Abstract
The E2 proteins of several papillomaviruses link the viral genome to mitotic chromosomes to ensure retention and the efficient partitioning of genomes into daughter cells following cell division. Bovine papillomavirus type 1 E2 binds to chromosomes in a complex with Brd4, a cellular bromodomain protein. Interaction with Brd4 is also important for E2-mediated transcriptional regulation. The transactivation domain of E2 is crucial for interaction with the Brd4 protein; proteins lacking or mutated in this domain do not interact with Brd4. However, we found that the C-terminal DNA binding/dimerization domain of E2 is also required for efficient binding to Brd4. Mutations that eliminated the DNA binding function of E2 had no effect on the ability of E2 to interact with Brd4, but an E2 protein with a mutation that disrupted C-terminal dimerization bound Brd4 with greatly reduced efficiency. Furthermore, E2 proteins in which the C-terminal domains were replaced with the dimeric DNA binding domain of EBNA-1 or Gal4 bound efficiently to the Brd4 protein, but the replacement of the E2 C-terminal domain with a monomeric red fluorescent protein did not rescue efficient Brd4 binding. Thus, E2 bound to Brd4 most efficiently as a dimer. To prove this finding further, the E2 DNA binding domain was replaced with an FKBP12-derived domain in which dimerization was regulated by a bivalent ligand. This fusion protein bound Brd4 efficiently only in the presence of the ligand, confirming that a dimer of E2 was required. Correspondingly, E2 proteins that could dimerize were able to bind to mitotic chromosomes much more efficiently than monomeric E2 polypeptides.
The papillomavirus E2 protein has several functions in the viral life cycle. It is the major viral transcriptional regulatory protein and, along with the E1 helicase, binds to the origin of replication to initiate DNA replication. The E2 protein also ensures that the infection is transmitted to daughter cells through many cell divisions by attaching the viral genomes to cellular mitotic chromosomes. For all papillomaviruses studied to date, the E2 transcriptional regulatory function involves interaction with the cellular protein Brd4 (12, 20, 27, 28, 34). Brd4 binds to acetylated chromatin via two bromodomains and regulates transcriptional elongation (13, 33). An E2-Brd4 complex binds to viral promoters and is involved in both the activation and the repression of viral transcription (26, 33, 34).
For a subset of papillomaviruses, the E2-Brd4 complex is also involved in genome tethering to mitotic chromosomes (5, 20, 35). The interaction of the bovine papillomavirus type 1 (BPV-1) E2 protein with both Brd4 and mitotic chromosomes has been well studied; BPV-1 E2 and Brd4 colocalize on both interphase chromatin and mitotic chromosomes in punctate speckles (21, 35). Furthermore, E2 greatly increases the affinity of interphase Brd4 for chromatin and relocalizes mitotic Brd4 from a diffuse cloud to punctate speckles on condensed chromosomes (21).
The interaction of BPV-1 E2 and Brd4 has been well studied. The E2 protein consists of a transactivation domain linked to a dimeric DNA binding domain by a flexible hinge region. The N-terminal domain has been shown previously to be capable of interacting with both mitotic chromosomes (4) and the Brd4 protein, and the region of interaction has been mapped to a face of the domain containing the conserved residues arginine 37 and isoleucine 73 (5, 28). The mutation of these residues to alanine completely abolishes the interaction of E2 with Brd4 and the association of E2 with mitotic chromosomes and results in a transcriptionally defective protein. The N-terminal half of the Brd4 protein contains the two bromodomains, and this region is similar to many other proteins in the same family (reviewed in reference 33). The C-terminal half of Brd4 is not very homologous to other BET (bromodomain-extraterminal domain) family members. Both N-terminal and C-terminal regions are important for the interaction with E2 (34, 35). A polypeptide derived from the C terminus of Brd4 can act as a dominant negative protein and can disrupt the interaction of E2 with Brd4 (20, 35).
In this study, we have further analyzed the association of the BPV-1 E2 protein with Brd4. We find that, although the primary region of interaction is the N-terminal domain of E2, the dimerization function of the C-terminal domain is required for efficient binding. Furthermore, the dimerization function of E2 is required to stabilize the interaction of Brd4 with mitotic chromosomes.
MATERIALS AND METHODS
Plasmids.
