typical LMV infection symptoms appeared in plants inoculated with LMV-4E0 7 to 14 days after inoculation, while as expected, nonrecombinant LMV caused symptoms only in the susceptible genotype.
According to their capabilities to restore the ability of LMV to cause symptoms in mo11 and mo12 genotypes, the “cap-binding” mutants could be separated into two main groups. The first group includes W123A and S223L, which have the same ability as eIF4E0 to restore LMV symptom expression (Fig. 3) and accumulation (Fig. 4B and Table 1) in mo11 and mo12 plants. The second group of “cap-binding” mutants includes W64A, W77L, R173A, and W182A, which displayed symptoms with a delay ranging from 3 to 16 days compared to LMV-4E0 in the mo1 genotypes Salinas 88 and Mantilia (Fig. 3) and Floribibb (data not shown). Symptoms were consistently restored by these mutants in Mantilia, whereas in Salinas 88, symptoms developed only once in three experiments for mutants R173A and W182A and twice for W64A. The absence or delay in symptom emergence for W64A, W182A, and R173A was correlated with a very weak accumulation of the corresponding recombinant LMV in Salinas 88 (Fig. 4B and Table 1). Unlike the previous recombinants, LMV-4E-W77L was more affected in Mantilia and Floribibb (mo11) than in Salinas 88 (mo12), as it accumulated at a level comparable to that of LMV-4E0 in Salinas 88 while no accumulation was detected in Mantilia (Fig. 4B and Table 1) and Floribibb (data not shown).
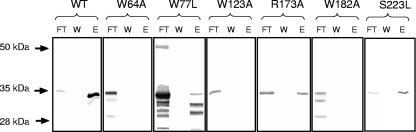
In summary, the “cap-binding” mutants W64A, W77L, W182A, and R173A were affected, although in various ways, in their abilities to restore LMV infectivity in the resistant genotypes, suggesting that amino acids W64, W77, W182, and R173 are probably involved in the function of eIF4E in the virus cycle. The abilities of these mutations to interfere with cap binding were confirmed by affinity chromatography to a cap analogue. For this purpose, mutant and nonmutant eIF4Es were produced in E. coli as described above. After purification by ion metal affinity chromatography on a Ni2+-bearing substrate, the cap-binding ability of each eIF4E protein was examined by affinity chromatography using m7GTP-Sepharose 4B (Fig. 6). The abilities of mutants W64A, W123A, and W182A to bind the cap analogue were completely abolished, confirming the role in cap binding of these amino acids located within the cap-binding pocket. W77L was aimed at one of the base-stacking tryptophans. Its association with free m7GTP was quantified using a spectroscopic method analogous to the one used for quantifying the pep4G-eIF4E interaction. Its ability to bind the cap analogue was affected (Kd = 1.0 ± 0.2 μM) compared to eIF4E0 (Kd = 0.25 ± 0.01 μM) but not completely abolished. Finally, the ability of R173A (Kd = 0.3 ± 0.05 μM) to bind the cap analogue was not affected despite its mutation in the cap-binding pocket at a position predicted to be indirectly involved in cap binding, based on homology with the murine eIF4E.
FIG. 6.
Effects of the mutations introduced on eIF4E cap binding. Mutant (W64A, W77L, W123A, R173A, W182A, and S223L) and wild-type (wt) lettuce eIF4E0 proteins were expressed as histidine fusions in E. coli, purified by Ni2+ affinity chromatography, assayed for cap binding by m7GTP affinity chromatography, and revealed by Western blotting using an antibody directed against lettuce eIF4E. FT, first washing of the m7GTP affinity chromatography, immediately after loading; W, last washing; E, eluate. The electrophoretic positions of molecular mass markers are shown on the left.
Therefore, in mutant R173A, the ability to rescue LMV accumulation in a mo1 background was absent despite a full cap-binding capacity. On the other hand, W123A was able to restore LMV accumulation while apparently completely devoid of any cap-binding capability. Together, these observations suggested that the role of eIF4E in the LMV cycle could be functionally uncoupled from cap binding.
Mutations at the outer surface of eIF4E affect its ability to restore LMV accumulation in a mo1 background.
In order to check the effect of the third class of mutations on the capability of the corresponding eIF4Es to restore LMV susceptibility in resistant lettuce, four recombinant viruses (LMV-4E-F65A, -R82L, -Y113A, and -A174P) were inoculated into susceptible, mo11 and mo12 lettuce plants and evaluated as described above. As before, no adverse effect of the insertion was detected in the susceptible variety Salinas (Table 1).
