Abstract
Whether chicken Mx inhibits influenza virus replication is an important question with regard to strategies aimed at enhancing influenza resistance in domestic flocks. The Asn631 polymorphism of the chicken Mx protein found in the Shamo (SHK) chicken line was previously reported to be crucial for the antiviral activity of this highly polymorphic chicken gene. Our aims were to determine whether cells from commercial chicken lines containing Asn631 alleles were resistant to influenza virus infection and to investigate the effects that other polymorphisms might have on Mx function. Unexpectedly, we found that the Asn631 genotype had no impact on multicycle replication of influenza virus (A/WSN/33 [H1N1]) in primary chicken embryo fibroblast lines. Furthermore, expression of the Shamo (SHK) chicken Mx protein in transfected 293T cells did not inhibit viral gene expression (A/PR/8/34 [H1N1], A/Duck/England/62 [H4N6], and A/Duck/Singapore/97 [H5N3]). Lastly, in minireplicon systems (A/PR/8/34 and A/Turkey/England/50-92/91 [H5N1]), which were highly sensitive to inhibition by the murine Mx1 and human MxA proteins, respectively, Shamo chicken Mx also proved ineffective in the context of avian as well as mammalian cell backgrounds. Our findings demonstrate that Asn631 chicken Mx alleles do not inhibit influenza virus replication of the five strains tested here and efforts to increase the frequency of Asn631 alleles in commercial chicken populations are not warranted. Nevertheless, chicken Mx variants with anti-influenza activity might still exist. The flow cytometry and minireplicon assays described herein could be used as efficient functional screens to identify such active chicken Mx alleles.
Mx proteins are interferon (IFN)-induced dynamin-like GTPases found in all vertebrate species examined so far, and they exhibit a range of antiviral capabilities (10). The Mx1 protein of the inbred A2G strain of mice confers resistance to doses of influenza virus that are lethal to other strains (20). A2G mice are homozygous for an intact Mx1 gene, while the majority of other laboratory strains carry defective alleles owing to a deletion or point mutation in the gene (36). The murine Mx1 protein is critical for effective innate immune defense against influenza (1, 37) and, strikingly, type I IFNs afford no protection in Mx1−/− mice (9). While murine Mx1 has specific activity against orthomyxoviruses, the human MxA protein has a broad antiviral spectrum against many families of RNA viruses, including Orthomyxoviridae, Rhabdoviridae, and Bunyaviridae (10). Murine Mx1 and human MxA mediate their anti-influenza effects via apparently different mechanisms. Mx1 is nuclear and inhibits primary transcription of the virus genome (17), while the cytoplasmic human MxA protein affects an ill-defined posttranscriptional step (29) probably via an interaction with NP (41). For other Mx proteins, such as the human MxB protein (30), rat Mx3 protein (24), and duck Mx (3), no antiviral activity has been detected. Lack of antiviral activity does not imply lack of function, as there is evidence that MxB may be involved in regulating nucleocytoplasmic trafficking and cell cycle progression (13).
The chicken Mx protein was originally found to lack antiviral activity (5), but more recently Ko et al. have reported the existence of antivirally active alleles in some breeds of chicken (14). Constitutive expression of the Mx allele in 3T3 cell lines from a Japanese Shamo (SHK) line of chickens resulted in a 10- to 100-fold reduction in influenza virus titers (14). Certain chicken Mx proteins have also been shown to inhibit replication of vesicular stomatitis virus (VSV) (14), and artificial mutation has demonstrated that the amino acid at position 631 of the chicken Mx protein is a crucial determinant of anti-VSV activity (Asn631 is active and Ser631 is inactive against VSV) (15). SHK Mx also has an Asn residue at position 631, and it has been inferred that this polymorphism is important for anti-influenza activity. The discovery that certain chicken Mx alleles could suppress influenza virus replication has led to considerable interest in the Asn631 allele frequency in commercial flocks and the prospect for breeding this allele back into the general chicken population (2, 19, 21, 35). However, the heterogeneity of the chicken Mx gene and the potential for other polymorphisms to influence its effectiveness means that it would be prudent to determine the inhibitory potential of a wide range of alleles.
The purpose of this study was to validate cell culture assays that could be used to identify Mx alleles from commercial lines of chickens with activity against influenza virus and to determine the functional contributions of polymorphisms in addition to that at position 631. Contrary to expectations, we found that chicken embryo fibroblasts (CEFs) with endogenous Ser631 or Asn631 Mx proteins were equally permissive to multicycle influenza virus replication. Transfected cells expressing SHK Mx sustained equivalent levels of influenza gene expression compared to cells transfected with a Ser631 allele or empty vector plasmid. Moreover, minireplicon systems in mammalian and avian cells were not inhibited by SHK Mx but were highly sensitive to murine Mx1 and human MxA, respectively. Our findings are in contrast to a previous report and indicate that the Shamo (SHK) chicken Mx protein (Asn631 allele) does not suppress influenza virus replication.
MATERIALS AND METHODS
Cells.
CEFs from five commercial pedigree chicken lines designated 8 (embryo 8.1), A (embryos A1 and A2), B (embryo B1), C (embryo C1), and D (embryo D1) were prepared from day 10 embryos (34). DF-1 (CRL-12203), MDCK (CCL-34), and 293T (CRL-11268) cells were obtained from the ATCC Cell Biology Collection and were grown according to ATCC guidelines.
Viruses.
Influenza A virus infections were performed with egg-grown stocks of the A/PR/8/34 (Cambridge) (2 × 108 PFU/ml), A/WSN/33 (4 × 107 PFU/ml), A/Duck/England/62 (H4N6) (2 × 108 PFU/ml), and A/Duck/Singapore/97 (H5N3) (3 × 107 PFU/ml) strains, which were kindly provided by Paul Digard (Cambridge) and Wendy Barclay (Imperial College London).
