Abstract
Gammaherpesvirus 68 (γHV68, or MHV68) is a naturally occurring rodent pathogen that replicates to high titer in cell culture and is amenable to in vivo experimental evaluation of viral and host determinants of gammaherpesvirus disease. However, the inability of MHV68 to transform primary murine B cells in culture, the absence of a robust cell culture latency system, and the paucity of MHV68-positive tumor cell lines have limited an understanding of the molecular mechanisms by which MHV68 modulates the host cell during latency and reactivation. To facilitate a more complete understanding of viral and host determinants that regulate MHV68 latency and reactivation in B cells, we generated a recombinant MHV68 virus that encodes a hygromycin resistance protein fused to enhanced green fluorescent protein as a means to select cells in culture that harbor latent virus. We utilized this virus to infect the A20 murine mature B-cell line and evaluate reactivation competence following treatment with diverse stimuli to reveal viral gene expression, DNA replication, and production of progeny virions. Comparative analyses of parental and infected A20 cells indicated a correlation between infection and alterations in DNA damage signaling following etoposide treatment. The data described in this study highlight the potential utility of this new cell culture-based system to dissect molecular mechanisms that regulate MHV68 latency and reactivation, as well as having the potential of illuminating biochemical alterations that contribute to gammaherpesvirus pathogenesis. In addition, such cell lines may be of value in evaluating targeted therapies to gammaherpesvirus-related tumors.
Gammaherpesvirus 68 (γHV68 or MHV68) is a rodent rhadinovirus found naturally in European bank voles and wood mice (6, 7). As a rodent pathogen, MHV68 infection of laboratory mice offers an experimental system to investigate aspects of gammaherpesvirus pathogenesis. Following experimental inoculation of laboratory mice, MHV68 undergoes productive replication at the site of inoculation, which facilitates dissemination to distal organs and is followed by quiescence or latency, a form of infection where viral gene expression is restricted and progeny virions are not produced (41, 51, 71). In the case of MHV68, latent infection is established in B lymphocytes, as well as epithelial cells, macrophages, and dendritic cells (19, 56, 58, 74). Latency is maintained for the lifetime of the host, and latently infected cells retain the capacity to reinitiate the productive replication cycle, a process termed reactivation (45, 51, 52).
The genetic malleability of MHV68 enables the identification of viral genes involved in specific aspects of viral pathogenesis by targeted mutagenesis (1). For example, such approaches have demonstrated the importance of the unique MHV68 M2 gene product in latency establishment, reactivation from latency, and promoting B-cell differentiation (24, 26), as well as defining the roles of conserved genes, such as orf73 (LANA homolog) in latency maintenance (22, 42) or orf72 (v-cyclin) in replication and reactivation (61, 68, 69). Paired with the capacity to generate distinct mutations in the host, or to infect inbred mouse strains with unique and disparate immunologic properties, MHV68 enables a critical evaluation of virus-host interactions that influence disease and makes MHV68 infection of mice a powerful system for characterizing gammaherpesvirus pathogenesis.
Despite the fundamental contributions of such in vivo experiments, the biochemical mechanisms by which MHV68 influences or alters host cells during latent infections, particularly the discrete molecular functions of viral genes in facilitating latent infection and recognizing and modulating host cell responses to various stimuli, remain largely undetermined. Such studies have been limited by the lack of a robust cell culture-based system for their evaluation in the context of a persisting viral infection. To date it has not been possible to transform primary murine B cells with MHV68, and MHV68 infection and maintenance in cultured B-cell lines is inefficient. Moreover, only one MHV68-positive tumor cell line (S11) has been isolated (63). Therefore, comparative analyses of how viral gene expression influences host B cells relative to uninfected cells are limited.
To facilitate MHV68 infection and maintenance in cultured cells, we generated a recombinant virus that encodes a hygromycin resistance protein fused to enhanced green fluorescent protein (EGFP). Infection of the mature murine B-cell line A20 followed by selection with hygromycin resulted in the establishment and maintenance of MHV68-positive cell lines (A20-HE) that can be carried indefinitely. Virus reactivation from A20-HE cells was induced by mitogenic stimulation, including phorbol-12-myristate-13-acetate (PMA) treatment and cross-linking surface immunoglobulin (Ig), and was characterized by increased detection of lytic cycle-associated viral antigens, viral DNA synthesis, and production of progeny virions. We also found that antimitogenic treatments, including the DNA damage-inducing drugs etoposide and camptothecin, elicited expression of viral replication-associated genes. These data suggest that MHV68 is primed to recognize and respond to diverse and apparently disparate cellular stimuli. As such, this tissue culture latency model offers a new opportunity to evaluate the MHV68-host relationship at a cellular level toward a broader understanding of the discrete biochemical events that regulate MHV68 latency and reactivation, thus providing a functional complement to in vivo pathogenesis analyses.
MATERIALS AND METHODS
Cell culture and viruses.
NIH 3T12 fibroblasts, NIH 3T3 fibroblasts, murine embryonic fibroblasts (MEFs), and Vero-cre cells (42) were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. A20, WEHI-231, and NSO cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 50 mM 2-mercaptoethanol (cRPMI). A20-HE cells were selected and cultured in cRPMI containing 300 μg/ml hygromycin B sulfate (Calbiochem). Wild-type MHV68 was strain WUMS (ATCC VR1465). Recombinant hygromycin phosphotransferase (Hpt)-EGFP-expressing MHV68 viruses (described in detail below) were propagated by bacterial artificial chromosome (BAC) transfection and expansion to P1 stocks in Vero-cre cells. Viral titers for stocks and for growth and reactivation assays were determined by plaque assays as described previously (61).
Generation of recombinant viruses.
The Hpt-EGFP (HE) fusion protein expression cassette (nucleotides [nt] 1 to 3130) was cloned by PCR from pHygEGFP (Clontech) with the following primers: NLHE_US, 5′-TTTACACCTTTACACTCAATATTGGCCATTAGCCA-3′; NLHE_DS, 5′-AAGCACTCTTGTCACATCCTTATCGATTTTACCAC-3′. The targeting construct was generated by overlap extension PCR to insert the HE expression cassette in the antisense orientation into the PmlI restriction site at nt 46347 in the MHV68 genome (see Fig. 2A, below) (70), a region previously defined as capable of supporting insertions (43). The flanking arms for recombination encompassed MHV68 nt 45377 to 47151. The targeting construct was cloned into pCR-Blunt (Invitrogen), digested with NsiI, and cloned into pGS284. Allelic exchange was performed utilizing the MHV68 BAC as described elsewhere (2, 42). HE-positive BAC colonies were identified by colony hybridization using a radiolabeled DNA probe from the Hpt-coding region of pHygEGFP. Candidate HE-positive MHV68 BAC clones initially were screened for genetic integrity by restriction digestion analysis using the parental wild-type (WT) BAC as a comparative control. Appropriate recombination for two independently derived clones, HE 1.1A and HE 2.1A, was further verified by Southern blotting (see Fig. 2A, below). Two μg of parental MHV68 BAC, HE 1.1A, and HE 2.1A DNA was digested overnight with BamHI, EcoRV, or HindIII and resolved by agarose gel electrophoresis. DNA was transferred to a nylon membrane and hybridized with a 32P-labeled, PCR-amplified probe homologous to the flanking arms of the targeting construct. The genomic position of restriction sites were as follows for WT or HE BACs, accounting for the 3,130-nt HE insertion: WT, BamHI at nt 42402 and 49939; HE, BamHI at nt 42402, 48386, and 53069; WT, EcoRV at nt 42579, 46420, and 50210; HE, EcoRV at nt 42579, 49550, and 53340; WT, HindIII at nt 45237 and 49653; HE, HindIII at nt 45237, 48725, and 52783.
FIG. 2.