In mo11 and mo12 genotypes, two mutations, Y113A and A174P, had no effect compared to wild-type eIF4E (Fig. 3 and 4C, Table 1, and data not shown for Floribibb). LMV-4E-R82L behaved essentially like LMV-4E0, except in one experiment of three, when it accumulated to low levels and did not induce symptoms in Salinas 88 and Mantilia (Table 1). LMV-4E-F65A was affected in its ability to induce symptoms in both Salinas 88 and Mantilia (Fig. 3 and Table 1). Furthermore, in Salinas 88, accumulation levels were highly variable from one plant to another (Fig. 4C), although neither modification nor deletion in the Ls-eIF4E mutant insert had occurred in the viral progeny in those plants. In summary (Table 1), amino acids F65 and, to a lesser extent, R82 seemed to be involved in the ability of eIF4E0 to complement LMV infection in resistant lettuce. Since cap binding could have been indirectly affected by these mutations, which occurred at the outer surface of eIF4E, we assayed the mutants' cap-binding abilities as described above. The mutants F65A and R82L were not adversely affected for cap binding (Kd = 0.75 ± 0.05 μM and Kd = 0.41 ± 0.03 μM, respectively), and the mutant A174P was significantly affected but still retained its ability to bind the cap analogue (Kd = 1,56 ± 0.17 μM) (Table 2).
The abilities of the mutations F65A, R82L, and A174P to interfere with VPg binding were tested in vitro. An ELISA was developed, based on the interaction of the recombinant protein Ls-eIF4E with VPg produced in E. coli (63). Plate wells were coated with purified eIF4E (wild-type or mutant form) and incubated with purified VPg. We checked that the amounts of eIF4E were equivalent for all proteins. Five independent experiments were done, showing that the optical-density signals obtained for the interaction between the wild-type Ls-eIF4E and VPg were not significantly different from those obtained with the F65A, R82L, and A174P mutants. These mutations did not abolish the interaction between Ls-eIF4E and VPg in this ELISA system (data not shown).
Fluorescence spectroscopy was performed to compare the strengths of VPg binding to these mutants on a more quantitative basis (40, 63). This method confirmed the ELISA in that the three F65A, R82L, and A174P mutants were able to bind VPg. However, their affinities for the virus protein were significantly decreased with respect to the wild-type form of eIF4E0. The replacements of F65 and R82 on the protein were associated with 10- and 5-fold increases of the dissociation constants, respectively (Table 2). This effect might be correlated with the biological observations mentioned above for F65A and, to a lesser extent, for R82L. Surprisingly, the effect of A174 substitution on the binding, although not correlated with a clear biological effect, was even more drastic. In vitro measurements do not mimic the plant biological environment. Moreover, dissociation constants in the micromolar range still reflect rather strong interactions. This might be sufficient to ensure the eIF4E-VPg complex-associated functions in planta. However, the F65A mutation, which affects eIF4E structure in another location, might induce the formation of a totally nonfunctional complex between eIF4E and VPg.
Assessment of the functionalities of eIF4E mutants by complementation in yeast.
The functionalities of the Ls-eIF4E mutants for translation initiation and/or other essential functions in vivo were evaluated in yeast (S. cerevisiae), using a rescue assay. For this purpose, the haploid yeast strain J055 expressing the human eIF4E was transformed with each of the mutant forms. Since the production of human eIF4E in this system is galactose dependent and undetectable in medium containing glucose, the strain allows the assessment of the functionality of any cDNA encoding a functional eIF4E under the control of a promoter active in glucose (24).
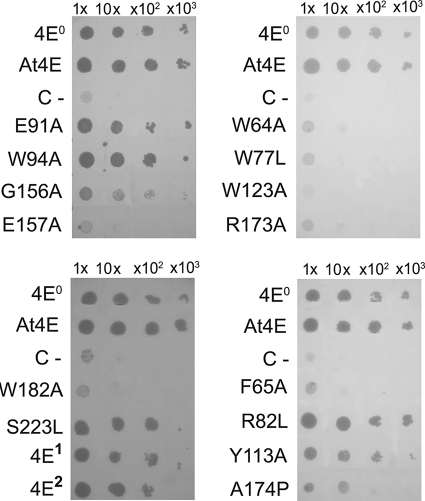
Figure 7 shows that the three natural Ls-eIF4E cDNAs (4E0, 4E1, and 4E2) that correspond, respectively, to eIF4E from susceptible (mo10) and resistant (mo11 and mo12) lettuce allowed the functional complementation of yeast clones growing on glucose, in a manner similar to that of A. thaliana eIF4E1 (At4E). Therefore, the Ls-eIF4E factors are able to fulfill all the functions related to growth in a heterologous species, including mRNA translation, and the amino acid differences in Ls-eIF4E1 and Ls-eIF4E2 do not impede essential eIF4E functions.
