Abstract
We previously described a T20-dependent human immunodeficiency virus type 1 variant from a patient on T20 therapy (3). This virus carries two mutations in the gp41 domain of the envelope protein (Env) that was proposed to undergo a premature conformational switch to the 6-helix bundle structure. The T20 peptide can rescue this hyperfusogenic Env protein by preventing the premature switch and preserving an earlier prefusion conformation, thus restoring virus infectivity and replication. In this study, we set out to critically test this mechanistic explanation with alternative effectors that may control the Env switch, including other fusion inhibitors and antibodies that target gp41.
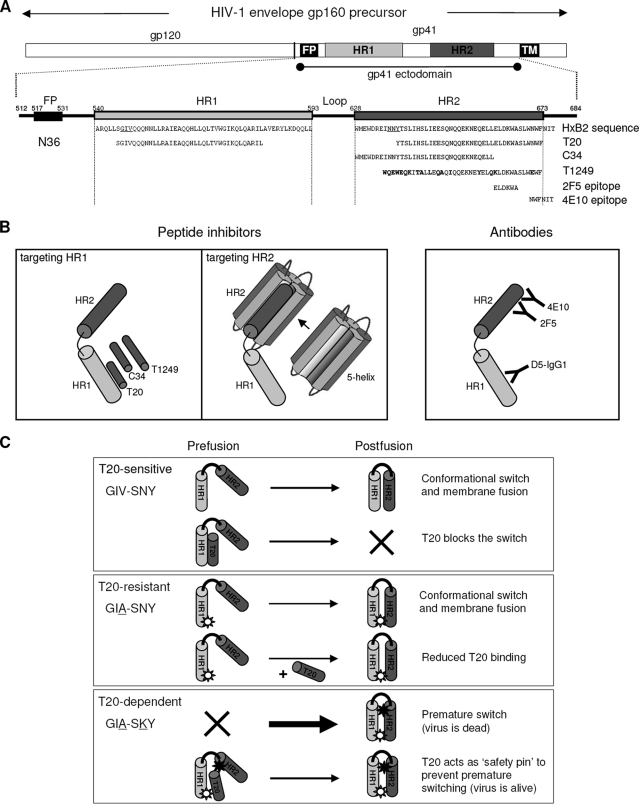
The fusion inhibitor T20 (Fuzeon) represents a new class of antivirals that inhibit viral entry (12, 23, 24). T20 is a 36-amino-acid peptide derived from the C-terminal region of HR2 (Fig. 1A). By binding to HR1, T20 blocks the formation of the 6-helix bundle, which is a prerequisite for membrane fusion and viral entry (Fig. 1B, left) (10, 24). T1249 is an improved 39-amino-acid peptide inhibitor (Fig. 1A). The C-terminal region of T1249 is almost identical to that of T20, but the N terminus differs in sequence and is extended by another 3 residues. T1249 is composed of sequences derived from human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (7, 21). Another HR2-derived inhibitor is C34 (Fig. 1A) (6, 14, 19). C34 includes residues located toward the N-terminal region of HR2, so it interacts with more-conserved HR1 regions, making it less susceptible to the evolution of drug-resistant viruses (5, 16). We also targeted the HR2 region of gp41 with the fusion inhibitor 5-helix protein (Fig. 1B, middle panel) (20), which is based on the 6-helix bundle crystal structure (6). The 5-helix protein contains five of these six helices joined by short peptide linkers, forming a stable 5-helix structure that exposes one HR2 binding site. We also used antibodies that target either the HR2 (2F5 and 4E10) or HR1 domain (D5-IGg1) of Env gp41 (Fig. 1B, right). The binding epitopes for 2F5 and 4E10 in HR2 are indicated in Fig. 1A.
FIG. 1.