Plasmids.
Plasmids pcDNA-PB1, -PB2, -PA, -NP, and -NS1 (6, 26), expressing the indicated proteins of A/PR/8/34 (H1N1), and pPOLI-CAT-RT (32) were kindly provided by Paul Digard. Plasmids PolI/II 50-92-PB1, -PB2, -PA, and -NP express proteins from A/Turkey/England/50-92/91 (H5N1) and were generously provided by Wendy Barclay (Imperial College London) (11). A plasmid expressing an influenza virus-based luciferase minireplicon RNA under the control of the human RNA polymerase I (PolI) promoter was constructed by exchanging the CAT gene of pPOLI-CAT-RT with a PCR-amplified luciferase gene. A plasmid of the same design but containing a chicken PolI promoter to drive an influenza-like luciferase minireplicon was also constructed. The chicken PolI promoter was PCR amplified from DF-1 genomic DNA using the primer ‘chPolIF’ (5′ TTT TCT CGA GGT GCT ACC GAC TCG CGC TC 3′) and a reverse primer ‘chPolI5VR’ (5′ ATG AAT TCA AGC TTA TTT AAT GAT AAA AAA CAC CCT TGT TTC TAC TAC AGA CGA ACA TAT AAG GCA TCC G 3′). This PCR product, corresponding to the chPolI promoter with the conserved 5′ terminal sequence of segment 8 of A/PR/8/34 (underlined) positioned at the PolI transcription start site, was cut with XhoI and HindIII (sites in bold in the primer sequences) and ligated into pBluescript SK (Stratagene). The sequence for the 3′ noncoding region followed immediately by the hepatitis delta virus genomic ribozyme sequence was then amplified from pPOLI-CAT-RT and ligated into the EcoRI and BamHI sites of this construct. Finally the firefly luciferase gene, amplified by PCR, was inserted in between the HindIII and EcoRI sites so that it was flanked by the 5′ and 3′ noncoding regions in the resulting vector pChLuc. Expression constructs for wild-type and mutant murine Mx1 and human MxA proteins [pcDNA3-mMx1(wt), pcDNA3-mMx1(K49A), pcDNA3-HA-MxA(wt), and pcDNA3-HA-MxA(T103A)] have been described previously (38, 39). MxA T103A (33) and Mx1 K49A (31) have single amino acid substitutions in their GTP binding domains that abolish GTPase and antiviral activity.
IFN treatment of CEFs.
The chicken IFN-α gene was amplified by PCR using the primers ‘cIFNF’ (5′ CAC AAC ACC GGT CCC ACC ATG GCT GTG CC 3′) and ‘cIFNR’ (5′ GCG TTT AGA TCT AAG TGC GCG TGT TGC CTG 3′) and a cDNA template derived from CEF mRNA. This PCR product was cloned using the AgeI and BglII restriction sites (shown in bold in the primer sequences) into the vector pEGFP-CI. The resulting construct was transfected into 293T cells. The supernatant from these cells was shown to be an effective inducer of chicken Mx transcription in CEFs by quantitative PCR (data not shown). Thereafter, this IFN-containing supernatant was added to CEF cultures at a concentration of 1:100 at 24 h prior to RNA extraction.
Cloning the chicken Mx gene.
The Mx genes from the various CEF lines were amplified by reverse transcriptase (RT)-PCR using SuperScript II RT (Invitrogen), oligo(dT)12-18, Pfx polymerase (Invitrogen), and primers ‘AvMxNewF’ (5′ TAG AAC AAA CCG GTA GAA CAG CAG AAC ATG AAC AAT CC 3′) and ‘AvMxNewR’ (5′ AGA TGA CTC GAG CTA CAG AGA CTT AAA GTC TAC CAG 3′).
The PCR-amplified Mx genes were cloned using external flanking AgeI and XhoI restriction sites (shown in bold in the above primers) into the plasmid pcDNA3GFP. This vector was derived from pcDNA3 (Invitrogen) by replacing the 501-bp NdeI-XhoI fragment (corresponding to the 3′ promoter and 5′ untranslated region of the vector) with the equivalent 1,109-bp fragment containing the green fluorescent protein (GFP) gene from the vector pEGFP-C1 (Clontech). Insertion of the Mx PCR products between the AgeI and XhoI sites of pcDNA3GFP substitutes the Mx cDNA in place of the GFP coding region to give the plasmid pchMx*, where the asterisk indicates the source of the cDNA sequence.
Mutagenesis to derive the Shamo (SHK) Mx allele.
DF-1 and Shamo (SHK) Mx sequences differ by only two nonsynonymous nucleotide changes (Table 1). Oligonucleotide primers ‘AvMxSHKMutF’ (5′ GCC CAA GAT ATA GTG GCT GGT ACC AAT AGT AGC ATT ACT GGA GAA CTA ATT TCC CTT G 3′) and ‘AvMxSHKMutR’ (5′ CAA GGG AAA TTA GTT CTC CAG TAA TGC TAC TAT TGG TAC CAG CCA CTA TAT CTT GGG C 3′) were designed to allow the mutagenesis of the DF-1 sequence to the Shamo (SHK) sequence. Two silent mutations to introduce a KpnI site (in bold type) facilitated screening (mutations are underlined). The pchMxDF-1 template was amplified and mutagenized in two sections via two separate PCRs, one using AvMxNewF/AvMxSHKMutR and the other using AvMxSHKMutF/AvMxNewR. PCR-mediated ligation of these two initial PCR products was followed by cloning to produce pchMxSHK.