FIG. 7.
Assessment of the abilities of Ls-eIF4E wild-type and mutant forms to promote mRNA translation in S. cerevisiae. The S. cerevisiae strain J055 was transformed with the Ura-selectable nonrecombinant vector p426GPD (negative control; C-), p426GPD containing cDNA encoding Ls-eIF4E derivatives, and A. thaliana eIF4E1 (At4E; positive control). The growth of pure (1×) and 10×, 100×, and 1,000× dilution cultures was assessed for each yeast clone following incubation at 30°C for 96 h in medium containing glucose (synthetic dropout glu −Ura −Trp). The same dilutions were assessed in parallel in medium containing galactose (SD gal/raf −Ura −Trp) to check that all the transformed cells grew equivalently in a nonselective medium (data not shown). At least four independent experiments were done and produced similar results.
Eight of the Ls-eIF4E mutants did not allow yeast growth (Fig. 7). Indeed, the eIF4G-binding mutant E157A; five cap-binding mutants, W64A, W77L, W123A, R173A, and W182A; and two outer-surface mutants, F65A and A174P, were not functional in supporting mRNA translation and growth in yeast.
Amino acids W64, W77, R173, and W182 seemed to be involved in both the function of eIF4E in the virus cycle and mRNA translation initiation (Table 3). Our affinity chromatography assays showed that these mutations interfered with cap binding, which could explain the lack of complementation for yeast mRNA translation. However, despite a full cap-binding capacity, the mutant R173A was not able to rescue both yeast growth and LMV accumulation, suggesting that another property of eIF4E could be affected to result in the loss of its main biological function. Furthermore, eIF4E1 and eIF4E2 failed to complement virus infection, although they were totally functional for yeast growth, unlike the mutants E157A, W123A, and A174P, which could not support yeast growth but allowed virus replication (Table 3). Together, all these observations suggested that the roles of eIF4E in the potyvirus cycle and in mRNA translation could be functionally uncoupled.
TABLE 3.
Summary of the biochemical and biological properties of Ls-eIF4E mutants
| Class of mutation | eIF4E form | In vitro cap bindinga,b | In vitro eIF4G bindingb | Virus rescuec | Yeast rescued |
|---|---|---|---|---|---|
| mo10 allele | eIF4E0 | + | + | + | + |
| mo11 allele | eIF4E1 | ± | NT | − | + |
| mo12 allele | eIF4E2 | ± | NT | − | + |
| eIF4G binding | E91A | + | ± | + | + |
| W94A | + | ± | + | + | |
| G156A | + | ± | + | + | |
| E157A | + | ± | + | −* | |
| Cap binding | W64A | − | NT | − | − |
| W77L | ± | NT | − | − | |
| W123A | − | NT | + | − | |
| R173A | + | NT | − | − | |
| W182A | − | NT | − | − | |
| S223L | + | NT | + | + | |
| Outer surface | F65A | ± | NT | − | − |
| R82L | + | NT | + | + | |
| Y113A | + | NT | + | + | |
| A174P | ± | NT | + | −* |
Estimated by cap affinity chromatography.
+, present; −, absent; ±, binding is affected, but not fully abolished; NT, not tested.
+, the eIF4E form is able to complement LMV infection in resistant plants; −, no complementation occurs.
10× serial dilutions of yeast strain J055 transformed with plasmids p426GPD-4E were plated on glucose selective medium. +, 100× or 1,000× dilutions grew on glucose-containing medium; -, these dilutions did not grow (behavior was identical to that of the negative control, cells transformed with the plasmid p426GPD); *, in only one experiment of 4 did cells grow for a 10× dilution, but none grew for the 100× and 1,000× serial dilutions.
DISCUSSION
This study, which combined biological and biochemical analyses, reported the mutagenesis of eIF4E0 isolated from a susceptible lettuce variety, and the ectopic expression of the mutant forms from the LMV genome, to identify eIF4E amino acids that are important for the virus cycle. In parallel, the yeast “knockout-and-rescue” system was used to assess the functionality of each of the mutant forms for mRNA translation initiation and/or other essential functions in vivo.