The series of pTZ plasmids that encoded the truncated and deleted E2 proteins and the E2 dimerization domain fusion proteins, pTZ E2-Gal4, and the pTZ Gal41-147 control have been described previously (18, 32). The following plasmids were all generated using standard cloning procedures, and details will be provided upon request. pTZ E2 W360G encodes the E2 protein with the W360G substitution (E2 W360G), as described previously (7). The pTZ E2-EBNA plasmid encodes residues 1 to 285 from E2, the simian virus 40 nuclear localization signal, and residues 450 to 641 of EBNA-1. pTZ EBNA-DBD encodes residues 450 to 641 of EBNA-1. The pTZ E2-RFP and pTZ RFP plasmids encode E2 residues 1 to 285 fused to the complete red fluorescent protein (RFP; 226 residues [derived from pDsRed-N1; Clontech]) and RFP alone, respectively. For the FKBP fusion proteins and controls, DNA encoding the FKBP domain was amplified from pC4-Fv1E (Ariad Pharmaceuticals) and fused to the 3′ end of the E2 gene region encoding residues 1 to 285, resulting in an E2 N-terminal domain and hinge region (NH)-FKBP fusion protein. A plasmid expressing the FKBP domain alone was also generated by the insertion of an initiation codon at the 5′ end of the FKBP gene. Each of the derived genes was subcloned, with the addition of a sequence encoding an N-terminal FLAG epitope, into the previously described pMEP-4 vector (23).
Establishment of cell lines expressing the E2 proteins.
Stable cell lines were generated by transfecting CV-1 cells with the pMEP-E2 expression plasmids by using Fugene (Roche). Cells containing the episomal plasmid were selected with 200 μg of hygromycin B/ml. Drug-resistant colonies were pooled after 2 weeks.
Immunoblotting.
For each sample, 2 μg of total protein was heated in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer at 100°C for 5 min. For the detection of E2 protein, samples were separated by SDS-PAGE and electrotransferred onto Immobilon-P polyvinylidene difluoride membranes (Millipore). E2 protein detection was performed according to standard protocols using an anti-FLAG M2 antibody followed by horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Pierce). Protein bands were detected using chemiluminescence reagent SuperSignal West Dura (Pierce).
Indirect immunofluorescence.
Cells were seeded directly onto glass slides (Superfrost Plus) and grown for 24 to 48 h. To enrich the mitotic cell population, cells were blocked in S phase by the addition of 2 mM thymidine for 14 to 16 h. The thymidine block was released by the removal of thymidine, and the cells were cultured for an additional 9 h. E2 expression was induced by the addition of 3 μM CdSO4 for the last 3 to 4 h. For the ligand-mediated dimerization experiments, 10 nM AP20187 (Ariad Pharmaceuticals) was added to the cells during the induction period. Cells were fixed directly in 4% paraformaldehyde in phosphate-buffered saline for 20 min at room temperature and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline for 15 min at room temperature. FLAG-tagged E2 proteins were detected using Sigma monoclonal anti-FLAG M2 (1:500). Fluorescein isothiocyanate-conjugated secondary antibody (Jackson Immunochemicals) was used at a 1:100 dilution. Brd4 was detected with a 1:500 dilution of the rabbit polyclonal antibody 2290 (9, 10) and with goat anti-rabbit immunoglobulin G conjugated to Texas Red (1:100 dilution; Jackson Immunochemicals). Slides were mounted in Vectashield mounting medium containing 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml. Fluorescence was detected using a Leica TCS-SP5 confocal imaging system.
Brd4-E2 binding assay.
The following procedure has been described previously (5). Purified Brd4 protein (0.64 μg in 5 μl) was bound to anti-FLAG M2 agarose by incubation in a mixture of 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% Tween-20, 1 mM dithiothreitol (DTT), and 1× complete EDTA-free protease inhibitor cocktail (Roche) at room temperature for 1 h. E2 proteins were synthesized and labeled with [35S]methionine by using Promega's TNT quick-coupled reticulocyte lysate system, and the concentrations of the proteins were normalized relative to one another based on the number of methionines. Equivalent amounts of 35S-labeled E2 proteins were incubated with Brd4-agarose beads for 1 h. For the ligand-mediated dimerization experiments, AP20187 (Ariad Pharmaceuticals) was added to the Brd4 incubation reaction mixtures at the concentrations indicated in the figure legends. Bound proteins were eluted in a solution of 2% SDS, 62.5 mM Tris-HCl (pH 6.8), and 25% glycerol and separated by SDS-PAGE. For quantitative assays, the amount of 35S-labeled E2 bound to Brd4 was measured directly by suspending the beads in CytoScint fluid (ICN Biomedicals) and counting in a Wallac Microbeta counter. For the antibody-mediated dimerization assay, 5 μl of B202 hybridoma supernatant was incubated with E2 protein lysates during the incubation with the Brd4-agarose beads. As a control, the B202 antibody was digested overnight with immobilized papain (Pierce) to cleave the antibody into the monovalent Fab and Fc fragments.
Gel filtration analysis.
Nuclear extracts from CV-1 cells expressing FLAG-tagged E2 proteins were collected in buffer C (20 mM HEPES [pH 7.9], 0.4 M KCl, 1.5 mM MgCl2, 25% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor cocktail) at 4°C for 30 min and subjected to dialysis against buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 150 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and 1 mM phenylmethylsulfonyl fluoride). Two-milligram samples of nuclear extracts were fractionated into 0.3-ml volumes by Superdex 200 10/300 GL column (GE Healthcare) chromatography using a running buffer (20 mM HEPES [pH 7.9], 20% glycerol, 150 mM KCl, 0.2 mM EDTA). The fractions were analyzed by immunoblotting using anti-FLAG M2 antibody. Gel filtration standards (Bio-Rad) were run in parallel to independently assess the sizes of the complexes.
