Abstract
T-cell receptors (TCRs) govern the specificity, efficacy, and cross-reactivity of CD8 T cells. Here, we studied CD8 T-cell clonotypes from Mane-A*10+ pigtail macaques responding to the simian immunodeficiency virus (SIV) Gag KP9 epitope in a setting of vaccination and subsequent viral challenge. We observed a diverse TCR repertoire after DNA, recombinant poxvirus, and live attenuated virus vaccination, with none of 59 vaccine-induced KP9-specific TCRs being identical between macaques. The KP9-specific TCR repertoires remained diverse after SIV or simian-human immunodeficiency virus challenge but, remarkably, exhibited substantially different clonotypic compositions compared to the corresponding populations prechallenge. Within serial samples from individual pigtail macaques, only a small subset (33.9%) of TCRs induced by vaccination were maintained or expanded after challenge. Most (66.1%) of the TCRs induced by vaccination were not detectable after challenge. Our results suggest that some CD8 T cells induced by vaccination are more efficient than others at responding to a viral challenge. These findings have implications for future AIDS virus vaccine studies, which should consider the “fitness” of vaccine-induced T cells in order to generate robust responses in the face of virus exposure.
The specificities of αβ T-cell receptors (TCRs) influence both immunodominance and the emergence of viral escape mutants in simian immunodeficiency virus (SIV)-infected rhesus macaques (29) and human immunodeficiency virus (HIV)-infected people (10, 22, 27, 42, 43). It is generally thought that broad TCR repertoires are advantageous, since they have an increased potential to recognize emerging viral escape variants (6, 11, 29). However, structural features of the bound epitope are a key determinant of repertoire selection (36) and thus the scope for altering clonotype recruitment in response to defined antigens by vaccination might be limited.
αβ TCRs are composed of two chains bearing membrane-proximal constant regions and variable regions that govern the interaction with major histocompatibility complex (MHC)-bound peptide antigen. The α and β chain complementarity-determining regions (CDRs) are critical for mediating contact between the TCR and peptide/MHC complex (17). In particular, the somatically recombined and highly variable CDR3 loops form the primary contact with the bound epitope (17). Studying CDR3 regions along with Vβ or Vα gene usage therefore provides substantial insight into the clonal architecture of a particular response. Molecular advances in the field of TCR repertoire analysis have enabled such detailed studies through the quantitative amplification and characterization of rearranged expressed TRB (and TRA) genes without bias (11, 29).
To date, studies of macaque TCRs expressed in SIV-derived epitope-specific CD8+ T-cell populations have been limited to rhesus macaque models (18, 27, 33). Further, data that inform on the evolution of TCR usage between vaccination and subsequent viral exposure are even more limited. This is a critical issue because the induction of specific T cells that most effectively control viral replication is a key goal of T-cell-based vaccination. A recent study of Vβ usage in rhesus macaques after vaccination and SHIV89.6 challenge showed that a focusing of Vβ usage occurred early after challenge (33). However, how well this translates to CDR3 sequences after challenge, and across other SIV models and macaque species, remains to be determined.
Pigtail macaques that possess the Mane-A*10 MHC class I allele develop an immunodominant response to the SIV Gag164-172 KP9 epitope following both vaccination and SIV or SHIV infection (9, 34). While the KP9-specific CD8 T-cell response is beneficial in the setting of infection with wild-type KP9 virus, viral escape occurs and abrogates the response (14). The induction of an immunodominant KP9-specific response in the setting of challenge with a virus already escaped at KP9 is counterproductive (13), presumably because it inhibits more useful subdominant responses (15, 16). Here we studied the TCR repertoire of KP9-specific CD8 T-cell populations in pigtail macaques in order to understand the nature of this important response at the molecular level.
MATERIALS AND METHODS
Pigtail macaque samples.