Among the six mutations designed to affect the cap-binding ability of lettuce eIF4E, four highly conserved tryptophans were changed to nonpolar residues. As expected, the cap-binding abilities of the mutants W64A, W123A, and W182A were significantly affected, and accordingly, these mutations impeded essential eIF4E functions in S. cerevisiae. In particular, we confirmed the importance for cap binding of W64 and W182 of the lettuce eIF4E, which correspond to W46 and W166 in yeast eIF4E, 2 residues required for cap binding (2). The mutagenesis of W77 into leucine, one of the residues directly involved in cap binding through base stacking and apparently present in all eukaryote eIF4Es (26), was associated with a strong decrease, but not a complete loss, of the cap-binding ability; however, it was sufficient to impede complementation in yeast. Our quantification experiment is in agreement with the 80% decrease in cap-binding ability previously reported for the equivalent mutation performed in human eIF4E (43). The S223L mutation did not affect the ability of eIF4E to bind the cap analogue. Phosphorylation of the homologous serine in mammalian eIF4E is thought to regulate cap binding in a controversial fashion (41, 70). However, this could not be addressed in our bacterial expression system, devoid of proper phosphorylation ability, but our chromatography data confirm that cap binding can occur even without a serine at this position. The R173A mutation did not affect cap binding, despite its predicted role in stabilizing the cap-binding pocket in the mouse eIF4E (38). To our knowledge, this property has not been confirmed experimentally, and our experiments show that replacement of R173 with an uncharged amino acid results in full capacity of Ls-eIF4E to bind the cap in vitro. However, the fact that this mutation abolishes yeast complementation suggests that this amino acid position is required for eIF4E activity in vivo.
Among the mutations designed to alter eIF4G binding, E91A, W94A, and G156A affected but did not abolish the interaction with pep4G or wheat eIF4G in our in vitro assays. These amino acids correspond, respectively, to E72, W75, and G139 in yeast eIF4E and were shown to be key residues involved in the eIF4G binding site, as their replacement by alanine or phenylalanine disrupted eIF4E-eIF4G association (54). Here, we showed that the equivalent residues in lettuce eIF4E are similarly important for eIF4G binding in vitro. However, these mutations did not affect the functionality of lettuce eIF4E in supporting yeast growth. The replacement of E157 by an alanine in E157A affected eIF4G binding and also abolished yeast growth, unlike the previous mutants. Surprisingly, mutagenesis of yeast eIF4E at the equivalent position (E140) had no effect on eIF4G binding in vitro (54). This apparent discrepancy between yeast and lettuce could be explained by differences in the biochemical assays used to test eIF4G binding in vitro.
The F65A mutation is close to the natural sequence variations linked to potyvirus resistance in several crop species (18, 48, 64, 73). This substitution had a negative effect on virus replication in plants, cap binding (3-fold reduction), VPg binding (10-fold reduction), and yeast complementation. F65 maps on the protein surface opposite to the opening of the cap-binding pocket. Modeling the lettuce eIF4E using the wheat eIF4E structure (42) suggests that the F65A substitution impacts the orientation of Y113 and, in turn, the bottom of the cap-binding pocket. This could be related to the lack of yeast complementation by F65A and, taken together, suggests a mechanism by which a mutation at the external surface of eIF4E can impact internal biochemical properties and the related in vivo properties of the protein. It is noteworthy that the Y113A mutation did not impact negatively on virus infection, while it is located in the same loop, H1-S3, as the natural variation found in mo11. This suggests that not all substitutions in this loop affect the ability of eIF4E to promote potyvirus infection.
Several possible roles of eIF4E in the potyvirus cycle have been proposed based on its known biological and biochemical features (18, 32). The main function of eIF4E is to bind the cap at the 5′ mRNA end, promoting its recruitment by the ribosomal machinery, although other functions in cell growth and the cell cycle have been identified (13). As the natural sbm1- and pvr1-encoded eIF4E variants associated with potyvirus resistance in pea and pepper are devoid of cap-binding ability (18, 27), a link, structural or functional, was suggested between these two properties. The experiments reported here functionally dissociated cap binding from the promotion of potyvirus infection, since the eIF4E W123A mutant clearly demonstrated that it could not bind the cap structure and complement yeast, whereas it was fully able to support LMV replication. In this respect, our results extend those of Kang et al. (27) showing that the pvr21- and pvr22-encoded eIF4E proteins maintain in vitro cap-binding activities. Therefore, although a number of natural eIF4E variants (18, 27) and in vitro mutants (this work) accumulate both defects, in that they fail both in binding cap structures and in supporting potyvirus infection, the function of eIF4E in the potyvirus cycle might be distinct from its physiological function of binding the cap structure at the 5′ ends of mRNAs to initiate translation.