RESULTS
The C-terminal domain of the E2 protein is required for efficient Brd4 binding.
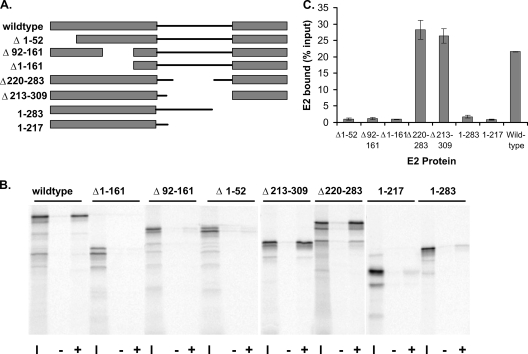
To determine which domains of the E2 protein are required for efficient Brd4 binding, a series of E2 proteins containing truncations or deletions were synthesized by in vitro transcription and translation. These proteins had deletions of either the transactivation domain (the protein lacking residues 1 to 52 [protein Δ1-52] and proteins Δ1-161 and Δ92-161), the hinge region (proteins Δ213-309 and Δ220-283), or the DNA binding/dimerization domain (the protein comprising residues 1 to 217 [protein 1-217] and protein 1-283). These 35S-labeled E2 proteins were assayed for their abilities to bind to Brd4 protein purified from insect cells. The Brd4 protein contained a C-terminal FLAG epitope tag enabling the resulting complexes to be isolated using anti-FLAG affinity beads. As shown in Fig. 1, both the transactivation and DNA binding domains were necessary for efficient binding to Brd4. Only E2 sequences from the hinge region were dispensable for Brd4 binding. Since it has already been clearly established that Brd4 interacts with the N-terminal transactivation domain of E2, we sought to determine the role of the C-terminal domain.
FIG. 1.
Efficient interaction of E2 and Brd4 requires both the transactivation and DNA binding/dimerization domains. (A) Diagram of truncated and deletion mutant E2 proteins used in the Brd4 binding assay. (B) In vitro-translated E2 proteins were tested for their abilities to bind Brd4. Aliquots (10 μl) of reticulocyte lysate (adjusted for the concentration of E2) were assayed for binding to 5 μl of Brd4 protein extract prebound to anti-FLAG immunobeads. Bound proteins were eluted and analyzed by SDS-PAGE. The input lanes (I) contain a 1/4 volume of lysate added to the binding reactions. The lanes labeled − show E2 bound in the absence of Brd4, and the lanes labeled + show E2 bound in the presence of Brd4 protein. (C) Percentage of E2 bound for each E2 protein, as obtained from two independent assays.
The DNA binding function of E2 is not required for efficient Brd4 binding.
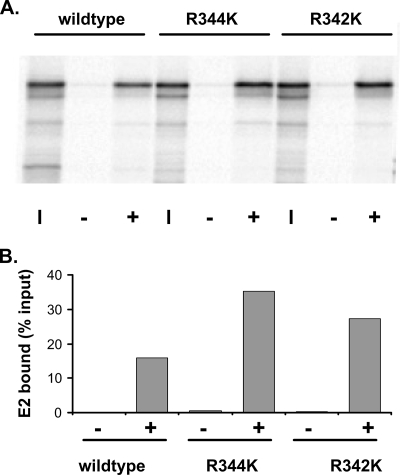
The E2 protein binds specifically to a 12-bp motif found in the regulatory elements of the viral genome (2). The DNA contact region has been clearly defined, and mutations in these crucial residues inactivate the DNA binding function of E2. DNA binding-defective proteins with mutations in two such residues, R344K and R342K (19), were tested for their abilities to bind Brd4. As shown in Fig. 2, both proteins bound Brd4 as efficiently as wild-type E2, indicating that DNA binding is not required for Brd4 binding. This finding is consistent with the observation that DNA binding is not required for the ability of E2 to bind mitotic chromosomes (30) or to enhance the association of Brd4 with chromatin (21).
FIG. 2.
The E2 DNA binding function is not required for Brd4 binding. (A) E2 proteins that are unable to bind specifically to DNA were tested in the Brd4 binding assay, as described in the legend to Fig. 1. The input lanes (I) contain lysate added to the reaction mixture at a ratio of 1/4. The lanes labeled − show E2 bound in the absence of Brd4, and the lanes labeled + show E2 bound in the presence of Brd4 protein. R344K and R342K, E2 mutant proteins with the R344K and R342K substitutions, respectively. (B) The amount of E2 bound was quantitated using a Molecular Dynamics Typhoon imager.
Antibody-mediated dimerization of the E2 protein enhances its ability to interact with Brd4.