Derivation of recombinant hygromycin-selectable MHV68 viruses and stably maintaining A20 cell lines. (A) A Hprt-EGFP (HE) fusion protein driven by the human cytomegalovirus immediate-early promoter was inserted into the MHV68 BAC between orfs27 and 29b by allelic exchange. Fidelity of recombination for two independently derived isolates (HE 1.1A and HE 2.1A) was analyzed relative to the parental MHV68 BAC by endonuclease restriction digestion and Southern blotting. (B) Analysis of recombinant virus replication. HE 1.1A, HE 2.1A, and WT viruses were generated by BAC transfection into a Cre recombinase-expressing cell line to remove the floxed BAC vector sequences. Recovered viruses were analyzed in multistep growth analyses following infection of primary MEFs at an MOI of 0.05 PFU/cell. Cells were harvested at the indicated times postinfection, and viral titers were determined by plaque assay. Results are means of triplicate samples. Error bars represent standard deviations. (C) Establishment of MHV68-stable A20 cell lines. A20 B cells were infected with HE 1.1A and HE 2.1A MHV68 viruses at an MOI of 10 PFU/cell and selected with hygromycin (300 μg/ml). Representative immunofluorescence and differential interference contrast images of derivative A20-HE1 and A20-HE2 cell lines captured at 100× magnification are shown.
Inducing stimuli and drugs.
PMA (Sigma-Aldrich) was reconstituted in ethanol. Etoposide, camptothecin, staurosporine, and cisplatin (Calbiochem) were reconstituted in dimethyl sulfoxide. Sodium butyrate and lipopolysaccharide (LPS; Sigma-Aldrich) were dissolved in phosphate-buffered saline (PBS). Donkey anti-mouse CD40 and mouse monoclonal anti-Fas were purchased from Becton Dickinson. Donkey anti-mouse IgG Fab was purchased from Jackson ImmunoResearch. All treatments were performed at the concentrations indicated in the Results section and figure legends.
Immunoblot analyses.
Cells were lysed with alternative RIPA buffer (150 mM NaCl, 20 mM Tris, 2 mM EDTA, 1% NP-40, 0.25% deoxycholate, 1 mM NaF, and 1 mM Na3VO4 supplemented with complete mini-EDTA-free protease inhibitors [Roche]) and quantitated using the Bio-Rad DC protein assay prior to resuspending in Laemmli sample buffer (32). Samples were heated to 100°C for 10 min and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Resolved proteins were transferred to nitrocellulose and identified by immunoblotting with chicken anti-ORF59 (62), rabbit anti-v-cyclin (67), rabbit anti-MHV68 (73), goat anti-CDK2 (sc-163-G; Santa Cruz Biotechnology), mouse anti-p53 (1C12), rabbit anti-phospho-S15 (S18 in mouse) p53, or rabbit anti-phospho-(S/T)Q (Cell Signaling Technology). Immobilized antigen and antibody were detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagents (Amersham/GE Healthcare) and exposed to film.
Immunofluorescence.
Untreated or treated cells at ca. 1 × 106/ml were adhered to poly-l-lysine (Sigma-Aldrich)-coated coverslips for 30 to 60 min at room temperature. Cells were fixed with 10% formalin, washed with PBS, permeabilized, and blocked with 0.1% Triton X-100 (TX100) in PBS containing 5% bovine serum albumin (blocking buffer). Primary antibodies (goat anti-GFP from Rockland Immunochemicals, to increase the sensitivity of detection for Hygro-EGFP, or polyclonal MHV68 antiserum [73]) were diluted in blocking buffer with 1% normal donkey serum and incubated for 1 h at 37°C. Cells were washed three times with 0.1% TX100 in PBS (wash buffer) and incubated with species-appropriate Alexa Fluor-conjugated secondary antibodies (488 for Hygro-EGFP or 568 for MHV68 antiserum; Molecular Probes/Invitrogen) diluted in blocking buffer at 37°C for 45 to 60 min. Cells were washed three times with wash buffer and once with PBS and mounted on slides using ProLong Anti-fade Gold reagent (Molecular Probes/Invitrogen). DNA was detected with 4′,6′-diamidino-2-phenylindole (DAPI) in the mounting medium. Images were captured using a Zeiss Axio A1 Imager fluorescence microscope.
RNA isolation and RT-PCR.
RNA was isolated from A20, WEHI-231, or NSO cells 24 h after infection at a multiplicity of infection (MOI) of 10 PFU/cell using guanidium isothiocyanate-phenol as previously described (3). Reverse transcription-PCR (RT-PCR) for spliced orf50 or orf73 transcripts was performed as previously described (3, 37). RNA was isolated from A20-HE1 or A20-HE2 cells prior to or at 8 h and 24 h after treatment with 20 ng/ml PMA, contaminating DNA was removed by DNase I digestion (Invitrogen), and RNA was reverse transcribed using SuperScript II (Invitrogen) as previously described (21). No-RT controls were carried out in parallel. Resulting cDNAs were diluted 1:4 in water. Two μl of cDNA was used in each PCR mixture. Primers for orf6, orf8, orf9, mK3, orf72, orf73, M1, M3, M4, M6, M8/orf57, and M9 were previously described (inner primer sets [71]). Primers for orf50 and GAPDH were previously described (21). Primers for orf25 were 25-1, 5′-CCCATGGCAGTATCTAAAGAGG-3′, and 25-2, 5′-CGGAGTTGCTCTAACAGTTGTG-3′. Primers for orf45 were 45-1, 5′-AAAGTCTGCTGGGTATCGTCAC-3′, and 45-2, 5′-ATGGGTCATCAACTCCAGACTC-3′. Cycling conditions were as follows: 94°C for 2 min, 22 cycles (A20-HE1) or 30 cycles (A20-HE2) at 94°C for 60 seconds, 62°C for 60 seconds, and 72°C for 60 seconds, and 72°C for 2 min. M2 was detected using nested primer sets (3). Round 1 PCR was performed as above for 30 cycles. Round 2 was performed using 5 μl of round 1 product as template for the round 2 reaction for 30 cycles as above. S11-derived cDNA was used as a positive control for M2 amplification.
Cell viability assays.
For acute titrations, cells were treated with dimethyl sulfoxide (vehicle), etoposide, or anti-Fas for 18 h in the presence or absence of 50 ng/ml cidofovir as indicated in Results and the figure legends. For recovery assays, cells were cultured in the presence of cidofovir for 4 h with vehicle or 25 μM etoposide, pelleted, and rinsed with PBS. Cells were pelleted again, resuspended in complete RPMI, plated, and cultured for 1 week in the presence of cidofovir. Cell viability was assessed using the CellTiter Glo viability assay (Promega) per the manufacturer's instructions.
RESULTS
Murine B-cell lines do not support lytic MHV68 replication.
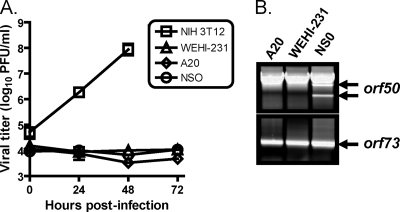
In an effort to establish latently infected B-cell lines, we infected the murine B-cell lines A20 and WEHI-231, the myeloma cell line NSO (which were previously demonstrated to support long-term, persistent MHV68 infection in culture [57]), and NIH 3T12 fibroblasts as a positive control for lytic virus replication. In these experiments, while NIH 3T12 cells supported robust viral replication, viral titers did not increase over time in the infected A20, WEHI-231, or NSO cell lines (Fig. 1A). RT-PCR for the spliced latency-associated orf73 transcript (3) indicated that all B-cell lines were infected (Fig. 1B). Moreover, RT-PCR did not detect spliced orf50 transcripts, a requirement to generate the functional Rta lytic transactivator protein (46), in the A20 or WEHI-231 cell lines. In contrast, spliced orf50 transcripts were detected following infection of the NSO myeloma cell line, consistent with previous data indicating that the NSO cell line supports persistent MHV68 replication (57). These data suggest that murine B-cell lines support latent infection by MHV68.
FIG. 1.
Murine B cells do not support MHV68 lytic replication. (A) Murine B-cell lines A20 and WEHI-231, myeloma cell line NSO, and NIH 3T12 fibroblasts were infected with MHV68 at an MOI of 10 PFU/cell. Cells were harvested at the indicated times postinfection, and viral titers were determined by plaque assay. Results are means of triplicate samples. Error bars represent standard deviations. (B) RT-PCR analyses of spliced orf50 (Rta) and orf73 (mLANA) transcripts 24 h postinfection. Spliced orf50 transcripts (indicated by arrows) are associated with and required for lytic replication. Spliced orf73 transcript are present during lytic and latent phases of infection.