In the infection cycle, it was proposed that eIF4E could play the same role as in translation of the cellular mRNAs (10), through interaction with VPg, which would functionally replace the cap in potyvirus mRNAs (30, 32). VPg has been shown to interact with eIF4E in several plant-potyvirus systems (7, 33, 63, 69, 75) and to play a key role in overcoming several unrelated host resistance genes from distinct plant families (8, 11, 44, 56, 57). This could be simply interpreted as the 5′ VPg of potyvirus RNAs functionally playing a role equivalent to the 5′ cap of cellular mRNAs, as recently shown for an animal calicivirus (21). The translation of several potyvirus RNAs is cap independent (6, 35, 50) but is nevertheless eIF4G dependent (17). The implication of eIF4G in plant virus infection was demonstrated in the case of natural resistance in rice against a sobemovirus, Rice yellow mottle virus (1), and in reduction of the accumulation of a cucumovirus, Cucumber mosaic virus, in A. thaliana cum2 mutants (78). Furthermore, we recently showed that in A. thaliana eIF4E and eIF4G factors are both recruited for potyvirus infection, including LMV (47).
A possible implication of eIF4E in the virus cycle could be to allow RNA circularization by interaction of the 5′ VPg with the 3′ poly(A), mediated by the same protein complex as in mRNA translation, namely, eIF4E-eIF4G-PABP. Besides a role in translation, genome circularization may be required for virus RNA replication or other processes of the virus cycle. Indeed, genome circularization is an important feature of the replication of picornaviruses (23), relatives of potyviruses that infect animal hosts. Three mutations of lettuce eIF4E that had adverse effects on the binding of pep4G and wheat eIF4G in vitro were associated with an apparently full functionality for the virus cycle and mRNA translation in yeast. This could suggest that genome circularization, if it is required for potyvirus translation and/or replication, might not need the assembly of an eIF4F complex through a physical interaction between eIF4E and eIF4G. Alternatively, assembly of eIF4F in vivo might be less affected than it is in vitro by the eIF4E mutations introduced, in relation, for instance, with different biochemical and/or structural environments. An interaction between PABP and the TuMV VPg-Pro was demonstrated in planta (34), suggesting that potyvirus RNA circularization could be a shortcut for eIF4F, leaving another functional meaning for the binding of eIF4E to VPg.
Through its interaction with VPg and possibly other host and virus factors, eIF4E might be involved in the control of the successive fates encountered by the viral RNA, such as intracellular and cell-to-cell trafficking or encapsidation. In pea and pepper, eIF4E assists potyvirus cell-to-cell movement (4, 18). In mo1 lettuce, grafting experiments showed that while the systemic movement of LMV is severely impaired, neither phloem loading nor phloem unloading is completely inhibited (19, 20).
These phenotypes could result either from a direct involvement of eIF4E in movement or from an effect on virus accumulation that might have pleiotropic effects on virus invasion, such as lack of replication in defined tissues and cells. The eIF4E mutants characterized in this work will be useful to unravel unknown aspects of the relationships between eIF4E and its cellular and viral ligands.
Acknowledgments
This work was partially supported by EPR Aquitaine (reference 20000307004), by Génoplante (NewVir GNP03 and Transvir GNP05003G), and by the National Agency for Research (Poty4E, reference ANR-05-BLAN-0302-01). V.N. was supported by a fellowship from the French ARN (Association pour la Recherche sur les Nicotianae).
We thank Claude Manigand (CNRS-Université Bordeaux 1-ENITAB, IECB) for the synthesis of the oligopeptide pep4G, John M. X. Hughes (Manchester University) for the yeast strain J055, and Karen Browning (University of Texas at Austin) for purified wheat eIF4G and rabbit antibodies to wheat eIF4G and eIF4E. We are grateful to Thierry Mauduit and Marylin Roncoroni for taking care of the plants and to all members of the “IPV” group for useful discussions.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Albar, L., M. Bangratz-Reyser, E. Hebrard, M. N. Ndjiondjop, M. Jones, and A. Ghesquiere. 2006. Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 47417-426. [DOI] [PubMed] [Google Scholar]
- 2.Altmann, M., I. Edery, H. Trachsel, and N. Sonenberg. 1988. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E. Effects on cap binding activity. J. Biol. Chem. 26317229-17232. [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22195-201. [DOI] [PubMed] [Google Scholar]