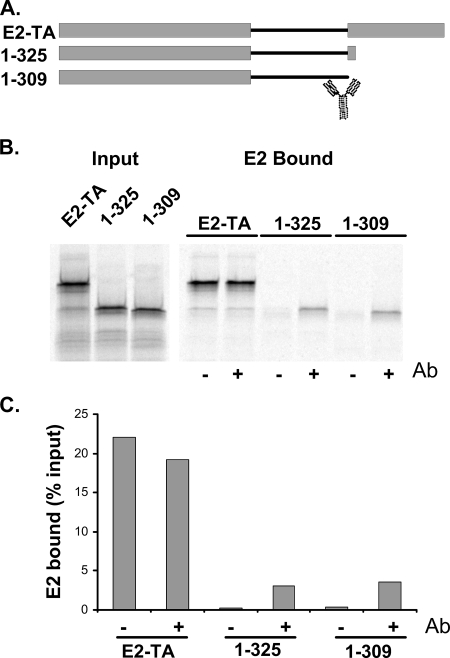
The epitope of the E2-specific monoclonal antibody B202 has been mapped to a region between amino acids 286 and 310 (E. Androphy, personal communication). Bivalent E2 antibodies have been used previously to mediate the dimerization of E2 (24), and so we tested the ability of the B202 antibody to increase Brd4 binding by dimerizing E2 proteins that were missing the C-terminal domain. As shown in Fig. 3, the addition of B202 to two different truncated proteins, proteins 1-309 and 1-325, increased binding to Brd4. Furthermore, the digestion of B202 with papain, which cleaves the antibody between the Fc and Fab fragments and results in monovalent recognition regions, was unable to enhance Brd4 binding (data not shown). Thus, the dimerization of the transactivation domain enhances the ability of E2 to bind Brd4.
FIG. 3.
Antibody-mediated dimerization enhances the ability of E2 to bind to Brd4. (A) Diagram of truncated E2 proteins used in the Brd4 binding assay. The cartoon of an antibody molecule indicates the position of the B202 epitope. E2-TA, wild-type E2 protein. (B) In vitro-translated E2 proteins were tested for their abilities to bind Brd4 in the absence or presence of the B202 antibody. The panel on the left shows results for a 1/4 volume of lysate added to the binding reactions. The panel on the right shows the bound proteins in the absence (−) and presence (+) of the B202 antibody (Ab). (C) The amount of E2 bound was quantitated using a Molecular Dynamics Typhoon imager.
Heterologous dimerization domains can substitute for the E2 C-terminal domain for Brd4 binding.
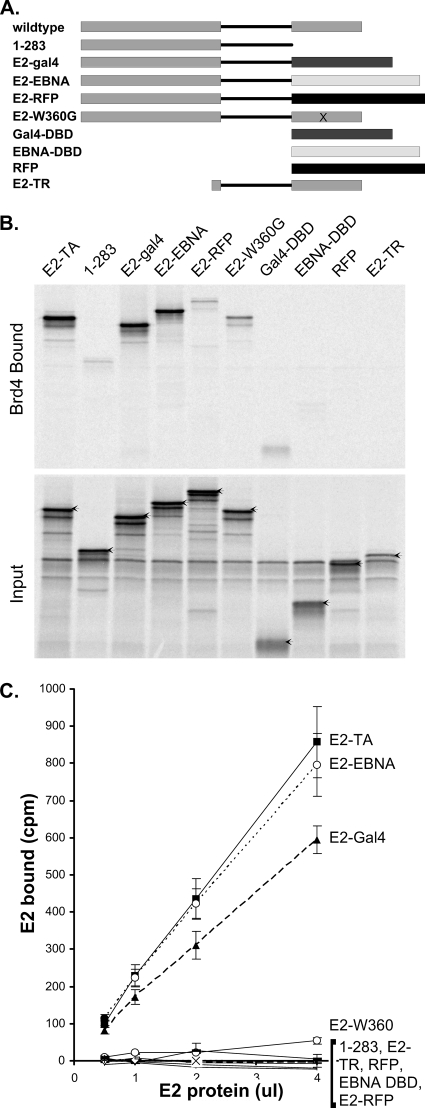
To prove further that the C-terminal function of E2 in enhancing Brd4 binding was due to dimerization, we replaced this domain with several heterologous domains that have been characterized as dimers or monomers. E2 was fused to the DNA binding domain of EBNA-1 (the Epstein-Barr virus tethering protein), which has been shown previously to have a structure very similar to that of the analogous E2 domain (6). A fusion between E2 and the Saccharomyces cerevisiae Gal4 protein DNA binding domain was also generated. This domain also forms a dimer, but the structure is quite different from those of the E2 and EBNA proteins. Finally, E2 was fused to a monomeric form of the RFP dsRed (17). As shown in Fig. 4, only the former two dimerization domains were able to restore the ability of an E2 NH protein (protein 1-283) to efficiently bind Brd4. The monomeric RFP was unable to enhance this interaction.
FIG. 4.