Generation of a recombinant selectable MHV68.
Although we detected orf73 transcription in the MHV68-infected A20 and WEHI-231 cell lines, several attempts to establish and maintain MHV68 B-cell lines were not successful. We reasoned that, analogous to Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (12, 18), MHV68 maintenance in transformed B cells may be inefficient due to the presumed lack of selection pressures on the cells for maintaining viral infection. To facilitate MHV68 maintenance in cultured cell lines, we generated a recombinant virus that encodes a hygromycin resistance protein fused to EGFP in the MHV68 genome between open reading frames (ORFs) 27 and 29b (Fig. 2A). We previously demonstrated that this locus in the MHV68 genome supports transgene insertions (30, 43). Appropriate recombination was verified by restriction digestion and Southern blotting (Fig. 2A). Lytic replication of two independently derived transgenic viruses (HE 1.1A and HE 2.1A) was analyzed relative to wild-type (WT) BAC-derived MHV68 over time following low multiplicity of infection of MEFs. Both HE 1.1A and HE 2.1A replicated equivalently to WT MHV68 in these experiments (Fig. 2B), indicating the transgenic viruses are fully replication competent.
Establishment of MHV68-maintaining B-cell lines.
We next tested the capacity of MHV68-HE viruses to infect and persist in NSO, A20, and WEHI-231 cell lines. Cells were adsorbed for 2 h with either HE or WT MHV68 viruses at multiplicities of 10 PFU per cell. Infected cells were then cultured in the presence of 300 μg/ml hygromycin B sulfate until WT-infected cells succumbed to selection and HE virus-infected, hygromycin-resistant cells expanded. HE virus-infected NSO cells were readily selected within 1 week postinfection, while selection of MHV68-infected A20 cells took approximately 3 weeks to select at the concentration of hygromycin used. Notably, we were unsuccessful in establishing MHV68-infected WEHI-231 cell lines. It is conceivable that WEHI-231 cells, which express surface IgM but are incapable of class-switch recombination (53), represent an immature B-cell differentiation stage that does not support MHV68 latency. This idea is supported in vivo by the preferential establishment of long-term MHV68 latency in memory B cells (20, 75). In initial analyses, NSO-HE cells maintained titers of >105 PFU/ml at cellular densities of ca. 2 × 105 to 5 × 105 cells per ml, and the supernatants recovered from the A20-HE infected cell lines exhibited titers that varied between 103 and 104 PFU/ml at densities of ca. 0.5 × 106 to 1.5 × 106 cells per ml. Representative differential interference contrast and EGFP expression images for selected HE 1.1A and HE 2.1A-infected A20 cell lines are shown (Fig. 2C). The A20-derived cell lines were pursued for further characterization. The MHV68-infected A20 cells lines are referred to as A20-HE1 or A20-HE2 (indicative of the BAC-derived recombinant MHV68 isolate used for infection). For the experiments presented in this report, cells were maintained in bulk populations.
Reactivation of A20-HE cells.
Herpesvirus infections are characterized by two very distinct stages: a lytic or productive replication phase, in which viral genes are abundantly and promiscuously expressed, the viral genome is replicated, and progeny virions are produced, and a latent or quiescent phase, in which the viral genome is maintained with limited viral gene expression. In general, herpesviruses maintain the capacity to reactivate, or reinitiate the lytic phase, from latency following exposure to a variety of stimuli. For gammaherpesviruses that latently infect B cells, these stimuli classically include phorbol esters, such as PMA (also known as TPA), promitogenic or differentiation-inducing stimuli such as BCR ligation, or inhibition of histone deacetylases by butyric acid (NaB). We therefore tested the capacity of A20-HE1 and A20-HE2 cells to respond to each of these treatments. We also treated cells with the bacterial cell wall mitogen LPS, which we previously demonstrated could enhance reactivation of explanted splenocytes from latently infected mice (44), and agonistic antibodies to the B-cell costimulatory molecule CD40, which is critical to memory B-cell development and to limiting persistent MHV68 replication in vivo (29, 75).
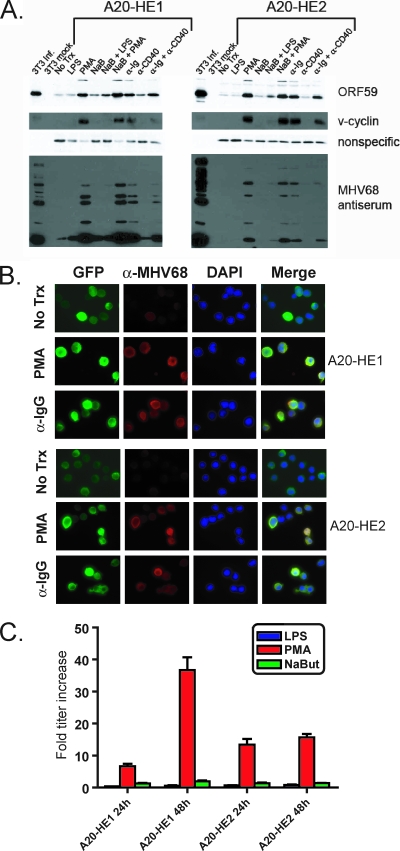
We initially analyzed A20-HE reactivation by immunoblotting with MHV68 antiserum and polyclonal antibodies to the viral DNA polymerase processivity factor encoded by ORF59. We also assessed induction of the MHV68 cyclin homolog (v-cyclin), which is critical for viral reactivation from latency (61, 69). A20-HE cells reactivated in response to PMA treatment and BCR cross-linking (anti-Ig) as evidenced by production of lytic antigens and detection of ORF59 and v-cyclin (Fig. 3A). Mock-infected and infected NIH 3T3 fibroblasts, which support lytic MHV68 replication, served as negative and positive controls for these experiments, respectively. LPS, NaB, and anti-CD40 treatments did not induce detectable expression of these antigens at the time points analyzed. Moreover, CD40 cross-linking appeared to reduce anti-Ig-induced reactivation in these assays, perhaps reflecting a role for CD40 signaling in promoting MHV68 latency and limiting persistent virus replication (29, 75). Parallel treatments of the MHV68-positive S11 lymphoma line did not result in any detectable increase in expression of the antigens described above (data not shown), an observation that reiterates the poor reactivatability of the S11 cell line (44).
FIG. 3.
Induction of A20-HE cell reactivation. (A) Immunoblot analysis for lytic cycle-associated antigens following B-cell-activating stimulation. A total of 5 × 105 A20-HE cells per ml were treated as follows: 10 μg/ml LPS, 20 ng/ml PMA, 2 mM NaB, 5 μg/ml anti-immunoglobulin G (α-Ig), 2.5 μg/ml anti-CD40, or the combinations indicated at the same concentrations. Mock-infected or infected (MOI, 5 PFU/cell, 24 h postinfection) NIH 3T3 fibroblasts served as respective positive and negative controls. Cells were harvested 24 h after treatment. Forty μg of total protein for A20-HE cell samples (12.5 μg for 3T3s) was resolved by SDS-PAGE followed by immunoblot analysis for the indicated proteins. The nonspecific band was included as a loading control for A20-HE samples. (B) Immunofluorescence analysis following stimulation. A20-HE cells were treated with the indicated stimuli as in panel A and processed for immunofluorescence microscopy 24 h after treatment. Cells were stained with antibodies to GFP and MHV68 antisera. DNA was detected with DAPI in the mounting medium. Magnification, ×100. (C) Virus production following stimulation. A20-HE cells were treated with the indicated stimuli as for panel A. Cells were harvested at the indicated times posttreatment, and viral titers were determined by plaque assay. Data represent the fold increase in titer following treatment relative to mock-treated controls. Results are means of triplicate samples. Error bars represent standard deviations.