- 4.Arroyo, R., M. J. Soto, J. M. Martinez-Zapater, and F. Ponz. 1996. Impaired cell-to-cell movement of Potato virus Y in pepper plants carrying the ya (pvr21) resistance gene. Mol. Plant-Microbe Interact. 9314-318. [Google Scholar]
- 5.Bannerot, H., L. Boulidard, J. Marrou, and M. Duteil. 1969. Etude de la tolérance au virus de la mosaïque de la laitue chez la variété Gallega de Invierno. Ann. Phytopathol. 1219-226. [Google Scholar]
- 6.Basso, J., P. Dallaire, P. J. Charest, Y. Devantier, and J. F. Laliberte. 1994. Evidence for an internal ribosome entry site within the 5′ non-translated region of Turnip mosaic potyvirus RNA. J. Gen. Virol. 753157-3165. [DOI] [PubMed] [Google Scholar]
- 7.Beauchemin, C., N. Boutet, and J. F. Laliberte. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgstrom, B., and I. E. Johansen. 2001. Mutations in pea seedborne mosaic virus genome-linked protein VPg after pathotype-specific virulence in Pisum sativum. Mol. Plant-Microbe Interact. 14707-714. [DOI] [PubMed] [Google Scholar]
- 9.Bos, L., N. Huijberts, and C. Cuperus. 1994. Further observations on variation of lettuce mosaic virus in relation to lettuce (Lactuca sativa), and a discussion of resistance terminology. Eur. J. Plant Pathol. 100293-314. [Google Scholar]
- 10.Browning, K. S. 2004. Plant translation initiation factors: it is not easy to be green. Biochem. Soc. Trans. 32589-591. [DOI] [PubMed] [Google Scholar]
- 11.Charron, C., M. Nicolai, J. L. Gallois, C. Robaglia, B. Moury, A. Palloix, and C. Caranta. 2008. Natural variation and functional analyses provide evidence for coevolution between plant eIF4E and potyviral VPg. Plant J. 5456-68. [DOI] [PubMed] [Google Scholar]
- 12.Clark, M. F., and A. N. Adams. 1977. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 34475-483. [DOI] [PubMed] [Google Scholar]
- 13.Culjkovic, B., I. Topisirovic, and K. L. Borden. 2007. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 665-69. [DOI] [PubMed] [Google Scholar]
- 14.Decroocq, V., O. Sicard, J. M. Alamillo, M. Lansac, J. P. Eyquard, J. A. Garcia, T. Candresse, O. Le Gall, and F. Revers. 2006. Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 19541-549. [DOI] [PubMed] [Google Scholar]
- 15.Dinant, S., and H. Lot. 1992. Lettuce mosaic virus: a review. Plant Pathol. 41528-542. [Google Scholar]
- 16.Duprat, A., C. Caranta, F. Revers, B. Menand, K. S. Browning, and C. Robaglia. 2002. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32927-934. [DOI] [PubMed] [Google Scholar]
- 17.Gallie, D. R. 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 7512141-12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, Z., E. Johansen, S. Eyers, C. L. Thomas, T. H. Noel Ellis, and A. J. Maule. 2004. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40376-385. [DOI] [PubMed] [Google Scholar]
- 19.German-Retana, S., E. Redondo, G. Tavert-Roudet, O. Le Gall, and T. Candresse. 2003. Introduction of a NIa proteinase cleavage site between the reporter gene and HC-Pro only partially restores the biological properties of GUS- or GFP-tagged LMV. Virus Res. 98151-162. [DOI] [PubMed] [Google Scholar]
- 20.German-Retana, S., J. Walter, and O. Le Gall. 2008. Lettuce mosaic virus: from pathogen diversity to host interactors. Mol. Plant Pathol. 9127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow, I., Y. Chaudhry, I. Gioldasi, A. Gerondopoulos, A. Natoni, L. Labrie, J. F. Laliberte, and L. Roberts. 2005. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 6968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 23.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, J. M., M. Ptushkina, M. M. Karim, N. Koloteva, T. von der Haar, and J. E. McCarthy. 1999. Translational repression by human 4E-BP1 in yeast specifically requires human eIF4E as target. J. Biol. Chem. 2743261-3264. [DOI] [PubMed] [Google Scholar]
- 25.Irwin, S. V., R. V. Kesseli, W. Waycott, E. J. Ryder, J. J. Cho, and R. W. Michelmore. 1999. Identification of PCR-based markers flanking the recessive LMV resistance gene mo1 in an intraspecific cross in lettuce. Genome 42982-986. [Google Scholar]
- 26.Joshi, B., K. Lee, D. L. Maeder, and R. Jagus. 2005. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, B. C., I. Yeam, J. D. Frantz, J. F. Murphy, and M. M. Jahn. 2005. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42392-405. [DOI] [PubMed] [Google Scholar]
- 28.Kang, B. C., I. Yeam, and M. M. Jahn. 2005. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43581-621. [DOI] [PubMed] [Google Scholar]