The efficient interaction of E2 and Brd4 requires E2 dimerization. (A) Diagram of truncated, deletion mutant, and fusion E2 proteins used in the Brd4 binding assay. DBD, DNA binding domain; E2-TR, E2 repressor protein. (B) In vitro-translated E2 proteins were tested for their abilities to bind Brd4. Aliquots (4 μl) of reticulocyte lysate (adjusted for the concentration of E2) were assayed for binding to 5 μl of Brd4 protein extract prebound to anti-FLAG immunobeads. Bound proteins were eluted and analyzed by SDS-PAGE. The input lanes contain a 1/4 volume of lysate added to the binding reactions. E2-TA, wild-type E2 protein. (C) The efficiency of E2-Brd4 binding was quantitated by adding increasing volumes of reticulocyte lysate (adjusted for the concentration of E2) to Brd4 beads. The amount bound was determined directly by scintillation counting.
An E2 protein defective in dimerization shows reduced ability to bind to the Brd4 protein.
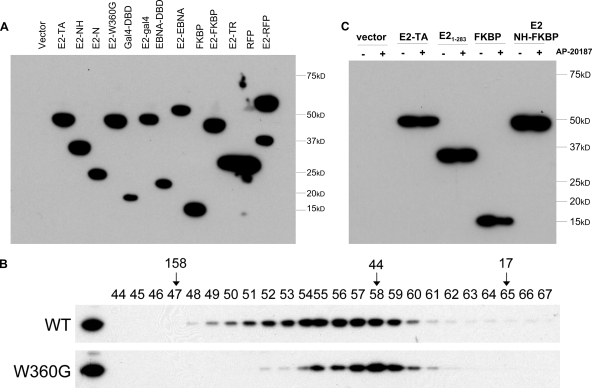
Residue W360 of the E2 protein has been shown to make crucial contacts in the dimerization interface, and mutations in this residue result in dimerization-defective proteins (24). Nuclear extracts of wild-type and E2 W360G proteins were analyzed by gel filtration to verify that the mutated protein was in fact a monomer (see Fig. 6). Then, an E2 protein containing the mutation W360G was tested for its ability to bind Brd4. Although not quite as defective as the E2 protein 1-283, E2 W360G was clearly quite defective in Brd4 binding (Fig. 4). This finding further supports the evidence that E2 binds Brd4 most efficiently as a dimer.
FIG. 6.
(A) FLAG-tagged E2 proteins were expressed in CV-1 cell lines, and the level of each protein was determined by immunoblot analysis using an anti-FLAG M2 antibody. E2-TA, wild-type E2 protein; E2-N, E2 N terminus; DBD, DNA binding domain; E2-TR, E2 repressor protein. (B) E2 W360G is defective in dimerization. Nuclear extracts from CV-1 cells expressing FLAG-tagged wild-type BPV-1 E2 (WT) and W360G proteins were fractionated by Superdex 200 10/300 GL column chromatography. The fractions were analyzed by immunoblotting using anti-FLAG M2 antibody. Gel filtration standards (Bio-Rad) were run in parallel to independently assess the sizes of the complexes and are indicated in kilodaltons. (C) FLAG-tagged E2-FKBP fusion proteins were expressed in CV-1 cell lines, and the level of each protein in the presence (+) or absence (−) of 10 nM AP20187 was determined by immunoblot analysis using an anti-FLAG M2 antibody.
Ligand-mediated dimerization of E2 restores its ability to bind efficiently to the Brd4 protein.
Several systems have been developed that utilize small-molecule “dimerizers” to bring together (or dimerize) two proteins simultaneously (8). The Argent system (Ariad Pharmaceuticals) is based on the human protein FKBP12 and its small-molecule ligands. FKBP is an abundant cytoplasmic protein that serves as the initial intracellular target for the natural-product immunosuppressive drugs FK506 and rapamycin. The FKBP system has been further optimized by the development of a synthetic homodimerizer, AP20187, which binds with subnanomolar affinity to FKBP proteins with a single amino acid substitution (F36V) while binding with 1,000-fold-reduced affinity to the wild-type protein.
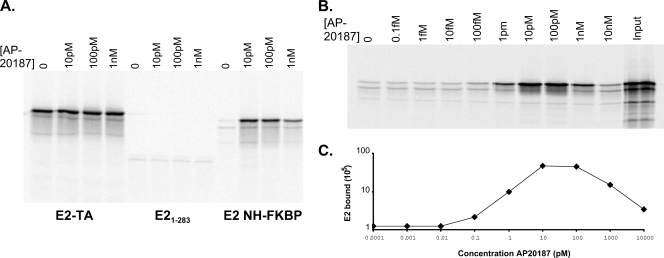
The FKBP-derived domain described above was fused in frame with the N-terminal domain and hinge sequences of the E2 protein. The resulting fusion protein was tested for its ability to bind Brd4 in the absence or presence of the AP20187 dimerizer. As shown in Fig. 5A, AP20187 had no effect on the ability of either the full-length E2 or the NH protein to interact with Brd4. However, 10 pM AP20187 was able to induce strong Brd4 binding of the fusion protein, further indicating that dimerization is required for efficient E2-Brd4 interaction. It was predicted that excess amounts of the synthetic dimerizer would inhibit the dimerization of the FKBP domain and subsequent Brd4 interaction, as the binding sites in each monomer would become saturated with excess AP20187. As shown in Fig. 5B, this scenario is exactly what was observed for the E2 NH-FKBP interaction; optimal binding peaked at AP20187 concentrations between 10 and 100 pM, and greater concentrations were inhibitory.