Eleven serial peripheral blood mononuclear cell (PBMC) samples stored in liquid nitrogen from five Mane-A*10+ pigtail macaques involved in previously published vaccine studies were studied as shown in Fig. 1a. Our institutional animal ethics committees approved all of our animal studies. All five macaques were juvenile, with similar ages of 3 to 4 years. Both animals in a study involving a recombinant vaccinia virus prime/recombinant fowlpox virus boost coexpressing the immunomodulatory molecule 4-1BBL (VV/FPV) followed by a SIVmac251 challenge were studied (32). We also studied two Mane-A*10+ animals that were involved in a DNA prime/recombinant fowlpox virus boost (DNA/FPV) vaccine study and challenged with SHIVmn229 (9). These two animals were selected from the four Mane-A*10+ animals receiving the same DNA/FPV vaccines based on high numbers of frozen PBMC available for study. The one Mane-A*10+ animal that received a live attenuated SIVmac239 strain with nef and the long terminal repeat deleted in a previous study was also analyzed for comparison (20). All vaccines expressed the wild-type KP9 epitope within SIV Gag. The DNA/FPV study vaccine strain also expressed SIV Pol and HIV type 1 AE subtypes Env, Tat, and Rev; the VV/FPV study vaccine strain also expressed SIV Pol (9, 32).
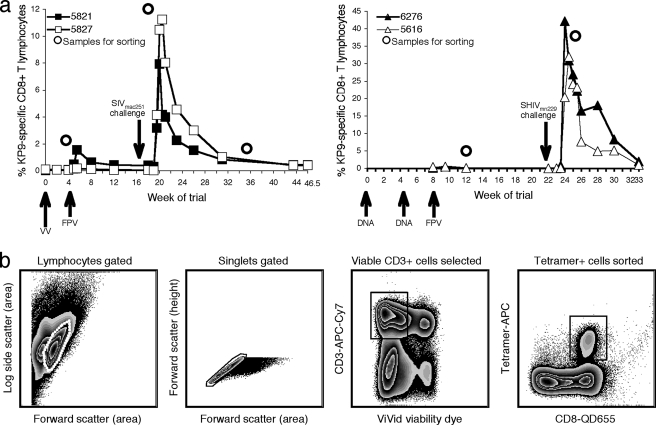
FIG. 1.
Samples and sorting strategy for TCR repertoire analysis. (a) Sorted KP9-specific CD8 T cells (indicated by dots) were derived from two animals receiving VV/FPV vaccines (total, six samples; left panel) and two animals receiving DNA/FPV vaccines (total, four samples; right panel). Vaccination time points are indicated by upward-pointing arrows, along with the vaccine modalities administered. Viral challenge is indicated by a downward-pointing arrow. (b) Gating strategy used to sort KP9-specific CD8 T cells. Thawed pigtail macaque PBMC samples were stained with CD3, CD8, KP9 tetramer, CD45RA, CD28, and ViVid viability dye. Viable single CD3+ tetramer+ lymphocytes, and singlets were sorted into RNAlater with a modified BD FACSAria under biosafety level 3 conditions and used for TCR repertoire analysis.
Sequencing of the pigtail macaque TRBC locus.
The constant region of the pigtail macaque TCR β chain (TRBC) was sequenced with primers based on an alignment of human and rhesus macaque TRBC sequences. Primers were directed to regions on either side of the binding site for the TRBC primer previously described for the unbiased analysis of expressed rhesus macaque TRB genes (11, 29). Pigtail macaque cDNA was amplified by PCR with the TCRB-CFwd2 (GAG GAC CTG AAA AAG GTG TTC) and TCRB-CRev (CAT AGA GGA TGG TGG CAG ACA) primers in a reaction mixture that included 10 μl 5× Phusion HF buffer (Finnzymes), 1 μl 10 mM deoxynucleoside triphosphates (dNTPs; Promega), and 1 U Phusion DNA polymerase (Finnzymes), generating a 448-bp product. Cycling parameters were 98°C for 30 s; 35 cycles of 98°C for 5 s, 50°C for 1 s, and 72°C for 15 s; and then a final extension at 72°C for 5 min. Amplicons were A tailed, ligated into the pGEM-T Easy vector (Promega) according to the manufacturer's protocol, and transformed into competent Escherichia coli JM109. Plasmid DNA from six clones was sequenced in both directions with the M13+ and M13− primers.