- 29.Kanyuka, K., A. Druka, D. Caldwell, A. Tymon, N. McCallum, R. Waugh, and M. Adams. 2005. Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 6449-458. [DOI] [PubMed] [Google Scholar]
- 30.Khan, M. A., H. Miyoshi, D. R. Gallie, and D. J. Goss. 2008. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 2831340-1349. [DOI] [PubMed] [Google Scholar]
- 31.Krause-Sakate, R., O. Le Gall, H. Fakhfakh, M. Peypelut, M. Marrakchi, C. Varveri, M. A. Pavan, S. Souche, H. Lot, F. M. Zerbini, and T. Candresse. 2002. Molecular characterization of Lettuce mosaic virus field isolates reveals a distinct and widespread type of resistance-breaking isolate: LMV-Most. Phytopathology 92563-572. [DOI] [PubMed] [Google Scholar]
- 32.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 121046-1051. [DOI] [PubMed] [Google Scholar]
- 33.Leonard, S., D. Plante, S. Wittmann, N. Daigneault, M. G. Fortin, and J. F. Laliberte. 2000. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 747730-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard, S., C. Viel, C. Beauchemin, N. Daigneault, M. G. Fortin, and J.-F. Laliberte. 2004. Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 851055-1063. [DOI] [PubMed] [Google Scholar]
- 35.Levis, C., and S. Astier-Manifacier. 1993. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes 7367-379. [DOI] [PubMed] [Google Scholar]
- 36.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 154990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcotrigiano, J., A.-C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3707-716. [DOI] [PubMed] [Google Scholar]
- 38.Marcotrigiano, J., A.-C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89951-961. [DOI] [PubMed] [Google Scholar]
- 39.Metz, A. M., R. T. Timmer, and K. S. Browning. 1992. Isolation and sequence of a cDNA encoding the cap binding protein of wheat eukaryotic protein synthesis initiation factor 4F. Nucleic Acids Res. 204096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michon, T., Y. Estevez, J. Walter, S. German-Retana, and O. Le Gall. 2006. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2731312-1322. [DOI] [PubMed] [Google Scholar]
- 41.Minich, W. B., M. L. Balasta, D. J. Goss, and R. E. Rhoads. 1994. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl. Acad. Sci. USA 917668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monzingo, A. F., S. Dhaliwal, A. Dutt-Chaudhuri, A. Lyon, J. H. Sadow, D. W. Hoffman, J. D. Robertus, and K. S. Browning. 2007. The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 1431504-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morino, S., H. Hazama, M. Ozaki, Y. Teraoka, S. Shibata, M. Doi, H. Ueda, T. Ishida, and S. Uesugi. 1996. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 239597-601. [DOI] [PubMed] [Google Scholar]
- 44.Moury, B., C. Morel, E. Johansen, L. Guilbaud, S. Souche, V. Ayme, C. Caranta, A. Palloix, and M. Jacquemond. 2004. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. 17322-329. [DOI] [PubMed] [Google Scholar]
- 45.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156119-122. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, J. F., R. E. Rhoads, A. G. Hunt, and J. G. Shaw. 1990. The VPg of tobacco etch virus RNA is the 49-kDa proteinase or the N-terminal 24-kDa part of the proteinase. Virology 178285-288. [DOI] [PubMed] [Google Scholar]
- 47.Nicaise, V., J. L. Gallois, F. Chafiai, L. M. Allen, V. Schurdi-Levraud, K. S. Browning, T. Candresse, C. Caranta, O. Le Gall, and S. German-Retana. 2007. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 5811041-1046. [DOI] [PubMed] [Google Scholar]
- 48.Nicaise, V., S. German-Retana, R. Sanjuan, M. P. Dubrana, M. Mazier, B. Maisonneuve, T. Candresse, C. Caranta, and O. LeGall. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 1321272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niedzwiecka, A., J. Marcotrigiano, J. Stepinski, M. Jankowska-Anyszka, A. Wyslouch-Cieszynska, M. Dadlez, A.-C. Gingras, P. Mak, E. Darzynkiewicz, and N. Sonenberg. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319615-635. [DOI] [PubMed] [Google Scholar]
- 50.Niepel, M., and D. R. Gallie. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J. Virol. 739080-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nieto, C., M. Morales, G. Orjeda, C. Clepet, A. Monfort, B. Sturbois, P. Puigdomenech, M. Pitrat, M. Caboche, C. Dogimont, J. Garcia-Mas, M. A. Aranda, and A. Bendahmane. 2006. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 48452-462. [DOI] [PubMed] [Google Scholar]
- 52.Pink, D. A. C., D. Kostova, and D. G. A. Walkey. 1992. Differentiation of pathotypes of lettuce mosaic virus. Plant Pathol. 415-12. [Google Scholar]