Immunofluorescence analyses (IFA) with MHV68 antiserum and anti-ORF59 (not shown) confirmed that PMA and anti-Ig cross-linking induced the production of lytic antigens (Fig. 3B). These stimuli also increase expression of the human cytomegalovirus immediate-early promoter-driven Hygro-EGFP fusion protein (note the increase in EGFP “bright” cells and the correlation with induction of MHV68 lytic antigen staining) as a potential correlate of virus reactivation. We note that for untreated A20-HE1 and A20-HE2 cells the levels of spontaneous reactivation, as quantitated by IFA with MHV68 antiserum, were 1 to 5% and 0.1 to 0.5%, respectively. Reactivating stimuli resulted in positive staining for ca. 80 to 95% of A20-HE1 cells and 50 to 80% of A20-HE2 cells by 24 h posttreatment. Additionally, we analyzed viral titers following treatment with LPS, PMA, or NaB (Fig. 3C). Consistent with the immunoblotting and IFA, PMA treatment elicited greater than 10-fold increases in viral titers relative to untreated controls for both A20-HE1 and A20-HE2 cells by 48 h posttreatment, while titers remained relatively unchanged following LPS or NaB treatments. Together, these data demonstrate that the A20 B-cell line is capable of maintaining hygromycin-selectable recombinant MHV68 genomes in a fully reactivation-competent state.
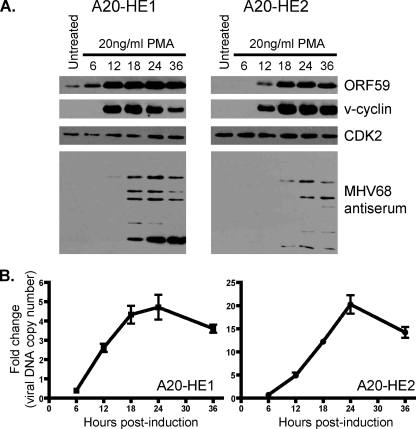
To evaluate the kinetics of MHV68 reactivation following PMA stimulation, we performed a time course analysis of viral antigen production and viral DNA synthesis. For both A20-HE1 and A20-HE2 cells, ORF59 and v-cyclin proteins were rapidly induced upon stimulation and increased in abundance over time (Fig. 4A). The low-level presence of ORF59 in unstimulated A20-HE1 cells was likely a consequence of spontaneous reactivation in the culture. For A20-HE1 cells, antigens recognized by the MHV68 polyclonal antiserum raised in MHV68 rabbits was maximal by 18 h posttreatment (as was v-cyclin detection), while detection of MHV68 antigens was maximal 24 to 36 h posttreatment for A20-HE2 cells. These findings suggest that A20-HE1 cells more readily respond to PMA-mediated induction. Replication of viral DNA, as assessed by quantitative PCR, demonstrated that DNA synthesis begins between 6 and 12 h poststimulation and peaks at ca. 24 h following PMA treatment (Fig. 4B), which correlated with maximal detection of MHV68 antigens by immunoblotting (Fig. 4A). We speculate that the apparently reduced levels of DNA replication for A20-HE1 cells were a consequence of the higher levels of spontaneous reactivation and thus increased basal levels of viral genomes present prior to PMA stimulation.
FIG. 4.
Induction time course following PMA treatment. A20-HE cells (5 × 105 per ml) were treated with 20 ng/ml PMA and harvested at the indicated times posttreatment. (A) Total protein (A20-HE1, 32 μg; A20-HE2, 50 μg) was resolved by SDS-PAGE followed by immunoblot analysis for the indicated antigens. Cellular CDK2 was used as a loading control. (B) Total DNA (200 ng) was analyzed by quantitative PCR for the presence of viral genome using orf50-specific primers. Data represent the fold increase following PMA treatment relative to untreated controls as a ratio of orf50 to GAPDH. Results are means of triplicate samples. Error bars represent standard deviations.
Analysis of viral transcription in A20-HE cells during reactivation.
As a complement to the experiments described above, we performed RT-PCR analyses of 21 viral genes in A20-HE cells at 8 h and 24 h following PMA stimulation (Fig. 5). Several viral genes with roles in virus replication were induced by PMA, including the single-strand DNA binding protein (orf6), the viral DNA polymerase (orf9), and the viral lytic transactivating protein Rta (orf50), as were genes for structural proteins, including the major capsid protein (orf25) and glycoprotein B (orf8). Other genes essential for viral replication that were induced by PMA included the nuclear phosphoprotein (orf45) (28) and the RNA export protein (orf57) (note that the oligo set used overlaps with the unique MHV68 gene M8).
FIG. 5.
Transcription of viral genes following PMA stimulation. RNA was isolated from A20-HE cells before or 8 h and 24 h posttreatment with 20 ng/ml PMA. RT-PCR was performed using primers for the indicated viral transcripts or cellular GAPDH as a control. The plus or minus signs indicate the presence or absence of RT in the cDNA reactions. The asterisk for M2 denotes that nested reactions were performed.
We also detected several genes whose expression during MHV68 latency in vivo was demonstrated by RT-PCR (41, 71) or RNase protection assays (4, 49). The MHV68 homologs of K3 (mK3) and the latency-associated nuclear antigen (mLANA; orf73) genes with roles in modulation of host immune or cell defense responses and contribution to the establishment of latent infection (5, 8-10, 21, 22, 42, 54, 55) were PMA inducible. Although levels were low prior to PMA stimulation, we noted that orf73 transcripts were readily detected following infection of A20 cells with WT MHV68 using splice-specific nested primer sets (Fig. 1). Also induced by PMA treatment were genes that influence host colonization (M4) (15) or reactivation upon explant of latently infected cells, including M1, v-cyclin (orf72), and v-bcl2 (M11) (13, 23, 38, 61, 69). PMA treatment also induced transcription of the multifunctional M2 gene, which influences both latency establishment and reactivation, particularly in B cells, perhaps through modulating host regulatory signaling cascades (24, 26, 34, 35, 40, 50). Finally, we detected induction of genes for M6 and M9, unique MHV68 genes whose functions are unknown.
We did not detect transcripts before or after PMA treatment for orf36, a viral kinase required for replication in primary macrophages (59), orf49, a coactivator for Rta-mediated gene expression (33), orf74, a viral G-protein-coupled receptor homolog (72), M3, a viral chemokine sequestration protein and determinant of pathogenesis (64-66), or the unique M12 gene (70). However, we acknowledge that failure to detect these transcripts may be due to limitations of the assay.
Induction of MHV68 gene expression by antimitogenic stimuli.
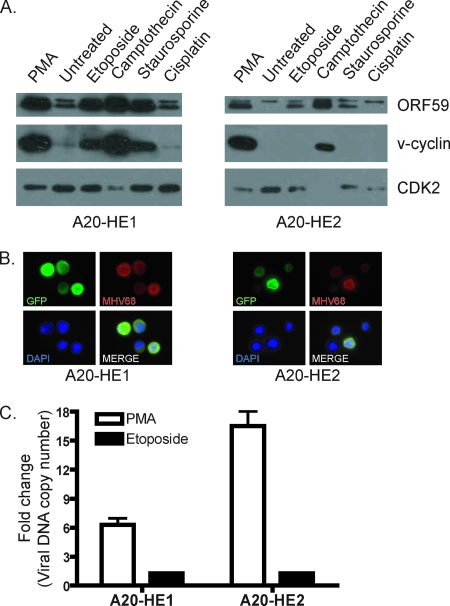
The cellular tumor suppressor protein p53 is a sequence-specific transcription factor that we recently demonstrated promotes MHV68 gene expression and lytic replication (21). Tarakanova and colleagues also reported that the DNA damage sensing kinase, ataxia telangiectesia mutated (ATM), an enzyme that phosphorylates and activates p53 and other targets, critically regulates MHV68 replication in macrophages (59). In addition, treatment of EBV-positive lymphoid and epithelial tumors with several p53-inducing chemotherapy drugs induced lytic viral gene expression (16, 17). Thus, we reasoned that MHV68 reactivation might be inducible by DNA damage/p53 activating stimuli. To test this hypothesis, we treated A20-HE cells with several known DNA damage and p53-inducing stimuli, including etoposide (topo-isomerase II inhibitor), camptothecin (topo-isomerase I inhibitor), staurosporine (general kinase inhibitor), and cisplatin (DNA damage by platinum adducts). Both A20-HE cell lines exhibited induction of ORF59 by immunoblot analysis following treatment with the topo-isomerase inhibitors and staurosporine, with the more reactivation-responsive A20-HE1 cells displaying more robust viral protein expression than A20-HE2 cells (Fig. 6A). Similarly, v-cyclin induction was readily detected for treated A20-HE1 cells, but only following camptothecin treatment of A20-HE2 cells at this time point, although 50 μM etoposide treatment did elicit detectable v-cyclin induction in separate experiments (data not shown). Consistent with these results, etoposide treatment increased production of MHV68 proteins in A20-HE cells as determined by IFA using MHV68 antiserum (Fig. 6B). Notably, the toxicity of the drugs tested in these experiments prevented analyses of viral replication by plaque assays. Quantitative PCR, however, indicated a less-than-twofold amplification of viral genomes over negative controls following 24 h of etoposide treatment, while PMA treatment elicited efficient viral DNA replication (Fig. 6C). These data suggest that MHV68 has the capacity to recognize and respond to DNA damage stimuli by initiating viral gene synthesis, but viral gene expression in response to such stimuli did not result in efficient virus replication.