Sample preparation and cell sorting.
Pigtail macaque PBMC samples (6 × 106 to 14 × 106 cells) were thawed rapidly in a 37°C water bath and resuspended in 11 ml RF-10 (RPMI medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin) containing 1 μl DNase. Cells were washed twice to remove all traces of dimethyl sulfoxide and then stained with 2 μl allophycocyanin-conjugated Mane-A*10/KP9 (1 μg, a saturating amount) tetramer for 15 min at 37°C. After a further wash in phosphate-buffered saline, cells were stained at 4°C for 30 min with a panel of surface markers (anti-CD28-phycoerythrin, anti-CD45RA-fluorescein isothiocyanate, anti-CD3-allophycocyanin-Cy7, and anti-CD8-quantum dot 655/705) plus 5 μl of the amine-reactive viability dye ViViD (Invitrogen) (28). Cells were then washed in RF-10 plus DNase and resuspended in 500 μl RF-10 plus DNase. We have previously shown that the Mane-A*10/KP9 tetramer demonstrates excellent separation of positive and negative cells and that this tetramer does not stain CD8 T cells in Mane-A*10-positive animals not exposed to SIV Gag or in Mane-A*10-negative animals (9, 13, 21, 32, 35). Recognition of KP9 is specific to this 9-mer peptide and does not overlap other SIV Gag epitopes described to date (14).
Sorting of viable, single, tetramer+ CD3+ lymphocytes was performed with a modified FACSAria (BD) under biosafety level 3 conditions. Up to 10,000 KP9-specific CD8+ T cells were sorted into 1.5-ml tubes containing 150 to 250 μl RNAlater (Ambion). Cell samples were then centrifuged briefly and immediately frozen at −80°C.
mRNA extraction and cDNA synthesis.
Cell samples were thawed and then pelleted out of RNAlater by centrifugation. mRNA was extracted with the Oligotex Direct mRNA Mini Kit (Qiagen) by following the manufacturer's protocol. cDNA synthesis was conducted with the 5′ SMART RACE (rapid amplification of cDNA ends) cDNA amplification kit (Clontech). The RACE cDNA reaction involved 3 to 5 μl mRNA together with 1 μl 10 μM SMART IIA oligonucleotide (Clontech) and 1 μl 10 μM 5′CDS primer (Clontech) placed at 70°C for 1 min and then at −20°C for 1 min. A 2-μl volume of 5× RT buffer (Clontech), 1 μl 20 mM dithiothreitol (Clontech), 1 μl RNaseOUT (Invitrogen), 1 μl 10 mM dNTPs (Invitrogen), and 200 U Superscript II RNase H− reverse transcriptase (Invitrogen) were then added, and the reaction mixture was placed at 42°C for 2 h. The reverse transcription reaction was stopped by the addition of 10 μl Tricine buffer (Clontech), followed by incubation at 72°C for 7 min. The cDNA was then either stored at −80°C or used immediately for the anchored PCR.
Anchored PCR for unbiased amplification of TRB gene products.
Expressed TRB gene products were amplified in a 50-μl reaction mixture comprising 7 μl 5′ RACE cDNA, 5 μl 10× PCR buffer (Clontech), 5 μl 10× universal primer mix (Clontech), 0.5 μl 25 μM MBC 2 primer (rhesus macaque TRBC specific; TGC TTC TGA TGG CTC AAA CAC AGC GAC CT), 0.5 μl 25 μM piggy MBC 2 primer (pigtail macaque TRBC specific; TGC TTC TGA TGG CTC AAA CAC AGC AAC CT), 1 μl 10 mM dNTPs (Invitrogen), and 1 μl AdvanTaq2 DNA polymerase enzyme cocktail (Clontech). Controls without a template were set up in parallel for each sample. Amplification conditions were as follows: 30 s at 95°C; 5 cycles of 95°C for 5 s and 72°C for 2 min; 5 cycles of 95°C for 5 s, 70°C for 10 s, and 72°C for 2 min; and then 30 cycles of 95°C for 5 s, 68°C for 10 s, and 72°C for 2 min.