FIG. 5.
(A) In vitro-translated E2 proteins were tested for their abilities to bind Brd4 in the presence of increasing amounts of AP20187. Aliquots (4 μl) of reticulocyte lysate (adjusted for the concentration of E2) were assayed for binding to 5 μl of Brd4 protein extract prebound to anti-FLAG immunobeads. Bound proteins were eluted and analyzed by SDS-PAGE. E2-TA, wild-type E2 protein. (B) The optimal amount of AP20187 required to enhance E2-FKBP E2 binding to Brd4 was determined as described in the legend to panel A. Bound E2-FKBP was quantitated using a Molecular Dynamics Typhoon imager. Levels of bound E2 are expressed as percentages relative to the input.
Dimerization enhances the association of the E2 protein with mitotic chromosomes.
We have previously demonstrated that the N-terminal domain is both necessary and sufficient for association with mitotic chromosomes (4). However, the N-terminal domain is found to be associated with mitotic chromosomes much less frequently than the full-length E2 protein, which we previously concluded was due to lower expression levels of the N-terminal domain than of the full-length protein. To investigate whether dimerization can also enhance mitotic chromosome binding activity, cell lines were established that expressed the E2 transactivation domain and NH, the E2 W360 dimerization-defective protein, and the fusion proteins consisting of the E2 transactivation domain and NH with the EBNA-1 DNA binding domain, the Gal4 DNA binding domain, dsRed, and the FKBP dimerization domain. Figure 6A shows the relative expression levels of these proteins as detected by immunoblot analysis, and Fig. 6B shows a gel filtration analysis that confirms that the E2 W360G protein was defective in dimerization in vivo. Cells expressing the E2-FKBP fusion proteins were also treated with 10 nM AP20187 to ensure that AP20187 did not have any effect on the overall levels of the proteins (Fig. 6C).
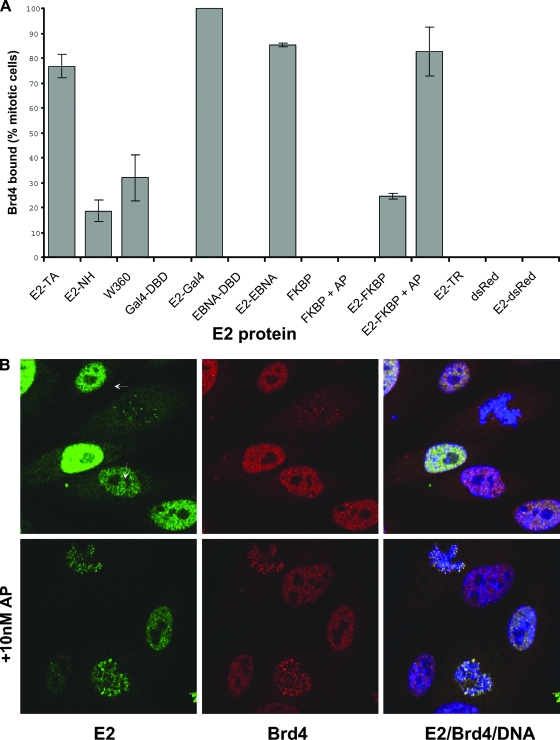
The localization of these E2 proteins, in complexes with the Brd4 protein, was examined by using indirect immunofluorescence with E2 and Brd4 antibodies. In the absence of E2, Brd4 is either undetectable or present as a diffuse cloud on mitotic chromosomes, but in the presence of E2, the two proteins colocalize in punctate speckles (21). The percentages of E2-expressing cells with E2 and Brd4 colocalized as punctate speckles on mitotic chromosomes were determined and are shown in the graph in Fig. 7A. Among cells expressing monomeric proteins containing the E2 N-terminal domain (the NH protein, E2 W360, E2-dsRED, and E2-FKBP), E2-Brd4 complexes were found in speckles bound to mitotic chromosomes in about 18 to 32% of cells. However, Brd4 was observed to bind much more efficiently to dimeric E2 proteins (the wild type, E2-Gal4, E2-EBNA, and E2-FKBP in the presence of AP20187), forming complexes in 76 to 100% of E2-expressing cells. Furthermore, not only was the percentage of cells with E2-Brd4 protein bound to mitotic chromosomes among cells expressing dimeric E2 proteins much higher than that among cells expressing monomeric proteins, but the number of E2-Brd4 speckles bound to each mitotic figure in cells with dimeric E2 proteins was also greatly increased compared to that in cells with monomeric proteins, although the sizes of the individual speckles remained similar. This pattern can be observed in the representative image in Fig. 7B, in which the addition of the AP20187 dimerizer is shown to greatly increase the number of E2-Brd4 speckles observed in each mitotic cell, as well as the number of positive mitotic cells. Thus, E2 dimerization increases the affinity of Brd4 binding in vitro but also greatly enhances the interaction of the E2-Brd4 complex with chromatin in vivo.