FIG. 6.
Induction of lytic antigens by DNA damage stimuli. (A) A20-HE cells (5 × 105 per ml) were treated with etoposide (25 μM), camptothecin (2 μM), staurosporine (20 μg/ml), or cisplatin (100 μM). PMA (20 ng/ml) and untreated cells served as respective positive and negative controls. Cells were harvested 24 h posttreatment. Total protein (50 μg) was resolved by SDS-PAGE followed by immunoblot analysis for the indicated viral or cellular (CDK2) antigens. (B) A20-HE cells were treated with 50 μM etoposide and processed for immunofluorescence microscopy 24 h posttreatment. Cells were stained with antibodies to GFP and MHV68 antisera. DNA was detected with DAPI in the mounting media. Magnification, ×100. (C) Total DNA (200 ng) was analyzed by quantitative PCR for the presence of viral genome using orf50-specific primers. Data represent the fold increase following 24 h of stimulation with PMA (20 ng/ml) or etoposide (25 μM) relative to vehicle-treated controls. Results are means of triplicate samples. Error bars represent standard deviations.
To assess whether the failure of etoposide to trigger viral DNA replication reflected increased sensitivity of the A20-HE cell lines to etoposide-induced cell death, we performed drug titration experiments (Fig. 7). At the concentrations tested, these analyses did not reveal a substantial difference in cellular viability due to etoposide treatment (Fig. 7A), which indicates that the presence of latent or induced viral genes did not acutely influence cellular survival or death. Moreover, these data indicate that the 25 μM concentration of etoposide used to induce gene expression in Fig. 6 was not overtly cytotoxic, and therefore the data suggest that failure to replicate viral DNA was not a consequence of induction of host cell death. A similar titration performed in the presence of the herpesvirus lytic replication inhibitor cidofovir, a nucleoside analog that inhibits DNA chain elongation, enhanced etoposide cytotoxicity for both infected and uninfected cells (Fig. 7B). These data further suggest that viral replication-dependent late gene synthesis and/or progeny virion production did not directly contribute to either cell death or survival in these experiments (Fig. 7B). Additionally, both parental A20 and A20-HE cells were equally sensitive to direct induction of apoptosis by stimulation with a Fas-specific monoclonal antibody (Fig. 7B).
FIG. 7.
Infection-related sensitivity to etoposide. A20 and A20-HE cells were treated with vehicle or etoposide at the indicated concentrations in the absence (A) or presence of cidofovir (50 ng/ml) (B). Cell viabilities were determined 18 h posttreatment and are expressed as the percentage of viable cells with treatment relative to vehicle-treated controls. Anti-Fas (2.5 μg/ml in the presence of 10 μg/ml cycloheximide) was included as a control for cell death induction in panel B. Results are means of triplicate samples. Error bars represent standard deviations.
Potential uncoupling of DNA damage signals in MHV68-infected B cells.
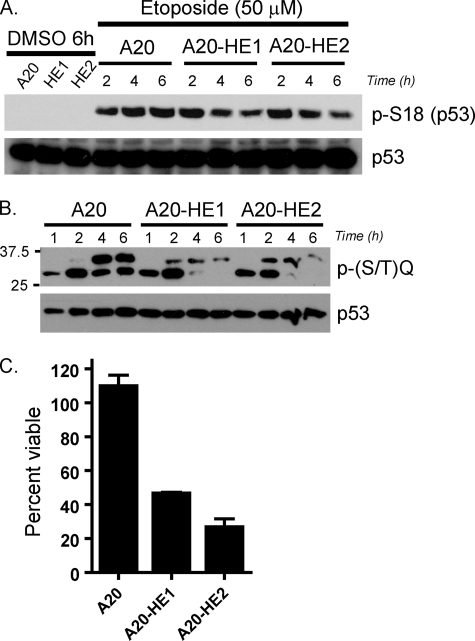
The capacity to specifically cleave, remove, and then repair large sections of chromosomes to generate functional antigen receptors is a defining characteristic of B cells and T cells. DNA damage response (DDR) proteins enforce cell cycle checkpoints to facilitate these events or trigger apoptosis or senescence when the process goes awry (27). As a pathological consequence, inactivation of specific DDR proteins in mice promotes the accumulation of chromosomal translocations between the IgH and c-myc loci, a growth-promoting mutation in humans that is strongly associated with Burkitt's lymphoma (11, 47). Given this disease association, the utilization of lymphocytes as latency reservoirs, and the finding that MHV68 gene expression was induced by DNA damage stimuli, we reasoned that the presence of MHV68 in B cells might influence the capacity of an infected cell to respond to DNA damage stimuli. To test this hypothesis, parental A20 cells and A20-HE1 and A20-HE2 cells were treated with etoposide to induce a DDR. Analysis of p53 phosphorylation on S18 by immunoblotting did not reveal any overt inhibition of p53 phosphorylation due to MHV68 infection and confirmed activation of upstream DNA damage-signaling cascades in treated cells (Fig. 8A), as did the presence of phosphorylated ATM and Chk2 (data not shown) regardless of infection status. In contrast, a time course immunoblot analysis using antisera specific for phosphorylated ATM or ATR (ATM and Rad3-related) target motifs [P-(S/T)Q] demonstrated that, although detection of undefined 35-kDa and 30-kDa protein species was similar in infected and uninfected cells at 1 and 2 h after etoposide treatment, infected cells exhibited reduced phosphorylation of the 35-kDa protein and complete disappearance of the 30-kDa protein at 4 and 6 h posttreatment, while these bands became more pronounced in uninfected A20 cells (Fig. 8B). Notably, detection patterns were similar in both uninfected and infected cells in both vehicle and cisplatin-treated samples (data not shown). These data suggest that, while the DDR is activated in MHV68-infected B cells, the propagation of the response is altered in the presence of MHV68.
FIG. 8.
Altered DNA damage signaling in MHV68-infected A20 cells. (A) A20 and A20-HE cells were treated with vehicle for 6 h or etoposide (50 μM) for the indicated times. Total protein was resolved by SDS-PAGE followed by immunoblot analyses for S18-phosphorylated p53 or total p53 as a loading control. (B) A20 and A20-HE cells were treated with etoposide (50 μM) for the indicated times. Total protein (50 μg) was resolved by SDS-PAGE followed by immunoblot analyses with phosphorylated-(S/T)Q motif antisera or total p53 as a loading control. (C) A20 and A20-HE cells were treated with vehicle or etoposide (25 μM) in the presence of cidofovir (50 ng/ml). After 4 h, cells were washed and then cultured in the presence of cidofovir. After 1 week, cell viabilities were determined and are expressed as the percentage of viable cells with treatment relative to vehicle-treated controls. Results are means of quadruplicate samples. Error bars represent standard deviations.