Purification, cloning, and sequencing of amplified TRB gene products.
PCR products were resolved on a 1% agarose Tris-acetate-EDTA gel, sized against a 100-bp DNA ladder (Fermentas), excised, and purified with a QIAquick gel extraction kit (Qiagen) according to the manufacturer's protocol. Purified products were then ligated into the pGEM-T Easy vector (Promega) by following the manufacturer's instructions. Ligation reaction mixtures were left to incubate at 4°C overnight and then transformed into DH5α MAX Efficiency competent E. coli (Invitrogen) and plated onto LB-100 μg/ml ampicillin plates prespread with isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Invitrogen). Ninety-six white (insert-containing) colonies were selected from each sample for a further round of PCR amplification and sequencing. Reaction mixtures were set up in a 96-well plate containing 25 μl PCR mix per well as follows: 2.5 μl 10× HiFi buffer (Invitrogen), 1 μl 50 mM MgSO4, 0.5 μl 10 mM dNTPs (Invitrogen), 1 μl each 5 μM M13+ primer and M13− primer, and 0.7 U Platinum Taq DNA polymerase (Invitrogen). Individual colonies were selected and dotted onto a fresh LB-100 μg/ml ampicillin plate. PCR conditions were 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 3 min. A final 3-min 72°C step completed the reaction. A 12.5-μl volume of each sample was then diluted 1:1 in nuclease-free water and sent for sequencing (Agencourt Bioscience Corporation).
Sequence analysis of amplified TRB gene products.
Sequence analysis of subcloned TRB gene products was conducted with Sequencher version 4.1.2 (Gene Codes Corporation). All 96 sequences from each sample were trimmed of ambiguous bases to a maximum of 400 bp. The trimmed sequences were then assembled by using the dirty-data algorithm, with a minimum match percentage of 93% and a minimum overlap of 20%. Amino acid translations were then used to identify sequence motifs defining the ends of the Vβ (CASS) and Jβ (XFGXG) regions. Any sequences less than 200 bp in length were discarded, as were any sequences with no discernible Vβ or Jβ regions and those with in-frame stop codons upstream of the CASS and XFGXG motifs. Vβ designations were based on comparison of the pigtail macaque amino acid sequences with 54 human TRBV amino acid sequences listed in IMGT, the international ImMunoGeneTics information system (23), accessible at http://imgt.cines.fr/textes/IMGTrepertoire/Proteins/protein/human/TRB/TRBV/Hu_TRBVallgenes.html; most designations were possible with just the 14 amino acids ending with the CA of the CASS motif. Similarly, Jβ designations were based on comparison of the pigtail macaque amino acid sequences with 13 human TRBJ sequences from IMGT, accessible at http://imgt.cines.fr/textes/IMGTrepertoire/Proteins/protein/human/TRB/TRBJ/Hu_TRBJallgenes.html. In cases where there was no exact match, the closest human Vβ or Jβ sequence was selected. Arden's nomenclature is used herein (1), with sequences converted from the IMGT nomenclature according to the table accessible at http://imgt.cines.fr/textes/IMGTrepertoire/LocusGenes/nomenclatures/human/TRB/TRBV/Hu_TRBVnom.html#3.
Comparison of antigen-specific CD8 T-cell repertoires.
Appropriate measurements of TCR repertoire diversity and similarity must account for differences in sample size and the dominance hierarchy of clonotypes (39). Taking the smallest sample size and repeatedly randomly drawing the same number of events from the larger samples (without replacement) can account for different sample sizes (39). These “selected” samples can then by analyzed, and the median result can be compared to the same analysis of the smallest sample. Simpson's diversity index is appropriate for the analysis of TCR repertoire diversity (26, 39); it is sensitive to dominant clonotypes and less sensitive to the number of clonotypes present. The Morisita-Horn index, originally used in ecological studies, is appropriate for determining the similarity of paired TCR repertoire samples (19, 38). Additionally, it is most sensitive to the clone sizes of the dominant clonotypes.