TABLE 1.
Amino acid polymorphisms in the Mx proteins from the CEF lines used in this studya
| Line | Amino acid in Mx protein at position:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 21 | 41 | 42 | 85 | 94 | 185 | 199 | 202 | 232 | 308 | 339 | 548 | 583 | 631 | |
| A1 | R | R | P | L | L | R | K | G | S | G | V | T | A | T | S |
| A2 | R | Q | P | L | L | R | K | G | T | G | I | A | A | T | S |
| 8.1 | R | R | P | L | L | R | K | G | S | G | V | T | V | T | S |
| B1 | R | R | P | L | L | R | K | G | S | G | V | T | A | T | N |
| C1 | R | R | P | L | L | R | K | G | S | R | V | T | V | T | N |
| D1 | R | Q | R | S | L | R | K | G | S | G | V | T | V | T | N |
| DF-1 | R | Q | R | S | L | R | K | G | S | G | V | T | V | T | N |
| SHKb | R | Q | R | S | L | R | K | S | T | G | V | T | V | T | N |
| WLRc | W | Q | R | S | L | Q | K | G | S | G | I | A | A | A | S |
| Anti-VSV active lineb | WLK-2 | WLK-2 | WLK-2 | WLK-2 | WLK-2 | HN | HN | ||||||||
The amino acid residues at the 15 reported polymorphic sites of the chicken Mx protein are given for the CEF and DF-1 cell lines, with reference to those of the Shamo (SHK) and White Leghorn (WLR) lines. Bold type indicates where the amino acid is the same as that found in SHK. The bottom row shows breed identifiers, as given in reference 14, for Mx alleles that differ from the SHK amino acid at the positions shown but which are active against VSV. (The allele WLK-2 is from a strain of White Leghorn, and the HN allele is from the Hinaidori breed.)
Data from reference 14.
Accession number Z23168.
Virus yield assay.
Second-passage CEFs were infected with A/WSN/33 at a multiplicity of infection (MOI) of 0.01. At the indicated times postinfection, supernatants were harvested and the virus yields titered on MDCK cells by plaque assay (23).
Luciferase minireplicon reporter assays.
293T or DF-1 cells were transfected using Fugene 6 reagent (Roche) in 24-well plates. Each well received 25 ng of each of the plasmids expressing the viral PB1, PB2, PA, and NP proteins, 100 ng of the luciferase reporter plasmid, 50 ng of a plasmid expressing secreted alkaline phosphatase (SEAP) (4), and 250 ng of an Mx-bearing plasmid or pcDNA3 control plasmid, unless otherwise stated. Forty-eight hours posttransfection, luciferase expression was assayed using a Bright-Glo luciferase assay system (Promega). SEAP activity was determined using the colorimetric assay described previously (4). Luciferase activity was normalized to the SEAP activity for each well to account for minor differences in transfection efficiency.
Detection of viral antigen in Mx-transfected 293T cells using flow cytometry.
293T cells were transfected with a DNA mix consisting of 1.5 μg of Mx-expressing plasmid (or pcDNA3) and 0.5 μg of pEGFP-CI. At 48 h posttransfection, the cells were infected with either A/PR8/34 (MOI of 10), A/Duck/England/62 (MOI of 1), or A/Duck/Singapore/97 (MOI of 0.5) for 1 h. Fourteen hours postinfection, the cells were trypsinized, fixed for 10 min using 2% formaldehyde in phosphate-buffered saline (PBS), permeabilized for 10 min using 0.2% Triton X-100 with 10% goat serum in PBS, and then blocked for 15 min using 10% goat serum in PBS. The cells were incubated at room temperature for 1 h with a 1:200 dilution (in 0.02% Triton X-100 and 2% goat serum in PBS) of rabbit polyclonal antisera to fowl plague virus (Rostock) viral ribonucleoprotein (vRNP) (22), kindly provided by Paul Digard (University of Cambridge). The cells were then washed three times in PBA (PBS, 0.1% bovine serum albumin, 0.01% sodium azide) before incubation with a 1:20 dilution of anti-rabbit R-phycoerythrin (PE)-conjugated antibody (Sigma P-9537) on ice for 30 min in the dark. Finally, the cells were washed an additional three times in PBA before detection of their fluorescence using a Becton Dickinson FACSCalibur. For each sample, 20,000 (A/PR/8/34) or 10,000 (A/Duck/England/62 and A/Duck/Singapore/97) GFP-positive cells were analyzed for their PE fluorescence using the FL2-H channel. Cell Quest 3.3 software was used to analyze the data.
Statistics.
Data were analyzed for statistical significance using a two-tailed Student's t test assuming populations of unequal variance and a conservative threshold for significance (P < 0.01). Data sets were compared to the pcDNA3 controls where appropriate. The number of independent data sets for each experiment is indicated in the figure legends.
RESULTS
Sequencing of Mx cDNA from a panel of chicken lines.
Primary CEF cultures were prepared from chick embryos from a selection of chicken lines. The primary CEF and DF-1 cells were treated with recombinant chicken IFN-α for 24 h prior to RT-PCR and sequencing of their Mx mRNAs. Sequence comparisons identified 12 amino acid polymorphisms relative to the reference sequence of the White Leghorn strain (WLR) (GenBank accession number Z23168) (Table 1). All of the observed polymorphisms have been reported previously (2, 14). Bold type in Table 1 indicates where the amino acid at that position is the same as that found for the Shamo (SHK) allele that represents the only chicken Mx allele for which anti-influenza activity has been demonstrated (14). Standard type therefore highlights where the sequence differs and could potentially affect Mx activity. Some of these differences have been observed in chicken Mx genes present in other breeds that have been shown to be active against VSV (14). Where this is the case, the bottom row of the table contains the identifier for the breeds of chicken reported to carry that polymorphism.