Given the apparent alterations in DDR signaling, we reasoned that the MHV68-infected A20 cell lines would exhibit reduced capacities to recover from transient DDR induction compared to the parental uninfected A20 cell line. To assess this possibility, A20 and A20-HE cells were pulsed with etoposide for 4 h, drug was washed out, and the cells were allowed to recover in culture. After 1 week, the viability of treated cells was measured in comparison to cells treated in parallel with vehicle alone. While uninfected A20 cells treated with etoposide recovered and maintained viability at levels comparable to the vehicle control cultures, both A20-HE1 and A20-HE2 cell lines exhibited dramatic reductions in viability following etoposide treatment (46% and 26%, respectively) relative to the vehicle control-treated cultures (Fig. 8C). These data indicate that the presence of MHV68 negatively impacts host cell viability following limited treatment with a chemotherapeutic drug in the presence of cidofovir.
DISCUSSION
In this report we describe the generation of a recombinant MHV68 that expresses a hygromycin phosphotransferase-EGFP fusion protein. We utilized this virus to establish long-term infection of the surface IgG-positive, mature murine B-cell line A20, and we designated the derivative lines A20-HE cells. We showed that MHV68 in A20-HE cells is maintained as a reactivation-competent entity, responding to PMA and BCR cross-linking with increased expression of lytic cycle-associated viral proteins, viral DNA synthesis, and production of progeny virus. We also examined the transcription of several viral genes involved in replication/reactivation and host modulation following PMA treatment. Additionally, we provided evidence that DNA damage-inducing drugs elicit expression of MHV68 replication-associated proteins but did not lead to detectable viral DNA replication. Finally, we demonstrated that the presence of MHV68 correlates with alterations in DNA damage signaling and increased sensitivity to death following limited treatment with etoposide. Taken together these data offer a general characterization of a novel MHV68 cell culture latency system and highlight its utility for understanding viral alterations in host cell homeostatic pathways during latency and reactivation.
Reactivating stimuli.
As an initial characterization, we tested the capacity of A20-HE cells to reactivate following treatment with a variety of different stimuli. In these experiments, robust lytic induction occurred following treatment with PMA and anti-Ig cross-linking, but not by sodium butyrate, LPS, or anti-CD40 cross-linking. By no means were these experiments definitive, and we consider them an initial depiction of potential responses to classically utilized stimulations that initiate lytic gammaherpesvirus replication from latently infected B-cell lines. Upon the generation of additional cell lines, however, we think it quite likely that MHV68-harboring cell lines will exhibit differential responsiveness to inducing stimuli, as has been repeatedly demonstrated for EBV cell lines.
Nonetheless, reactivation in response to PMA and anti-Ig is certainly analogous to other gammaherpesvirus latency systems, as is the low level of spontaneous reactivation we observed in A20-HE cells. We think it interesting that the differences in spontaneous reactivation and responsiveness to stimulation observed for A20-HE1 and A20-HE2 cell lines reflect slightly altered homeostatic set points in cellular and/or viral gene expression. This might also be due to differences in viral genome copy number. The quantitative PCR presented in Fig. 4 and 6 indicated ca. 10-fold more viral genome in A20-HE1 than A20-HE2 cells without stimulation (data not shown), although the contribution to such an increase by the higher level of spontaneous reactivation cannot be accounted for in this assay.
In addition to analyzing viral replication following PMA stimulation, we analyzed the basal and induced transcription of genes with known roles in lytic replication, latency, and reactivation, as well as several unique MHV68 genes with unknown functions. The roles of transcribed genes with previously described functions in vivo were described above. Given the previously mentioned percentages of spontaneous reactivation and the detection of transcripts from viral genomic regions that encode structural genes without induction, we think it premature to comment on potential latency programs in A20-HE cells. However, some interesting observations deserve additional comment. Consistent with a role in episome maintenance, we readily detected spliced orf73 transcripts by nested RT-PCR early after nonproductive infection of A20 cells. Importantly, however, it should be noted that orf73 transcripts are not detected in all latently infected cell types in vivo, suggesting that such a function may not be an absolute requirement for long-term latency (3, 41, 49, 71). Indeed, the PMA-inducible nature of orf73 transcription may reflect a prominent role in facilitating reactivation in a manner analogous to that described during lytic replication in fibroblasts (21).
Detection of M2 in this assay necessitated nested RT-PCR (3), which indicates that M2 expression is low and might otherwise be deleterious to the host cell or to viral replication, perhaps through uncoupling of DNA damage signals (34), as we generally described in Fig. 8. In agreement with this idea, we are unable to reliably detect M2 protein expression prior to or following PMA induction. Consistent with requirements for M2 functions in B cells, the M2 transcripts detected were spliced, which would enable production of the presumed functional M2 polypeptide. Spliced M2 transcripts are not produced during lytic infection of murine fibroblasts (unpublished data) but are present in the S11 tumor cell line and in some latently infected cells in vivo (3, 25).
Finally, the relative abundance of the M9 transcript in noninduced cells is of some interest and is consistent with previous analyses of MHV68 latency programs that demonstrated transcription from the M9 locus in vivo (4, 41, 49, 71) and in S11 cells (25). Whether the detected RT-PCR products reflect M9 transcripts or transcription of another gene remains to be determined. As such, cloning and mapping these transcripts will be required and may reveal a critical molecule involved in MHV68 latency in B cells. In general, it is important to consider that certain viral genes may be required or transcribed only in particular cell types, in specific cellular differentiation states, or in response to explicit reactivation stimuli. Potential assay limitations aside, failure to detect genes such as M3, orf36, orf49, orf74, or M12 may echo this point. These ideas are further emphasized by the apparent restriction of M2 splicing to B cells and the requirement for ORF36 for MHV68 replication in macrophages (59), and most convincingly by the cell-type-specific transcription patterns demonstrated in vivo by Marques and colleagues (41). MHV68-infected A20 cells, which are surface IgG positive, conceivably represent a memory B-cell-restricted transcriptional program for latency and reactivation.
Induction of viral gene expression by DNA damage-inducing drugs.
In experiments presented here, we demonstrated that stimulating infected A20 cells with several cytotoxic drugs that induce DDR promoted viral gene expression, a finding that demonstrates a parallel functional response to EBV-infected tumors (16, 17). For all but cisplatin, these treatments are capable of inducing double-strand DNA breaks and p53 activation, presumably via the ATM pathway. Cisplatin, however, a chemotherapeutic agent that forms platinum adducts on DNA and promotes single-strand DNA breaks that lead to p53 activation via ATR signaling pathways, did not result in viral gene expression (ORF59, v-cyclin, or lytic antigens) in these assays. These characteristics suggest that ATM-directed signaling pathways, and not ATR, are recognized for reactivation by MHV68 in latently infected B cells, although additional experimentation is required to definitively make this point. These findings support our previous demonstration that etoposide treatment enhanced orf50 transcription and v-cyclin translation in MHV68-infected 3T3 fibroblasts (21). The data also correlate with the demonstration that the efficiency of MHV68 replication in primary macrophages depends on the presence of ATM. Although the mechanism of such dependence is not yet clear, numerous reports have demonstrated herpesvirus recruitment of and/or requirement for DNA damage-related signaling molecules within replication complexes (31, 36, 39). A20-HE cells offer a new opportunity to probe the functional significance of DDR proteins in herpesvirus reactivation.
Although utilization of DDR proteins for replication now seems a common herpesvirus theme, it is interesting that DDR signals would directly induce gammaherpesvirus gene expression given the latency tropism of gammaherpesviruses for lymphocytes. Indeed, the capacity to break and remove large regions of “unnecessary” DNA is part and parcel of a B cell's capacity to generate functional antigen receptors and secreted antibodies, and aberrations in DDR pathways can have pathological consequences (27). DNA damage-signaling checkpoints through activated ATM halt cell cycle progression to limit potential mutagenic recombinations while these processes occur (11). Failure to appropriately recombine leads to signal amplification, sustained p53 activation, and ultimately apoptosis. Hence, we think it quite likely that pathogens that establish long-term, latent infections in B cells are primed to respond to, and perhaps alter or suppress, DNA damage-signaling events.