Alanine scan to assess recognition of mutated KP9 epitopes.
Nine peptides containing alanine substitutions across the KP9 epitope were custom ordered (GL Biochem, Shanghai). The alanine residue at position 5 of the wild-type epitope was replaced with a lysine. These mutated epitopes, along with previously described escape mutant peptides K165R (KRFGAEVVP) and P172S (KKFGAEVVS) (14), were used to restimulate PBMC from Mane-A*10+ animals known to respond to KP9 at various concentrations in a standard 6-h gamma interferon (IFN-γ) intracellular cytokine staining assay as previously described (8).
Nucleotide sequence accession numbers.
The Macaca nemestrina TRBC sequences have been lodged with GenBank and assigned accession numbers EU493254 and EU493255.
RESULTS
Repertoire studies of KP9-specific CD8 T cells in pigtail macaques.
TCR specificities have a significant bearing on the efficacy of a CD8 T-cell response. We were interested in assessing the modulation of TCR repertoires over time, particularly between vaccination and infection. A total of 11 postvaccination and postchallenge PBMC samples from five Mane-A*10+ pigtail macaques were analyzed. Four macaques received prime/boost vaccines (two animals received DNA/FPV vaccines, and two received VV/FPV vaccines, all expressing wild-type SIV Gag), and one macaque received a live attenuated SIV for comparison (9, 20, 32). The levels of KP9-specific CD8 T cells during the course of prime/boost vaccinations and viral challenge are shown in Fig. 1a.
Viable KP9-tetramer-positive cells were successfully sorted from all thawed, uncultured PBMC samples as depicted in Fig. 1b. Sample profiles, including sort yields and the eventual numbers of TCR clones analyzed, are provided in Table 1. Yields of viable KP-specific CD8 T cells from prechallenge samples were lower due to the reduced frequency of Kp9-specific CD8 T-cell populations present at these times.
TABLE 1.
TCR repertoire analysis
| Animal | Vaccine, challenge virus (reference) | Sample | Time point (wk) | % Tetramera | No. of cellsb | TCR clone no.c |
|---|---|---|---|---|---|---|
| 5821 | VV/FPV-4-1BBL, SIVmac251 (32) | Prechallenge | 5 | 0.47 | 237 | 81 |
| Postchallenge acute | 19.5 | 3.17 | 10,000 | 83 | ||
| Postchallenge chronic | 36 | 1.4 | 2,500 | 50 | ||
| 5827 | VV/FPV-4-1BBL, SIVmac251 (32) | Prechallenge | 5 | 0.15 | 800 | 65 |
| Postchallenge acute | 19.5 | 4.15 | 5,000 | 84 | ||
| Postchallenge chronic | 36 | 1.7 | 5,000 | 70 | ||
| 5616 | DNA/FPV, SHIVmn229 (9) | Prechallenge | 12 | 0.14 | 530 | 75 |
| Postchallenge acute | 25 | 24.19 | 5,000 | 79 | ||
| 6276 | DNA/FPV, SHIVmn229 (9) | Prechallenge | 12 | 0.14 | 300 | 93 |
| Postchallenge acute | 25 | 26.91 | 10,000 | 75 | ||
| M18 | Live-attenuated SIV (20) | Postchallenge acute | 7 | 0.21 | 700 | 63 |
The tetramer responses listed were measured in fresh whole blood at the original trial time points, except for the animal M18 sample response and the animal 5821 and 5827 postchallenge chronic sample responses, which were measured in thawed PBMC samples.
Shown is the number of cells obtained by sorting and subsequently used for RNA extraction and TCR repertoire analysis.
Ninety-six colonies were screened for each sample. The TCR clone number is the number of colonies that yielded viable TCR sequence and were therefore included in the analysis.