FIG. 7.
(A) The E2 proteins indicated were expressed in CV-1 cells, and the localization of each E2 protein in a complex with Brd4 was detected by indirect immunofluorescence. The percentage of mitotic E2-expressing cells that had recruited Brd4 in speckles to the mitotic chromosomes is shown, along with the standard deviation of results derived from several experiments. The bars labeled +AP show results for cells treated with 10 nM AP20187. E2-TA, wild-type E2 protein; DBD, DNA binding domain; E2-TR, E2 repressor protein. (B) Representative indirect immunofluorescence images of E2-FKBP proteins (green), including proteins in complexes with Brd4 (red), on mitotic chromosomes in the presence or absence of 10 nM AP20187. Cellular chromosomes are stained with DAPI (blue). Mitotic cells are indicated by arrows.
DISCUSSION
The transactivation domain of the papillomavirus E2 protein is crucial for interaction with the Brd4 protein; proteins lacking this domain do not interact with Brd4, and the R37-I73 mutation disrupts binding (5, 27, 28). However, we demonstrate here that the isolated transactivation domain has much weaker affinity for Brd4 than the full-length protein. This finding seems at odds with the results of previous studies in which we demonstrated that the E2 transactivation domain is sufficient for interaction with mitotic chromosomes (4). However, in those studies, the domain was found to be associated with mitotic chromosomes in only a low percentage of cells, which we concluded was due to poor protein expression.
In this study, further analysis of the E2-Brd4 interaction demonstrated that neither the hinge region of E2 nor the DNA binding function is required for Brd4 interaction but that the dimerization function of the C-terminal domain is necessary for strong binding. Both the in vivo chromosomal binding and in vitro Brd4 binding role of the dimeric DNA binding domain of E2 can be fulfilled by the corresponding dimeric DNA binding domain of Gal4 or EBNA-1, but not by the monomeric RFP. Efficient Brd4 binding, both in vivo and in vitro, could also be retained by the fusion of E2 to an FKBP12-derived domain in which dimerization could be regulated by a small-molecule ligand.
In further support of the finding that E2 dimerization is important for efficient Brd4 binding, Kurg et al. generated a novel E2 single-chain heterodimer protein which contained one transactivation domain linked to a dimeric DNA binding domain (16). This protein can bind Brd4, but much less efficiently than the wild-type E2 protein. It can also activate transcription in a Brd4-dependent manner, but the requirement for Brd4 is also much less than that of the wild-type protein.
The E2 transactivation domain has also been implicated in self-interaction, and the looping of DNA containing E2 binding sites has been observed previously by electron microscopy for BPV-1, human papillomavirus type 16 (HPV-16), and HPV-11 E2 proteins (3, 11, 15, 29). For HPV-16 E2, self-interaction involves residues R37 and I73, which are also crucial for Brd4 binding (1, 5, 20), and thus, self-interaction may modulate Brd4 binding. However, the precise interactions between N-terminal domains of different E2 proteins are not consistent and may result from nonspecific crystal packaging (14). A structure of the BPV-1 N-terminal domain revealed a disulfide bond between cysteine residues at positions 57 in the two monomers, which interfered with the E1 interaction face of the domain (25). A different interface was also observed previously in the protein crystal lattice obtained for the HPV-11 E2 protein (31). Thus, while self-interaction through the N-terminal domain may be important for E2 function, more studies are required to determine the precise details for each E2 protein.
We propose that the dimerization of E2 through the C-terminal domain enables two transactivation domains to interact with two copies of the Brd4 protein. This pattern would increase the local concentration of E2 interaction surfaces and thereby increase Brd4 binding. In support of an E2-Brd4 heterotetramer, a recent crystallographic structure of the HPV-16 E2 transactivation domain in a complex with a C-terminal 20-amino-acid peptide from Brd4 demonstrated that two Brd4 peptides were found sandwiched between two E2 N-terminal domains (1). The dimerization of the E2 protein through the C-terminal domain would greatly stabilize such a tetramer complex. It has also recently been shown that Brd2, a protein related to Brd4, forms dimers mediated through one of the bromodomains (bromodomain 1) (22, 33). If Brd4 were to dimerize in a similar fashion, E2 and Brd4 could also form higher-order complexes whereby each dimeric E2 protein could interact with the C-terminal tails from two different dimeric Brd4 proteins. This cooperative binding would increase the local binding of E2 and Brd4 and potentially allow the formation of large complexes consisting of a lattice of multiple E2 and Brd4 dimers, which could explain why the Brd4-E2 complex is observed as punctate speckles on mitotic chromosomes (21) whereas, in the absence of E2, Brd4 forms a diffuse coating (10).