(A) Schematics of gp160, the gp120 and gp41 subunits (top), and a close-up of the gp41 ectodomain (bottom). This figure was adapted from reference 3. The positions and amino acid residues of peptide-based fusion inhibitors and epitope recognition sites for the antibodies used are indicated. Amino acid residues in boldface indicate deviations from the prototype HxB2 sequence. The GIV sequence in HR1 which includes amino acid residue 38 of gp41 is underlined. (B) Target sites of the different peptide fusion inhibitors and antibodies tested (drawing not to scale). For simplicity, we show only one gp41 molecule instead of the gp41 trimer. (C) Proposed model for T20-dependent viral entry. Each box depicts one of three scenarios: T20-sensitive (GIV-SNY), T20-resistant (GIA-SNY), or T20-dependent (GIA-SKY) viral entry. A simplified gp41 ectodomain comprising only one subunit of HR1 (dark-gray cylinder) and HR2 (light-gray cylinder) joined by a loop region (black curved line) is used to depict the prefusion and postfusion states of the peptide. The thickness of the arrows represents the speed of the conformational switch between pre- and postfusion conformations; thicker arrows indicate faster speed. A star with a white circle within it represents the GIA mutation in HR1, and an all-black star represents the SKY mutation in HR2. Explanations for each reaction are provided on the right.
T20-resistant HIV-1 variants have been described for patients failing T20 therapy (3, 9, 17, 18, 22, 25). Sequence analysis revealed the acquisition of mutations within a stretch of three HR1 amino acids, glycine-isoleucine-valine (the GIV motif underlined in Fig. 1A; HxB2 amino acid positions 547 to 549 of gp160). These HR1 mutations disrupt T20 binding, thus providing a mechanism for resistance (Fig. 1C, middle panel). However, these mutations also affect the HR1-HR2 interaction, and hence, T20-resistant viruses usually have decreased fitness (2, 3, 13). Recently, we described the evolution of a drug-dependent HIV-1 variant in a patient that failed T20 therapy (3). This virus acquired the T20 resistance mutation GIA in HR1 (GIV to GIA; mutated amino acids are underlined) and a subsequent change in the 3-amino-acid SNY sequence of the HR2 domain (SNY to SKY). This HR1-HR2 double mutant (with the GIA and SKY mutations) dominated the viral population after 32 weeks of therapy, and it was not only highly resistant to T20 but was in fact critically dependent on this peptide for its replication.
We proposed a mechanistic model for drug-dependent viral entry (Fig. 1C, bottom panel) (2, 3). Briefly, resistance to T20 is caused by the GIA mutation in HR1, which weakens the interaction with T20 (resistance) and HR2 (6-helix bundle formation). The reduced HR1-HR2 affinity negatively impacts Env-mediated fusion and HIV-1 fitness (2, 3, 13). T20 dependence is caused by the SKY mutation in HR2, which stabilizes the HR1-HR2 interaction (3). However, the SKY mutation creates a hyperfusogenic Env gp41 that may prematurely undergo the conformational switch, which effectively kills virus infectivity. T20 is able to prevent this premature switch by preserving an earlier prefusion conformation, enabling gp41 to undergo the conformational switch at the correct moment in the fusion process. T20 control should be transient, as the peptide should leave the complex to allow the subsequent HR1-HR2 interaction. According to this mechanism of T20 dependence, any compound that transiently interferes with the HR1-HR2 interaction should be able to support the replication of the GIA-SKY mutant virus.