The Mx amino acid sequence of line D1 was identical to that of DF-1 cells, both of which carry the Asn631 allele and are the most closely related to the Shamo (SHK) line (differing only at amino acids 199 and 202). Two of the CEF lines, designated A1 and B1, were found to have Mx amino acid sequences identical to each other except for the amino acid at position 631. Where these Mx alleles differed from the SHK allele (i.e., at positions 199 and 202 for D1 and additionally at positions 21, 41, 42, and 548 for A1 and B1), the same changes had been observed in chicken Mx alleles shown to be active against VSV.
The CEF lines were maintained in parallel with identical passage histories, and second-passage cells were used for the subsequent virus replication experiments.
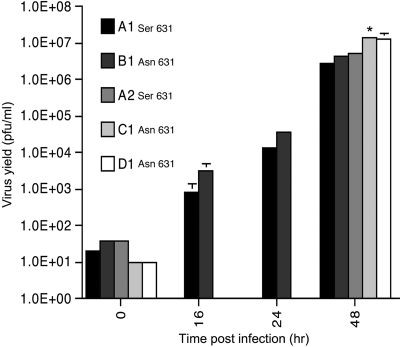
Growth of influenza A/WSN/33 in CEF cells with Asn631 or Ser631 alleles of Mx.
The permissiveness of the different CEF lines to replication of the laboratory strain of influenza A/WSN/33 was determined by multicycle yield assays. The yields of virus (and the rates of virus replication) were determined by a subsequent plaque assay on MDCK cells (Fig. 1). For lines A1 and B1, which differ only at amino acid 631 of the chicken Mx coding region, viral titers were determined at 0, 16, 24, and 48 h postinfection. Titers increased logarithmically over the course of the assay. There was no significant difference in virus replication in primary CEF cells carrying the Ser631 allele of chicken Mx (line A1) compared to replication in cells carrying the Asn631 allele (line B1). In fact, the titers for line B1 were consistently higher over the course of the assay. Virus replication was also determined in lines A2 (Ser631) and C1 and D1 (both Asn631). The 48-h titer for line C1 was significantly greater than those for lines A1, B1, or A2 (P < 0.01). Thus, the presence of the Asn631 allele of chicken Mx did not inhibit the multicycle replication of influenza A/WSN/33 in primary CEF cells. For cell lines carrying either the Ser631 or Asn631 Mx alleles, IFN pretreatment of the CEFs with recombinant chicken IFN-α completely suppressed multicycle viral replication (titers were reduced by >5 log units), such that it was not possible to determine the growth kinetics of the virus under these conditions (data not shown). These results demonstrate that the Asn631 allele is neither necessary nor sufficient to confer protection to CEFs against infection with influenza A/WSN/33.
FIG. 1.
Multicycle influenza virus (A/WSN/33) replication in CEF lines. CEFs were infected with A/WSN/33 (MOI of 0.01). Supernatants were harvested at the times indicated, and the virus yields were titered on MDCK cells. The means (and standard deviations) of three independent experiments are shown. The amino acids at position 631 of the Mx alleles of the different lines are indicated. *, titer for C1 significantly different from A1, B1, or A2 (P < 0.01).
Replication of influenza strains A/PR/8/34, A/Duck/England/62, and A/Duck/Singapore/97 is not inhibited in 293T cells expressing chicken Mx.
The activity of the Mx genes in the cultured CEFs could have been confounded for two reasons: (i) none of the genes were identical to the Shamo (SHK) sequence, and so they could potentially have been inactive due to the combined effect of the other polymorphisms; and (ii) expression of the Mx genes is dependent on IFN induction and could have been prevented by the IFN-suppressive activity of the virus. In order to uncouple the effect of chicken Mx from its regulatory dependence on IFN and to isolate it from the influence of other IFN-responsive genes, the Mx genes were placed under the control of the CMV-IE promoter in the plasmid pcDNA3. The Mx gene from DF-1 cells was cloned and altered by site-specific mutagenesis to convert it to the identical amino acid sequence found in the SHK line. This represented the putative functional chicken Mx gene (SHK). A Ser631 allele was cloned from line 8.1 (Table 1) as a presumed nonfunctional chicken Mx gene.
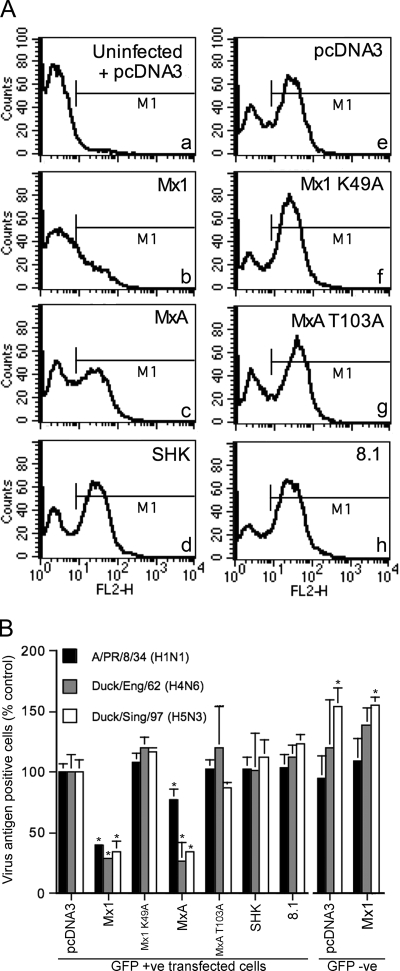
A flow cytometry-based assay was used to determine the effect of chicken Mx on viral gene expression for three different influenza virus strains. 293T cells were cotransfected with the pcDNA3-based Mx expression plasmids and a plasmid carrying the GFP (pEGFP-C1) in a ratio of 3:1. Preliminary experiments showed that when these plasmid ratios were used, over 95% of GFP-positive cells expressed the cotransfected plasmid. Forty-eight hours posttransfection, the cells were infected with either A/PR/8/34 (H1N1), A/Duck/England/62 (H4N6), or A/Duck/Singapore/97 (H5N3) at a multiplicity that achieved approximately 60% infection, as determined by FACS analysis using anti-influenza RNP-specific antibody and PE-conjugated secondary antibody. Cells were gated for green fluorescence to isolate the transfected from the nontransfected populations and then analyzed for PE fluorescence in the FL2-H channel (Fig. 2).