Functional consequences of MHV68 infection to DDR.
In its simplest form the function of a DDR is to recognize, respond to, and repair genetic lesions that might otherwise promote heritable mutations. Central to this idea is that failed repair and prolonged activation of DDR molecules should promote cell death or permanent cell cycle exit to ultimately limit the propagation of potentially harmful mutations. Therapeutic applications of these principals involve DDR induction paired with pharmacologic targeting of specific molecules in the DDR, thereby blocking the response, to enhance the efficacy of several cancer treatments (for a description of an experimental application, see reference 48). Although uninfected and infected A20 cells did not differ dramatically in their acute susceptibility to etoposide treatment (Fig. 7), a recovery experiment, wherein both parental and infected A20 cells received a short burst of etoposide prior to long-term culture, suggested that MHV68-infected B-cell tumor lines are less capable of recovering normal growth characteristics following limited drug treatment (Fig. 8C). This finding is analogous to EBV-positive gastric carcinomas (17) and Kaposi's sarcoma-associated herpesvirus-positive lymphomas (14) being more susceptible than uninfected cells to treatments with 5-fluorouracil and cisplatin or glycyrrhizic acid, respectively.
In the EBV studies, pairing chemotherapy with antiviral drugs substantially enhanced cell death in culture and tumor reduction in vivo. Thus, we note that reduced viability of A20-HE cells was observed in the presence of cidofovir, which we included in the experiment to limit death due to reactivation, even though 24-h treatment with etoposide did not induce viral DNA replication (Fig. 6C). Although we do not yet know the mechanism of death for MHV68-infected cells in these assays, we propose two possibilities which are not mutually exclusive. First, it is conceivable that viral gene expression alone is toxic to the induced host cell. In agreement with this idea, we previously demonstrated that mLANA-null MHV68 exhibited a robust increase in lytic viral gene expression even when viral replication was inhibited by cidofovir treatment (21), and the “hyperexpression” phenotype in these experiments correlated with dramatically enhanced cell death. The second hypothesis posits that virus-mediated alterations in the normal cellular response to damaged DNA, as demonstrated in Fig. 8, result in failure to repair damaged DNA, the physiologic consequence of which is cell death.
As discussed above, we think it likely that viral responsiveness to DNA damage signals offers a potential mechanism for reactivation when a host cell is unsuitable for long-term latency. As such, uncoupling the DDR may diffuse in the short-term strong apoptotic signals that would otherwise limit progeny virion production. Additionally, as MHV68 requires ATM and uses a viral kinase encoded by ORF36 to induce H2AX phosphorylation for efficient replication in primary macrophages, alterations in the DDR cascade may reflect viral commandeering and utilization of host DNA damage kinases and repair proteins to facilitate viral replication (59), essentially rendering them nonfunctional to the host cell. Understanding the biochemical basis for dysregulated signaling might further define mechanisms of gammaherpesvirus-induced transformation when the host cell is not killed as a result. Perhaps such events occur and promote MHV68 reactivation in the transition from viral tRNA-positive lesions in atypical lymphoid hyperplasia to tRNA-negative lymphomas observed following MHV68 infection of β2-microglobulin knockout mice (60). A promutagenic, but noncytopathic, reactivation event might offer a potential explanation for the proposed “hit-and-run” model of MHV68-associated transformation.
Potential utility of a cell culture-based system to study MHV68 latency and reactivation.
The data presented and discussed here provide a basic characterization of MHV68 maintenance and reactivation following infection and selection of a mature murine B-cell line by recombinant MHV68. A20-HE cells not only offer a much more robust and malleable system than the MHV68-positive S11 lymphoma line for evaluating viral genes and cellular signaling cascades that influence reactivation, but also they offer the capacity for direct evaluations of virus-specific alterations in cellular homeostasis by comparison to the parental, uninfected A20 line. The derivation of additional cell lines will further benefit these definitions. Such comparative systems potentially offer important new tools for not only identifying the basic biochemical pathways influenced by gammaherpesvirus infection with ready translation into natural primary infections, but also evaluating means of specifically targeting these pathways in rational therapeutic applications aimed at minimizing off-target toxicity. These cellular reagents additionally offer the potential for in vivo modeling by injection into an immunocompetent syngeneic host where, unlike modeling human gammaherpesvirus tumor therapy in SCID mice, these experiments would allow the simultaneous evaluation of the host immune response in facilitating clearance and/or tumor escape mechanisms upon incomplete reactivation in response to DNA damage induction therapy.
Acknowledgments
This work was supported by NIH R01 CA52004. J.C.F. was supported by a Leukemia and Lymphoma Society Postdoctoral Fellowship for part of this work. S.H.S. is supported by NIH RO1 grants CA52004, CA58524, CA95318, and AI05807.
We thank members of the Mocarski and Speck labs for helpful comments on this research.
Footnotes
Published ahead of print on 21 May 2008.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13111-121. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 746964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, R. D., III, S. Dickerson, and S. H. Speck. 2006. Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection. J. Virol. 802055-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E. S., M. L. Lutzke, R. Rochford, and H. W. T. Virgin. 2005. Alpha/beta interferons regulate murine gammaherpesvirus latent gene expression and reactivation from latency. J. Virol. 7914149-14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, N. J., J. S. May, and P. G. Stevenson. 2005. Gamma-herpesvirus latency requires T cell evasion during episome maintenance. PLoS Biol. 3e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84111-113. [DOI] [PubMed] [Google Scholar]
- 7.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24468. [PubMed] [Google Scholar]
- 8.Boname, J. M., B. D. de Lima, P. J. Lehner, and P. G. Stevenson. 2004. Viral degradation of the MHC class I peptide loading complex. Immunity 20305-317. [DOI] [PubMed] [Google Scholar]
- 9.Boname, J. M., J. S. May, and P. G. Stevenson. 2005. The murine gamma-herpesvirus-68 MK3 protein causes TAP degradation independent of MHC class I heavy chain degradation. Eur. J. Immunol. 35171-179. [DOI] [PubMed] [Google Scholar]
- 10.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15627-636. [DOI] [PubMed] [Google Scholar]
- 11.Callen, E., M. Jankovic, S. Difilippantonio, J. A. Daniel, H. T. Chen, A. Celeste, M. Pellegrini, K. McBride, D. Wangsa, A. L. Bredemeyer, B. P. Sleckman, T. Ried, M. Nussenzweig, and A. Nussenzweig. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell 13063-75. [DOI] [PubMed] [Google Scholar]
- 12.Chen, L., and M. Lagunoff. 2005. Establishment and maintenance of Kaposi's sarcoma-associated herpesvirus latency in B cells. J. Virol. 7914383-14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clambey, E. T., H. W. T. Virgin, and S. H. Speck. 2000. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J. Virol. 741973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curreli, F., A. E. Friedman-Kien, and O. Flore. 2005. Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. J. Clin. Investig. 115642-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, A. G., N. J. Moorman, D. O. Willer, and S. H. Speck. 2006. The M4 gene of gammaHV68 encodes a secreted glycoprotein and is required for the efficient establishment of splenic latency. Virology 344520-531. [DOI] [PubMed] [Google Scholar]
- 16.Feng, W. H., G. Hong, H. J. Delecluse, and S. C. Kenney. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 781893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, W. H., B. Israel, N. Raab-Traub, P. Busson, and S. C. Kenney. 2002. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 621920-1926. [PubMed] [Google Scholar]
- 18.Fingeroth, J. D., M. E. Diamond, D. R. Sage, J. Hayman, and J. L. Yates. 1999. CD21-Dependent infection of an epithelial cell line, 293, by Epstein-Barr virus. J. Virol. 732115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 1651074-1081. [DOI] [PubMed] [Google Scholar]
- 20.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 1961363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest, J. C., C. R. Paden, R. D. Allen III, J. Collins, and S. H. Speck. 2007. ORF73-null murine gammaherpesvirus 68 reveals roles for mLANA and p53 in virus replication. J. Virol. 8111957-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 843405-3416. [DOI] [PubMed] [Google Scholar]