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 21 May 2008.
REFERENCES
- 1.Abbate, E. A., C. Voitenleitner, and M. R. Botchan. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24877-889. [DOI] [PubMed] [Google Scholar]
- 2.Androphy, E. J., D. R. Lowy, and J. T. Schiller. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 32570-73. [DOI] [PubMed] [Google Scholar]
- 3.Antson, A. A., J. E. Burns, O. V. Moroz, D. J. Scott, C. M. Sanders, I. B. Bronstein, G. G. Dodson, K. S. Wilson, and N. J. Maitland. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403805-809. [DOI] [PubMed] [Google Scholar]
- 4.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 with mitotic chromosomes. Virology 270124-134. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, W. J. Furey, A. M. Edwards, and L. Frappier. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 8339-46. [DOI] [PubMed] [Google Scholar]
- 7.Corina, K., S. R. Grossman, J. Barsoum, S. S. Prakash, E. J. Androphy, and R. B. Pepinsky. 1993. The tryptophan bridge is a critical feature of the papillomavirus E2 DNA binding domain. Virology 391-396. [DOI] [PubMed]
- 8.Crabtree, G. R., and S. L. Schreiber. 1996. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem. Sci. 21418-422. [DOI] [PubMed] [Google Scholar]
- 9.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 1008758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Ramon, E. E., J. E. Burns, W. Zhang, H. F. Walker, S. Allen, A. A. Antson, and N. J. Maitland. 2008. Dimerization of the human papillomavirus type 16 E2 N terminus results in DNA looping within the upstream regulatory region. J. Virol. 824853-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19523-534. [DOI] [PubMed] [Google Scholar]
- 14.Janin, J., F. Rodier, P. Chakrabarti, and R. P. Bahadur. 2007. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr. D 631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight, J. D., R. Li, and M. Botchan. 1991. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc. Natl. Acad. Sci. USA 883204-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurg, R., H. Tekkel, A. Abroi, and M. Ustav. 2006. Characterization of the functional activities of the bovine papillomavirus type 1 E2 protein single-chain heterodimers. J. Virol. 8011218-11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matz, M. V., A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17969-973. [DOI] [PubMed] [Google Scholar]
- 18.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, A. A., and G. Myers. 1997. The E2 proteins: an update, p. III54-III99. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, NM.
- 20.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, Y., T. Umehara, K. Nakano, M. K. Jang, M. Shirouzu, S. Morita, H. Uda-Tochio, H. Hamana, T. Terada, N. Adachi, T. Matsumoto, A. Tanaka, M. Horikoshi, K. Ozato, B. Padmanabhan, and S. Yokoyama. 2007. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2824193-4201. [DOI] [PubMed] [Google Scholar]
- 23.Penrose, K. J., and A. A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 746031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prakash, S. S., S. R. Grossman, R. B. Pepinsky, L. A. Laimins, and E. J. Androphy. 1992. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 transactivator. Genes Dev. 6105-116. [DOI] [PubMed] [Google Scholar]
- 25.Sanders, C. M., D. Sizov, P. R. Seavers, M. Ortiz-Lombardia, and A. A. Antson. 2007. Transcription activator structure reveals redox control of a replication initiation reaction. Nucleic Acids Res. 353504-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweiger, M.-R., M. Ottinger, J. You, and P. M. Howley. 2007. Brd4-independent transcriptional repression function of the papillomavirus E2 proteins. J. Virol. 819612-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweiger, M.-R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus e2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senechal, H., G. G. Poirier, B. Coulombe, L. A. Laimins, and J. Archambault. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 35810-17. [DOI] [PubMed] [Google Scholar]
- 29.Sim, J., S. Ozgur, B. Y. Lin, J. H. Yu, T. R. Broker, L. T. Chow, and J. Griffith. 2008. Remodeling of the human papillomavirus type 11 replication origin into discrete nucleoprotein particles and looped structures by the E2 protein. J. Mol. Biol. 3751165-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 722079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Y., R. Coulombe, D. R. Cameron, L. Thauvette, M. J. Massariol, L. M. Amon, D. Fink, S. Titolo, E. Welchner, C. Yoakim, J. Archambault, and P. W. White. 2004. Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J. Biol. Chem. 2796976-6985. [DOI] [PubMed] [Google Scholar]
- 32.Winokur, P. L., and A. A. McBride. 1996. The transactivation and DNA binding domains of the BPV-1 E2 protein have different roles in cooperative origin binding with the E1 protein. Virology 22144-53. [DOI] [PubMed] [Google Scholar]
- 33.Wu, S. Y., and C. M. Chiang. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 28213141-13145. [DOI] [PubMed] [Google Scholar]
- 34.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]