FIG. 2.
Flow cytometric detection of influenza vRNPs in transfected 293T cells. 293T cells were cotransfected with Mx-expressing plasmids (or pcDNA3) (1.5 μg) and pEGFP-C1 (0.5 μg) and infected after 48 h with A/PR/8/34, A/Duck/England/62, or A/Duck/Singapore/97. Fourteen hours postinfection, the cells were stained for vRNP and analyzed by flow cytometry. Panel A shows the level of A/PR/8/34 antigen in GFP-positive cells that had been cotransfected with the indicated plasmids. Panel “a” shows the background staining detected in uninfected, pcDNA3-transfected cells and was used to set the fluorescence threshold marker (M1), which demarcates between antigen-positive and -negative cells. In panel B, the data are derived from independent experiments (A/PR/8/34, n = 5; A/Duck/England/62, n = 3; A/Duck/Singapore/97, n = 3) and the bar heights represent the percentages of antigen-positive cells expressed relative to that for the pcDNA3 control group for each virus. The means (and standard deviations) are shown for either GFP-positive or GFP-negative cell populations from wells that had been transfected with the constructs as shown. *, P of <0.01 relative to GFP-positive pcDNA3-transfected cells.
Transfected, uninfected cells showed low-level background fluorescence in the FL2-H channel (Fig. 2A, panel a). Infected cells transfected with the pcDNA3 control plasmid showed two distinct peaks corresponding to the uninfected and infected populations (Fig. 2A, panel e). Substituting the various Mx plasmids for the pcDNA3 plasmid allowed the effect of the Mx genes on viral gene expression to be determined. This is shown in the representative histograms in Fig. 2A and for replicate experiments for each of the three viruses in Fig. 2B. In cells transfected with murine Mx1, there was an approximately threefold reduction (P < 0.01) in the proportion of antigen-positive cells relative to cells transfected with pcDNA3 for all three virus strains tested. For human MxA, there was a reduction of similar magnitude in the proportion of antigen-positive cells for both of the avian strains A/Duck/England/62 and A/Duck/Singapore/97 and a less-pronounced but nevertheless significant (P = 0.002) reduction for A/PR/8/34. As expected, these effects were abrogated by the K49A and T103A mutations, respectively. However, transfection of cells with the SHK or 8.1 chicken Mx expression plasmids did not affect the proportion of antigen-positive cells compared to transfection with empty vector for infections with either A/PR/8/34, A/Duck/England/62, or A/Duck/Singapore/97.
In the nonproductively transfected subpopulations, i.e., GFP-negative cells, the antigen staining was either equivalent (A/PR/8/34 and A/Duck/England/62) or enhanced (A/Duck/Singapore/97) (P < 0.01) relative to the pcDNA3-transfected controls (Fig. 2B). This confirmed that the inhibition of viral replication demonstrated in this assay for murine Mx1 and human MxA was specific to the cells that had expressed these plasmids.
Activity of chicken Mx in influenza minireplicon reporter assays.
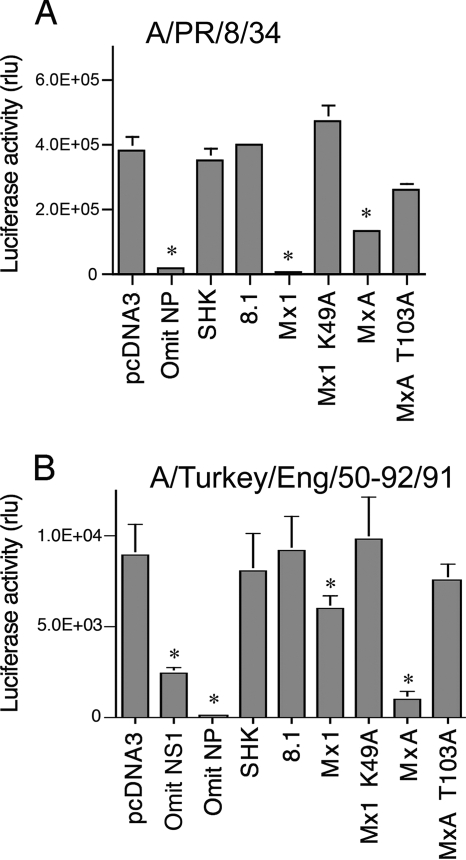
Orthomyxovirus minireplicon assays measure the transcriptional activity of the viral polymerase and are capable of sensitive detection of the antiviral activity of Mx genes (7, 12, 41, 42). In these assays, cells are transfected with plasmids that express the polymerase/NP replication complex proteins (3P/NP) and a minigenome reporter plasmid that produces a negative-sense luciferase gene bounded by the viral promoter sequences. Transcription and replication of this reporter RNA by the viral polymerase result in the synthesis of a viral mRNA-like transcript that is translated to produce the luciferase enzyme. Cotransfection of the Mx expression plasmids allows the effect of the Mx genes on this process to be determined.