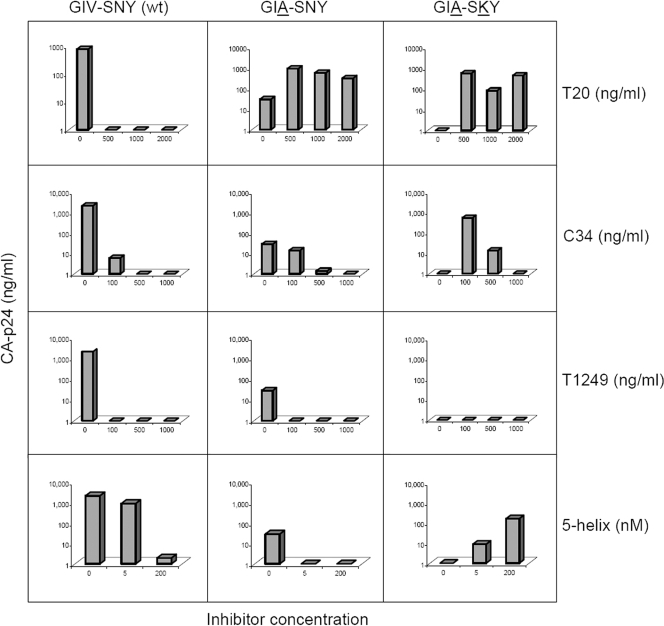
To test if this T20-dependent virus can be induced by other gp41 fusion inhibitors, we performed replication assays in the presence of C34, T1249, the 5-helix protein, and T20 as the positive control (Fig. 2). Besides the T20-dependent GIA-SKY mutant construct, two other viruses, the GIV-SNY (wild-type; T20 sensitive) and GIA-SNY (single mutant; T20 resistant) constructs, were used. Viral DNA constructs were transfected into the SupT1 T-cell line and cultured in the presence or absence of fusion inhibitors. We only show CA-p24 values for day 6 posttransfection, which best represent the relative differences in replication capacity. As expected, replication of the wild-type virus was strongly inhibited by all inhibitors, and the GIA-SNY single mutant was resistant to T20 (3, 6a, 22). This mutant displayed some resistance to C34 but was completely sensitive to T1249 and 5-helix. The GIA-SKY double mutant was clearly dependent on T20 as previously described (3). The C34 peptide was able to induce GIA-SKY mutant replication at concentrations up to 500 ng/ml, but higher concentrations inhibited replication. We previously reported a similar phenomenon at high T20 levels (3). C34 is somewhat more potent than T20, as it binds the deep pocket in HR1 (5). Our model suggests that the peptide needs to be released at some stage during the fusion process to enable the HR1-HR2 interaction, 6-helix bundle formation, and membrane fusion, and it is likely that peptide release is blocked at high peptide concentrations. T1249 did not induce GIA-SKY mutant replication, presumably due to its higher affinity for HR1. If the peptide remains bound to HR1, it will block all further conformational steps. 5-Helix, which targets the HR2 region of gp41, was also able to activate the GIA-SKY mutant in a dose-dependent manner.
FIG. 2.
Replication of the wild-type (wt) and mutant viruses. SupT1 cells were transfected with 0.5 μg of DNA from the molecular clones indicated above the graph. We show CA-p24 values (y axis) for day 6 posttransfection, which best represent the relative differences in replication capacity in the absence of inhibitor, and the resistance/dependence phenotypes observed for the wild type and mutants at different drug concentrations (x axis). The results shown are from a representative experiment; similar results were obtained in separate duplicate transfection experiments.
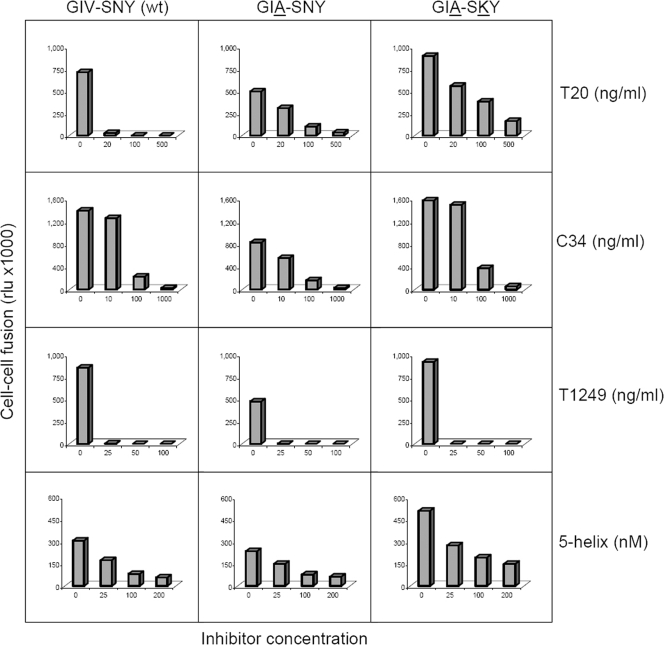
We performed a cell-cell fusion assay with the variant Env molecules (Fig. 3). Our hypothesis is that this measures Env activity before a potential premature switch can occur because Env molecules are engaged in the fusion process as soon as they appear at the cell surface (3). One cell expresses the wild-type or mutant Env protein and the other cell the appropriate receptors (CD4 and CXCR4), and fusion was scored by syncytium formation. A long terminal repeat-luciferase reporter was introduced in the acceptor cell that is activated upon cell fusion by the Tat protein expressed in the donor cell. Compared to the fusion activity for wild-type Env, we consistently measured reduced fusion activity (syncytium and luciferase counts) for the GIA-SNY single mutant and increased activity for the GIA-SKY double mutant, which confirms its hyperactivity (3).