- 53.Provvidenti, R., and R. O. Hampton. 1992. Sources of resistance to viruses in the Potyviridae. Arch. Virol. Suppl. 5189-211. [DOI] [PubMed] [Google Scholar]
- 54.Ptushkina, M., T. von der Haar, S. Vasilescu, R. Frank, R. Birkenhager, and J. E. McCarthy. 1998. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 174798-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajamaki, M. L., and J. P. Valkonen. 1999. The 6K2 protein and the VPg of Potato virus A are determinants of systemic infection in Nicandra physaloides. Mol. Plant-Microbe Interact. 121074-1081. [DOI] [PubMed] [Google Scholar]
- 57.Rajamaki, M. L., and J. P. Valkonen. 2002. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol. Plant-Microbe Interact. 15138-149. [DOI] [PubMed] [Google Scholar]
- 58.Redondo, E., R. Krause-Sakate, S. J. Yang, H. Lot, O. Le Gall, and T. Candresse. 2001. Lettuce mosaic virus (LMV) pathogenicity determinants in susceptible and tolerant lettuce varieties map to different regions of the viral genome. Mol. Plant-Microbe Interact. 14804-810. [DOI] [PubMed] [Google Scholar]
- 59.Reichmann, J. L., S. Lain, and J. A. Garcia. 1992. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 731-16. [DOI] [PubMed] [Google Scholar]
- 60.Revers, F., H. Lot, S. Souche, O. Le Gall, T. Candresse, and J. Dunez. 1997. Biological and molecular variability of Lettuce mosaic virus isolates. Phytopathology 87397-403. [DOI] [PubMed] [Google Scholar]
- 61.Robaglia, C., and C. Caranta. 2006. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 1140-45. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez, C. M., M. A. Freire, C. Camilleri, and C. Robaglia. 1998. The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13465-473. [DOI] [PubMed] [Google Scholar]
- 63.Roudet-Tavert, G., T. Michon, J. Walter, T. Delaunay, E. Redondo, and O. Le Gall. 2007. Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 881029-1033. [DOI] [PubMed] [Google Scholar]
- 64.Ruffel, S., M. H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 321067-1075. [DOI] [PubMed] [Google Scholar]
- 65.Ruffel, S., J.-L. Gallois, B. Moury, C. Robaglia, A. Palloix, and C. Caranta. 2006. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 872089-2098. [DOI] [PubMed] [Google Scholar]
- 66.Ryder, E. J. 1970. Inheritance of resistance to common lettuce mosaic. J. Am. Soc. Hortic. Sci. 95378-379. [Google Scholar]
- 67.Ryder, E. J. 1991. Salinas 88 lettuce. Hortscience 26439-440. [Google Scholar]
- 68.Sato, M., K. Nakahara, M. Yoshii, M. Ishikawa, and I. Uyeda. 2005. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 5791167-1171. [DOI] [PubMed] [Google Scholar]
- 69.Schaad, M. C., R. J. Anderberg, and J. C. Carrington. 2000. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273300-306. [DOI] [PubMed] [Google Scholar]
- 70.Scheper, G. C., B. van Kollenburg, J. Hu, Y. Luo, D. J. Goss, and C. G. Proud. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 2773303-3309. [DOI] [PubMed] [Google Scholar]
- 71.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shukla, D. D., C. W. Ward, and A. A. Brunt. 1994. The Potyviridae. CAB International, Wallingford, United Kingdom.
- 73.Stein, N., D. Perovic, J. Kumlehn, B. Pellio, S. Stracke, S. Streng, F. Ordon, and A. Graner. 2005. The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42912-922. [DOI] [PubMed] [Google Scholar]
- 74.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wittmann, S., H. Chatel, M. G. Fortin, and J. F. Laliberté. 1997. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 23484-92. [DOI] [PubMed] [Google Scholar]
- 76.Yamanaka, T., T. Imai, R. Satoh, A. Kawashima, M. Takahashi, K. Tomita, K. Kubota, T. Meshi, S. Naito, and M. Ishikawa. 2002. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J. Virol. 762491-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeam, I., J. R. Cavatorta, D. R. Ripoll, B. C. Kang, and M. M. Jahn. 2007. Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 192913-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshii, M., M. Nishikiori, K. Tomita, N. Yoshioka, R. Kozuka, S. Naito, and M. Ishikawa. 2004. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 786102-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshii, M., N. Yoshioka, M. Ishikawa, and S. Naito. 1998. Isolation of an Arabidopsis thaliana mutant in which accumulation of cucumber mosaic virus coat protein is delayed. Plant J. 13211-219. [DOI] [PubMed] [Google Scholar]
- 80.Yoshii, M., N. Yoshioka, M. Ishikawa, and S. Naito. 1998. Isolation of an Arabidopsis thaliana mutant in which the multiplication of both cucumber mosaic virus and turnip crinkle virus is affected. J. Virol. 728731-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]