We determined the activity of chicken Mx in minireplicon systems based on A/PR/8/34 (H1N1) and A/Turkey/England/50-92/91 (H5N1) (Fig. 3). The A/Turkey/England/50-92/91 assays were performed in the CEF cell line DF-1 in order to provide chicken host cell factors which might be required for the function of chicken Mx. DF-1 cells are IFN competent, and IFN is induced in CEFs by transfection of plasmid DNA (28). For the DF-1 minireplicon transfections, we therefore cotransfected a plasmid that expressed the A/PR/8/34 NS1 gene, which has been shown to effectively inhibit induction of IFN in CEFs. Omission of the NS1 plasmid from the transfection mix caused a significant decrease in luciferase levels, consistent with partial suppression of the system by the induction of IFN (Fig. 3B).
FIG. 3.
Activity of chicken Mx proteins in influenza virus minireplicon systems. (A) Effect of chicken Mx on an A/PR/8/34 minireplicon system. 293T cells were cotransfected with plasmids encoding A/PR/8/34 PB1, PB2, PA, and NP, a huPolI Luc minigenome plasmid, and a SEAP-expressing plasmid, together with pcDNA3 or a plasmid expressing the indicated Mx protein (chicken Mx proteins SHK [Asn631] and 8.1 [Ser631], murine Mx1 and its mutant Mx1 K49A, and human MxA and its mutant MxA T103A). The ratio of pcDNA3/Mx plasmid to each of the 3P/NP plasmids was 10:1. Forty-eight hours posttransfection, luciferase activity was measured and is shown as SEAP-normalized relative light units (rlu). The means (and standard deviations) of three independent experiments are shown. *, P of <0.01 relative to pcDNA3. (B) Effect of chicken Mx on the Turkey/England/50-92/91 minireplicon system in chicken DF-1 cells. DF-1 cells were transfected with plasmids encoding A/Turkey/England/50-92/91 PB1, PB2, PA, and NP (25 ng each), A/PR/8/34 NS1 (125 ng), a chicken PolI Luc minigenome plasmid (100 ng), and a SEAP-expressing plasmid (50 ng) together with pcDNA3 or a plasmid expressing the indicated Mx protein (125 ng). The ratio of pcDNA3/Mx plasmid to each of the 3P/NP plasmids was 5:1. Forty-eight hours posttransfection, luciferase activity was measured and is shown as relative light units (rlu). The means (and standard deviations) of six independent experiments are shown. *, P of <0.01 relative to pcDNA3.
In both the A/PR/8/34 and A/Turkey/England/50-92/91 systems, omission of the nucleoprotein-expressing plasmid caused a profound reduction in luciferase, showing that the luciferase expression was contingent on the presence of a functional viral replication complex. For both minireplicon systems, murine Mx1 and human MxA caused significant inhibition. Transfection of murine Mx1 expression plasmid reduced the level of luciferase activity by over 95% in the A/PR/8/34 minireplicon, while only a modest (<2-fold) inhibition was achieved in the A/Turkey/England/50-92/91 assay. In contrast, human MxA caused greater inhibition in the A/Turkey/England/50-92/91 system. Luciferase levels were restored by the Mx1 K49A and MxA T103A mutants, except where some residual inhibition was apparent for the T103A mutant in the A/PR/8/34 system. This might be related to the finding that human MxA promotes apoptosis in A/PR/8/34-infected cells (25), a property which is not dependent on GTP binding or hydrolysis (27) and would therefore be expected to be retained in MxA T103A.
We found that the chicken SHK or 8.1 Mx expression plasmids did not significantly affect luciferase activity, neither for A/PR/8/34 minireplicon systems nor for A/Turkey/England/50-92/91 systems, even when these were carried out in DF-1 cells (Fig. 3 and data not shown). Western blotting confirmed that the failure of SHK Mx to inhibit the minireplicon systems was not due to poor expression (data not shown).
DISCUSSION
The serious implications of the ongoing H5N1 avian influenza epizootic have raised the issue of whether it might be possible to generate influenza-resistant chickens either by selective breeding or genetic modification approaches. This would reduce the threat of avian influenza to the poultry industry and reduce the potential for chickens to act as bridging hosts for the emergence of new pandemic strains of influenza. One focus for selective breeding approaches has been the Asn631 polymorphism of the chicken Mx protein, which was reported by Ko et al. to confer antiviral activity on the Mx protein (14).
There have hitherto been no reports analyzing the susceptibilities of cells derived from any breed of chicken carrying the Asn631 polymorphism. To this end, we initially tested the permissiveness of CEF cell lines derived from three commercial breeds of chicken that were homozygous for the Asn631 polymorphism and compared them with two lines that were homozygous for Ser631. No evidence for resistance to infection by influenza virus A/WSN/33 was observed. IFN treatment of Asn631 or Ser631 allele cell lines completely prevented virus replication (data not shown), indicating that the Asn631 allele is not necessary to establish a potent antiviral state in response to IFN. This is to be expected given the presence of other potentially active IFN-responsive antiviral genes, such as protein kinase R and the 2′-5′ oligoadenylate synthetase/RNase L system, but it is in contrast to the curiously overriding importance of murine Mx1 in mouse cells (9, 16, 40). Our conclusion was that either the Mx alleles of these cell lines were inactive due to the other polymorphisms in the gene, that they were ineffective due to the virus's ability to suppress IFN-responsive gene expression, or that A/WSN/33 was not susceptible to chicken Mx for some other reason. To both uncouple Mx expression from its dependence on IFN and develop an efficient assay for determining the functionality of different chicken Mx alleles, we used a transfection/infection FACS assay for viral gene expression and several variants of the widely used minireplicon assay for orthomyxovirus replication and transcription. Although we could readily demonstrate antiviral activity for murine Mx1 and human MxA in these assays, no inhibition was observed for the prototype functional chicken Mx allele identical to that of the Shamo (SHK) breed.