FIG. 3.
Cell-cell fusion assay results. SupT1 cells were transfected with the molecular clones indicated above the graph. One day later, transfected cells were mixed with SupT1 cells containing a Tat-responsive long terminal repeat-luciferase reporter gene construct. The cell-cell fusion activity from a luciferase assay performed on the cell lysate is expressed in relative light units (rlu) on the y axis. These results are from a representative experiment. Similar results were obtained in two independent experiments. Inhibitor concentrations are indicated on the x axis, and the type of inhibitor is noted on the right. wt, wild type.
Next, we tested the effects of the inhibitors T20, C34, T1249, and 5-helix in the cell-cell fusion assay. Not surprisingly, the wild-type Env was inhibited by T20, and the GIA-SNY mutant was relatively T20 resistant. The GIA-SKY mutant displayed a similar resistance phenotype as that displayed by the GIA mutant. These results indicate that the GIA mutation in HR1 is responsible for reduced T20 affinity (resistance) and reduced HR2 affinity (fusion activity) and that the SKY mutation in HR2 creates a hyperfusogenic Env. The latter property may seem to contradict the observation of impaired virus replication, but a premature switch will result in dead Env spikes on the surfaces of virus particles. T20 may prevent such premature inactivation and thus rescue virus infectivity. C34 gave a resistance pattern very similar to that of T20, and this was expected because C34 has a similar mode of action. T1249 inhibited all HIV-1 variants tested, confirming the superior inhibitory activity of this inhibitor, particularly because lower concentrations of the drug were used. 5-Helix, which targets the HR2 region, gave very similar results for wild-type, GIA-SNY, and GIA-SKY viruses, indicating that these viruses are not resistant to 5-helix. This is not surprising because the GIA mutation is located outside the actual 5-helix target region. Because 5-helix is unrelated structurally to T20 and targets a different gp41 region, this result supports our T20 dependence model. It suggests that inhibitors that impede the formation of the 6-helix bundle provide an advantage to the hyperfusogenic Env protein, presumably by preventing the premature switch.
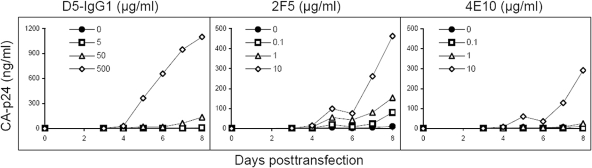
Env-targeted antibodies, which are much larger in size than peptide fusion inhibitors, provide an alternative tool to probe the mechanism of T20 dependence. We used the well-characterized antibodies D5-IgG1 (15), which targets HR1 and 2F5 (11), and 4E10 (4), which targets HR2 (Fig. 1B). The GIA-SKY molecular clone was transfected into the SupT1 T-cell line, and viral spread in the culture was monitored in the presence or absence of different concentrations of antibody. Viral replication curves were measured over an 8-day period (Fig. 4). D5-IgG1, which was suggested to target transient gp41 structural intermediates similar to the ones induced by the binding of fusion inhibitors T20, C34, and T1249 (15), had the greatest induction effect. The 2F5 and 4E10 antibodies, which recognize epitope sites toward the C-terminal end of HR2, were able to induce GIA-SKY virus replication. The 4E10 epitope binding site only partially overlaps HR2, which may explain its reduced induction capacity relative to that of 2F5.