- 23.Gangappa, S., L. F. van Dyk, T. J. Jewett, S. H. Speck, and H. W. t. Virgin. 2002. Identification of the in vivo role of a viral bcl-2. J. Exp. Med. 195931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herskowitz, J., M. A. Jacoby, and S. H. Speck. 2005. The murine gammaherpesvirus 68 M2 gene is required for efficient reactivation from latently infected B cells. J. Virol. 792261-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 967508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby, M. A., H. W. T. Virgin, and S. H. Speck. 2002. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. J. Virol. 761790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankovic, M., A. Nussenzweig, and M. C. Nussenzweig. 2007. Antigen receptor diversification and chromosome translocations. Nat. Immunol. 8801-808. [DOI] [PubMed] [Google Scholar]
- 28.Jia, Q., V. Chernishof, E. Bortz, I. McHardy, T. T. Wu, H. I. Liao, and R. Sun. 2005. Murine gammaherpesvirus 68 open reading frame 45 plays an essential role during the immediate-early phase of viral replication. J. Virol. 795129-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, I. J., E. Flano, D. L. Woodland, F. E. Lund, T. D. Randall, and M. A. Blackman. 2003. Maintenance of long term gamma-herpesvirus B cell latency is dependent on CD40-mediated development of memory B cells. J. Immunol. 171886-892. [DOI] [PubMed] [Google Scholar]
- 30.Krug, L. T., J. M. Moser, S. M. Dickerson, and S. H. Speck. 2007. Inhibition of NF-κB activation in vivo impairs establishment of gammaherpesvirus latency. PLoS Pathog. 3e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudoh, A., M. Fujita, L. Zhang, N. Shirata, T. Daikoku, Y. Sugaya, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 2808156-8163. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S., H. J. Cho, J. J. Park, Y. S. Kim, S. Hwang, R. Sun, and M. J. Song. 2007. The ORF49 protein of murine gammaherpesvirus 68 cooperates with RTA in regulating virus replication. J. Virol. 819870-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, X., M. T. Pickering, N. H. Cho, H. Chang, M. R. Volkert, T. F. Kowalik, and J. U. Jung. 2006. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J. Virol. 805862-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, X., Y. C. Shin, R. E. Means, and J. U. Jung. 2004. Inhibition of interferon-mediated antiviral activity by murine gammaherpesvirus 68 latency-associated M2 protein. J. Virol. 7812416-12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 1025844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, S., I. V. Pavlova, H. W. T. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 742029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh, J., Q. Huang, A. M. Petros, D. Nettesheim, L. F. van Dyk, L. Labrada, S. H. Speck, B. Levine, E. T. Olejniczak, and H. W. T. Virgin. 2005. A surface groove essential for viral Bcl-2 function during chronic infection in vivo. PLoS Pathog. 1e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 811934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madureira, P. A., P. Matos, I. Soeiro, L. K. Dixon, J. P. Simas, and E. W. Lam. 2005. Murine gamma-herpesvirus 68 latency protein M2 binds to Vav signaling proteins and inhibits B-cell receptor-induced cell cycle arrest and apoptosis in WEHI-231 B cells. J. Biol. Chem. 28037310-37318. [DOI] [PubMed] [Google Scholar]
- 41.Marques, S., S. Efstathiou, K. G. Smith, M. Haury, and J. P. Simas. 2003. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J. Virol. 777308-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 7710295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser, J. M., J. W. Upton, R. D. Allen III, C. B. Wilson, and S. H. Speck. 2005. Role of B-cell proliferation in the establishment of gammaherpesvirus latency. J. Virol. 799480-9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moser, J. M., J. W. Upton, K. S. Gray, and S. H. Speck. 2005. Ex vivo stimulation of B cells latently infected with gammaherpesvirus 68 triggers reactivation from latency. J. Virol. 795227-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B 356569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavlova, I. V., H. W. T. Virgin, and S. H. Speck. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J. Virol. 775731-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramiro, A. R., M. Jankovic, E. Callen, S. Difilippantonio, H. T. Chen, K. M. McBride, T. R. Eisenreich, J. Chen, R. A. Dickins, S. W. Lowe, A. Nussenzweig, and M. C. Nussenzweig. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhardt, H. C., A. S. Aslanian, J. A. Lees, and M. B. Yaffe. 2007. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11175-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 754955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues, L., M. Pires de Miranda, M. J. Caloca, X. R. Bustelo, and J. P. Simas. 2006. Activation of Vav by the gammaherpesvirus M2 protein contributes to the establishment of viral latency in B lymphocytes. J. Virol. 806123-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6276-282. [DOI] [PubMed] [Google Scholar]
- 52.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2403-409. [DOI] [PubMed] [Google Scholar]
- 53.Spillmann, F. J., and M. Wabl. 2004. Endogenous expression of activation-induced cytidine deaminase in cell line WEHI-231. J. Immunol. 1731858-1867. [DOI] [PubMed] [Google Scholar]
- 54.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 978455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8+ T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3733-740. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 1871941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1993. Interactions of murine gammaherpesvirus 68 with B and T cell lines. Virology 193825-833. [DOI] [PubMed] [Google Scholar]
- 58.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73(Pt 12)3275-3279. [DOI] [PubMed] [Google Scholar]
- 59.Tarakanova, V. L., V. Leung-Pineda, S. Hwang, C. Yang, K. Matatall, M. Basson, R. Sun, H. Piwnica-Worms, B. P. Sleckman, and H. W. Virgin. 2007. Gammaherpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarakanova, V. L., F. Suarez, S. A. Tibbetts, M. A. Jacoby, K. E. Weck, J. L. Hess, S. H. Speck, and H. W. T. Virgin. 2005. Murine gammaherpesvirus 68 infection is associated with lymphoproliferative disease and lymphoma in BALB β2 microglobulin-deficient mice. J. Virol. 7914668-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Upton, J. W., and S. H. Speck. 2006. Evidence for CDK-dependent and CDK-independent functions of the murine gammaherpesvirus 68 v-cyclin. J. Virol. 8011946-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Upton, J. W., L. F. van Dyk, and S. H. Speck. 2005. Characterization of murine gammaherpesvirus 68 v-cyclin interactions with cellular cdks. Virology 341271-283. [DOI] [PubMed] [Google Scholar]
- 63.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 706516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. I. Virgin. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 746741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Berkel, V., B. Levine, S. B. Kapadia, J. E. Goldman, S. H. Speck, and H. W. t. Virgin. 2002. Critical role for a high-affinity chemokine-binding protein in gamma-herpesvirus-induced lethal meningitis. J. Clin. Investig. 109905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Berkel, V., K. Preiter, H. W. T. Virgin, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 734524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Dyk, L. F., J. L. Hess, J. D. Katz, M. Jacoby, S. H. Speck, and H. I. Virgin. 1999. The murine gammaherpesvirus 68 v-cyclin gene is an oncogene that promotes cell cycle progression in primary lymphocytes. J. Virol. 735110-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Dyk, L. F., H. W. T. Virgin, and S. H. Speck. 2003. Maintenance of gammaherpesvirus latency requires viral cyclin in the absence of B lymphocytes. J. Virol. 775118-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dyk, L. F., H. W. T. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 747451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Virgin, H. W. T., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 715894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Virgin, H. W. T., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 732321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wakeling, M. N., D. J. Roy, A. A. Nash, and J. P. Stewart. 2001. Characterization of the murine gammaherpesvirus 68 ORF74 product: a novel oncogenic G protein-coupled receptor. J. Gen. Virol. 821187-1197. [DOI] [PubMed] [Google Scholar]
- 73.Weck, K. E., A. J. Dal Canto, J. D. Gould, A. K. O'Guin, K. A. Roth, J. E. Saffitz, S. H. Speck, and H. W. Virgin. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat. Med. 31346-1353. [DOI] [PubMed] [Google Scholar]
- 74.Weck, K. E., S. S. Kim, H. I. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 733273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willer, D. O., and S. H. Speck. 2005. Establishment and maintenance of long-term murine gammaherpesvirus 68 latency in B cells in the absence of CD40. J. Virol. 792891-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]