One possible explanation for the disparity between our data and those of Ko et al. (14) is the use of different strains of influenza virus. Certain strains of influenza virus are markedly less sensitive to murine Mx1 and human MxA than other strains, a property that segregates with the NP gene (7). It is conceivable that the A/Hong Kong/483/97 strain used by Ko et al. is sensitive to inhibition by chicken Mx, while the five influenza strains used in this study are resistant.
Here we have found various Asn631 alleles to be inactive against influenza virus. However, it remains possible that certain variants of the chicken Mx protein might have anti-influenza activity. In the case of the murine Mx1 gene, many mutations have been identified that disrupt the antiviral properties of the protein (8). Furthermore, a single amino acid substitution (E645R) in the human MxA protein (within the same domain as position 631 of the chicken Mx protein) abolished anti-VSV but not anti-influenza activity, which indicates that virus-specific effector regions exist (43). In view of these observations and the high degree of polymorphism found with the chicken Mx gene (both in coding [19, 35] and in the promoter/5′ untranslated region [18]), there may be value in large-scale screening of naturally occurring chicken Mx alleles for anti-influenza activity. We envisage that the minireplicon assays employed in this study could prove particularly efficient for this purpose.
Interestingly, mammalian strains of influenza virus are in general less sensitive than avian strains to the mammalian Mx proteins Mx1 and MxA (7). It was postulated that mammalian strains might be adapted to the Mx proteins of their mammalian hosts, while avian strains have not undergone this selection and thus have not adapted to counter the effects of mammalian Mx. If, as our evidence suggests, chicken Mx is indeed inactive against both mammalian and avian influenza virus strains, then this might reflect the long period of adaptation in the avian host, from which all influenza virus strains ultimately originated. Whether some chicken Mx genes exhibit activity against other avian viral pathogens, e.g., Newcastle disease and infectious bursal disease viruses, remains to be investigated.
In summary, in three assay systems for influenza virus replication, of at least equal validity to the previously used 3T3 cell line approach, the Asn631 allele of chicken Mx proved ineffective. It is important to resolve the disparity between our results and those of Ko et al. (14) and to determine whether any anti-influenza virus Mx variants exist that could form the basis of selective breeding programs. Transient and constitutive transfection assays can assist in this quest, but ultimately only challenge studies on birds carrying these identified alleles will definitively demonstrate the antiviral role of Mx in chickens.
Acknowledgments
This work was supported by the Cambridge Infectious Diseases Consortium (CIDC), which is funded jointly by DEFRA and HEFCE as part of the Veterinary Training Research Initiative (VTRI), and BBSRC grant no. BSB/B/00301.
We are grateful to Aviagen for their cooperation in supplying the CEFs used in this study. We thank Otto Haller, Willem Rens, and Barbara Blacklaws for their helpful advice and Paul Digard and Wendy Barclay for the reagents they supplied.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Arnheiter, H., S. Skuntz, M. Noteborn, S. Chang, and E. Meier. 1990. Transgenic mice with intracellular immunity to influenza virus. Cell 6251-61. [DOI] [PubMed] [Google Scholar]
- 2.Balkissoon, D., K. Staines, J. McCauley, J. Wood, J. Young, J. Kaufman, and C. Butter. 2007. Low frequency of the Mx allele for viral resistance predates recent intensive selection in domestic chickens. Immunogenetics 59687-691. [DOI] [PubMed] [Google Scholar]
- 3.Bazzigher, L., A. Schwarz, and P. Staeheli. 1993. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck Mx protein. Virology 195100-112. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 661-10. [DOI] [PubMed] [Google Scholar]
- 5.Bernasconi, D., U. Schultz, and P. Staeheli. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 1547-53. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco, M., M. J. Amorim, and P. Digard. 2004. Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic 5979-992. [DOI] [PubMed] [Google Scholar]
- 7.Dittmann, J., S. Stertz, D. Grimm, J. Steel, A. Garcia-Sastre, O. Haller, and G. Kochs. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of the Mx-GTPase. J. Virol. 823624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garber, E. A., D. L. Hreniuk, L. M. Scheidel, and L. H. van der Ploeg. 1993. Mutations in murine Mx1: effects on localization and antiviral activity. Virology 194715-723. [DOI] [PubMed] [Google Scholar]
- 9.Grimm, D., P. Staeheli, M. Hufbauer, I. Koerner, L. Martinez-Sobrido, A. Solorzano, A. Garcia-Sastre, O. Haller, and G. Kochs. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. USA 1046806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller, O., S. Stertz, and G. Kochs. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 91636-1643. [DOI] [PubMed] [Google Scholar]
- 11.Howard, W., A. Hayman, A. Lackenby, A. Whiteley, B. Londt, J. Banks, J. McCauley, and W. Barclay. 2007. Development of a reverse genetics system enabling the rescue of recombinant avian influenza virus A/Turkey/England/50-92/91 (H5N1). Avian Dis. 51393-395. [DOI] [PubMed] [Google Scholar]