FIG. 4.
Replication of the T20-dependent GIA-SKY mutant in the presence of gp41-targeted antibodies. SupT1 cells were transfected at day 0 with the GIA-SKY molecular clone, and viral replication was monitored over an 8-day period via CA-p24 production (y axis). The closed-circle curves represent replication in the absence of antibody; the other curves were obtained with the indicated antibodies, which were all tested at three different concentrations. The results shown are from a representative experiment; similar results were obtained in two separate transfection experiments.
It may perhaps be surprising that gp41-binding antibodies are also able to support the replication of the T20-dependent virus because an intrinsic prediction of the mechanistic model is that these reagents should not only bind to HR2 but also be released in time to allow membrane fusion. This release may be expected to be slower for high-affinity antibodies than for the small-peptide drugs. It may be of interest to address these issues in a follow-up study that focuses on the actual on-and-off rates of the peptide and antibody reagents in combination with the wild-type Env protein and the typical GIA-SKY mutant. This may also provide novel kinetic insight into the mechanism of HIV-1 membrane fusion. In fact, a recent study indicated that the 2F5 and 4E10 antibodies are also rather special in that they recognize a fusion-intermediate state of gp41 (8).
We proposed several possible mechanistic models for the unique T20-dependent replication phenotype (3). First, it could be envisaged that the T20 peptide is actively involved in the formation of the 6-helix bundle structure of the GIA-SKY Env. Thus, the wild-type T20 peptide could replace one or multiple mutant HR2 domains to enable the formation of this fusion-competent structure. This scenario can now be rejected, as structurally unrelated peptides and antibodies are also able to activate the GIA-SKY virus. Another scenario suggests that T20 dependence may mimic the process of enhancement of virus infectivity that is often observed in antibody neutralization experiments. However, such enhancement does not explain the nearly complete lack of infectivity of the GIA-SKY mutant in the absence of T20. Thus, the results presented in this study and a recent virus evolution study (1) exclusively support the proposed mechanistic model of drug dependence that is illustrated in Fig. 1C.
Acknowledgments
We thank Trimeris and Roche for providing us with the T20 and T1249 peptides, Merck Research Laboratories (Michael Miller) for providing us with the D5-IgG1 antibody, and Min Lu for generously providing us with the C34 and 5-helix peptide as well as the 2F5 and 4E10 antibodies. We thank Rogier Sanders for critically reading the manuscript and Stef Heynen for technical assistance.
This research was supported in part by grant 2005021 from the AIDS Fund (Amsterdam, The Netherlands).
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Baldwin, C. E., and B. Berkhout. 2006. Second site escape of a T20-dependent HIV-1 variant by a single amino acid change in the CD4 binding region of the envelope glycoprotein. Retrovirology 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., and B. Berkhout. 2007. HIV-1 drug-resistance and drug-dependence. Retrovirology 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 7812428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22163-173. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 9515613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 6a.Eggink, D., C. E. Baldwin, Y. Deng, J. P. M. Langedijk, M. Lu, R. W. Sanders, and B. Berkhout. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 826678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eron, J. J., R. M. Gulick, J. A. Bartlett, T. Merigan, R. Arduino, J. M. Kilby, B. Yangco, A. Diers, C. Drobnes, R. DeMasi, M. Greenberg, T. Melby, C. Raskino, P. Rusnak, Y. Zhang, R. Spence, and G. D. Miralles. 2004. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 1891075-1083. [DOI] [PubMed] [Google Scholar]
- 8.Frey, G., H. Peng, S. Rits-Volloch, M. Morelli, Y. Cheng, and B. Chen. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA 1053739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilby, J. M., J. P. Lalezari, J. J. Eron, M. Carlson, C. Cohen, R. C. Arduino, J. C. Goodgame, J. E. Gallant, P. Volberding, R. L. Murphy, F. Valentine, M. S. Saag, E. L. Nelson, P. R. Sista, and A. Dusek. 2002. The safety, plasma pharmacokinetics, and antiviral activity of subcutaneous enfuvirtide (T-20), a peptide inhibitor of gp41-mediated virus fusion, in HIV-infected adults. AIDS Res. Hum. Retrovir. 18685-693. [DOI] [PubMed] [Google Scholar]
- 10.Kilgore, N. R., K. Salzwedel, M. Reddick, G. P. Allaway, and C. T. Wild. 2003. Direct evidence that C-peptide inhibitors of human immunodeficiency virus type 1 entry bind to the gp41 N-helical domain in receptor-activated viral envelope. J. Virol. 777669-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunert, R., F. Ruker, and H. Katinger. 1998. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum Retrovir. 141115-1128. [DOI] [PubMed] [Google Scholar]
- 12.Lazzarin, A., B. Clotet, D. Cooper, J. Reynes, K. Arasteh, M. Nelson, C. Katlama, H. J. Stellbrink, J. F. Delfraissy, J. Lange, L. Huson, R. DeMasi, C. Wat, J. Delehanty, C. Drobnes, and M. Salgo. 2003. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 3482186-2195. [DOI] [PubMed] [Google Scholar]
- 13.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 784628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 959134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, M. D., R. Geleziunas, E. Bianchi, S. Lennard, R. Hrin, H. Zhang, M. Lu, Z. An, P. Ingallinella, M. Finotto, M. Mattu, A. C. Finnefrock, D. Bramhill, J. Cook, D. M. Eckert, R. Hampton, M. Patel, S. Jarantow, J. Joyce, G. Ciliberto, R. Cortese, P. Lu, W. Strohl, W. Schleif, M. McElhaugh, S. Lane, C. Lloyd, D. Lowe, J. Osbourn, T. Vaughan, E. Emini, G. Barbato, P. S. Kim, D. J. Hazuda, J. W. Shiver, and A. Pessi. 2005. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc. Natl. Acad. Sci. USA 10214759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otaka, A., M. Nakamura, D. Nameki, E. Kodama, S. Uchiyama, S. Nakamura, H. Nakano, H. Tamamura, Y. Kobayashi, M. Matsuoka, and N. Fujii. 2002. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew. Chem. Int. Ed. Engl. 412937-2940. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Alvarez, L., R. Carmona, A. Ocampo, A. Asorey, C. Miralles, D. C. Perez, M. Pinilla, G. Contreras, J. A. Taboada, and R. Najera. 2006. Long-term monitoring of genotypic and phenotypic resistance to T20 in treated patients infected with HIV-1. J. Med. Virol. 78141-147. [DOI] [PubMed] [Google Scholar]
- 18.Poveda, E., B. Rodes, C. Toro, L. Martin-Carbonero, J. Gonzalez-Lahoz, and V. Soriano. 2002. Evolution of the gp41 env region in HIV-infected patients receiving T-20, a fusion inhibitor. AIDS 161959-1961. [DOI] [PubMed] [Google Scholar]
- 19.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291884-888. [DOI] [PubMed] [Google Scholar]
- 21.Schneider, S. E., B. L. Bray, C. J. Mader, P. E. Friedrich, M. W. Anderson, T. S. Taylor, N. Boshernitzan, T. E. Niemi, B. C. Fulcher, S. R. Whight, J. M. White, R. J. Greene, L. E. Stoltenberg, and M. Lichty. 2005. Development of HIV fusion inhibitors. J. Pept. Sci. 11744-753. [DOI] [PubMed] [Google Scholar]
- 22.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 9112676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 919770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, L., A. Pozniak, A. Wildfire, S. A. Stanfield-Oakley, S. M. Mosier, D. Ratcliffe, J. Workman, A. Joall, R. Myers, E. Smit, P. A. Cane, M. L. Greenberg, and D. Pillay. 2005. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob. Agents Chemother. 491113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]