- 12.Janzen, C., G. Kochs, and O. Haller. 2000. A monomeric GTPase-negative MxA mutant with antiviral activity. J. Virol. 748202-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King, M. C., G. Raposo, and M. A. Lemmon. 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. USA 1018957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, J. H., H. K. Jin, A. Asano, A. Takada, A. Ninomiya, H. Kida, H. Hokiyama, M. Ohara, M. Tsuzuki, M. Nishibori, M. Mizutani, and T. Watanabe. 2002. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 12595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko, J. H., A. Takada, T. Mitsuhashi, T. Agui, and T. Watanabe. 2004. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Anim. Genet. 35119-122. [DOI] [PubMed] [Google Scholar]
- 16.Koerner, I., G. Kochs, U. Kalinke, S. Weiss, and P. Staeheli. 2007. Protective role of beta interferon in host defense against influenza A virus. J. Virol. 812025-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krug, R. M., M. Shaw, B. Broni, G. Shapiro, and O. Haller. 1985. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J. Virol. 56201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X. Y., L. J. Qu, Z. C. Hou, J. F. Yao, G. Y. Xu, and N. Yang. 2007. Genomic structure and diversity of the chicken Mx gene. Poult. Sci. 86786-789. [DOI] [PubMed] [Google Scholar]
- 19.Li, X. Y., L. J. Qu, J. F. Yao, and N. Yang. 2006. Skewed allele frequencies of an Mx gene mutation with potential resistance to avian influenza virus in different chicken populations. Poult. Sci. 851327-1329. [DOI] [PubMed] [Google Scholar]
- 20.Lindenmann, J. 1962. Resistance of mice to mouse-adapted influenza A virus. Virology 16203-204. [DOI] [PubMed] [Google Scholar]
- 21.Livant, E. J., S. Avendano, S. McLeod, X. Ye, S. J. Lamont, J. C. Dekkers, and S. J. Ewald. 2007. MX1 exon 13 polymorphisms in broiler breeder chickens and associations with commercial traits. Anim. Genet. 38177-179. [DOI] [PubMed] [Google Scholar]
- 22.Mahy, B. W. J., A. R. Carroll, J. M. T. Brownson, and D. J. McGeoch. 1977. Block to influenza virus replication in cells preirradiated with ultraviolet light. Virology 83150-162. [DOI] [PubMed] [Google Scholar]
- 23.Matrosovich, M., T. Matrosovich, W. Garten, and H. D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, E., G. Kunz, O. Haller, and H. Arnheiter. 1990. Activity of rat Mx proteins against a rhabdovirus. J. Virol. 646263-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mibayashi, M., K. Nakad, and K. Nagata. 2002. Promoted cell death of cells expressing human MxA by influenza virus infection. Microbiol. Immunol. 4629-36. [DOI] [PubMed] [Google Scholar]
- 26.Mullin, A. E., R. M. Dalton, M. J. Amorim, D. Elton, and P. Digard. 2004. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J. Gen. Virol. 853689-3698. [DOI] [PubMed] [Google Scholar]
- 27.Numajiri, A., M. Mibayashi, and K. Nagata. 2006. Stimulus-dependent and domain-dependent cell death acceleration by an IFN-inducible protein, human MxA. J. Interferon Cytokine Res. 26214-219. [DOI] [PubMed] [Google Scholar]
- 28.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 771501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlovic, J., O. Haller, and P. Staeheli. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 662564-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlovic, J., T. Zurcher, O. Haller, and P. Staeheli. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 643370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitossi, F., A. Blank, A. Schroder, A. Schwarz, P. Hussi, M. Schwemmle, J. Pavlovic, and P. Staeheli. 1993. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 676726-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pleschka, S., R. Jaskunas, O. G. Engelhardt, T. Zurcher, P. Palese, and A. Garcia-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 704188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponten, A., C. Sick, M. Weeber, O. Haller, and G. Kochs. 1997. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 712591-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekellick, M. J., and P. I. Marcus. 1986. Induction of high titer chicken interferon. Methods Enzymol. 119115-125. [DOI] [PubMed] [Google Scholar]
- 35.Seyama, T., J. H. Ko, M. Ohe, N. Sasaoka, A. Okada, H. Gomi, A. Yoneda, J. Ueda, M. Nishibori, S. Okamoto, Y. Maeda, and T. Watanabe. 2006. Population research of genetic polymorphism at amino acid position 631 in chicken Mx protein with differential antiviral activity. Biochem. Genet. 44437-448. [DOI] [PubMed] [Google Scholar]
- 36.Staeheli, P., R. Grob, E. Meier, J. G. Sutcliffe, and O. Haller. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 84518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staeheli, P., O. Haller, W. Boll, J. Lindenmann, and C. Weissmann. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44147-158. [DOI] [PubMed] [Google Scholar]
- 38.Stertz, S., J. Dittmann, J. C. Blanco, L. M. Pletneva, O. Haller, and G. Kochs. 2007. The antiviral potential of interferon-induced cotton rat Mx proteins against orthomyxovirus (influenza), rhabdovirus, and bunyavirus. J. Interferon Cytokine Res. 27847-855. [DOI] [PubMed] [Google Scholar]
- 39.Stertz, S., M. Reichelt, J. Krijnse-Locker, J. Mackenzie, J. C. Simpson, O. Haller, and G. Kochs. 2006. Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J. Interferon Cytokine Res. 26650-660. [DOI] [PubMed] [Google Scholar]
- 40.Tumpey, T. M., K. J. Szretter, N. Van Hoeven, J. M. Katz, G. Kochs, O. Haller, A. Garcia-Sastre, and P. Staeheli. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 8110818-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turan, K., M. Mibayashi, K. Sugiyama, S. Saito, A. Numajiri, and K. Nagata. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 32643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, F., O. Haller, and G. Kochs. 2000. MxA GTPase blocks reporter gene expression of reconstituted Thogoto virus ribonucleoprotein complexes. J. Virol. 74560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 111657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]