As TCR repertoire analysis of pigtail macaques had not previously been performed, it was necessary to sequence a segment of the TRB constant region to ensure successful, unbiased PCR amplification of expressed TRB gene products from the sorted cells. The rhesus and pigtail macaque sequences were more than 97% homologous. Two single-nucleotide substitutions were discovered in clones from both of two pigtail macaques studied that were not seen in rhesus macaque sequences. Significantly, one of these substitutions occurred within the region targeted by the published rhesus macaque TRBC primer (12, 29). Our TRBV studies were therefore conducted with an additional primer specific for the pigtail macaque variant. Eleven PBMC samples from five separate pigtail macaques were studied, yielding a total of 818 viable TRB sequences from 1,056 clones sequenced.
Analysis of KP9-specific TCRs in pigtail macaques.
The TCR sequences generated from the sorted KP9-specific CD8 T-cell populations were analyzed by comparing their amino acid sequences with human Vβ and Jβ sequences. In cases where the homology was low, the closest match to human sequences was applied. For a summary of the CDR3 sequences, Vβ and Jβ designations, and clonotype frequencies of all of the KP9-specific populations studied, see Table S1 in the supplemental material.
The most striking observation from this complex data set was the lack of any clear TCR selection patterns across the outbred Mane-A*10+ macaques, either after vaccination or in response to SIV or SHIV challenge. Multiple TCR clonotypes within the KP9-specific population were found, displaying highly variable Vβ usage, CDR3 lengths, and Jβ usage. Most samples showed diverse clonotypic repertoires, except for the prechallenge sample from animal 6276, in which only one KP9-specific clonotype was detected (6276 clonotype 12). This result is likely due to the low number and frequency of tetramer+ cells obtained from this sample. However, this clonotype (6276 clonotype 12) was the only clonotype found in more than one animal; clonotype 41 from macaque 5821 (found only at the acute postchallenge time point) was identical, even at the nucleotide level (data not shown).
Evolution of KP9-specific clonotypes between vaccination and challenge.
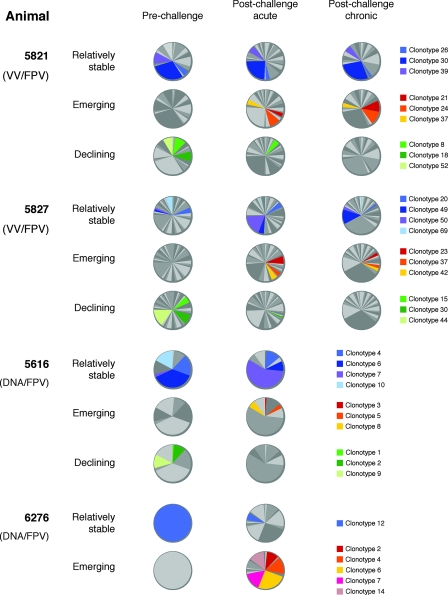
One advantage of working with macaques compared to murine studies is the ready availability of longitudinal blood samples from the same animal. Over time, the KP9-specific TCR repertoires in each animal altered dramatically. The observed repertoire modulations can be loosely divided into three categories: (i) clonotypes that were present at all of the time points analyzed, both after vaccination and after challenge (relatively stable); (ii) clonotypes that were not detectable after vaccination but subsequently emerged after challenge (emerging); and (iii) clonotypes that were present after vaccination but were subsequently lost after challenge (declining). Examples of such modulations are given in Fig. 2. Although some of the “relatively stable” clonotypes maintained fairly constant frequencies across time points (e.g., animal 5821 clonotype 30, which accounted for 29.6, 24.1, and 28% of the clonotypes), others varied greatly in frequency (e.g., animal 5827 clonotype 50, which accounted for 1.5, 17.9, and 2.9% of the clonotypes). Emerging clonotypes were not elicited by vaccination, appearing only at postchallenge time points (e.g., 5827 clonotype 42, which accounted for 0, 4.8, and 2.9% of the clonotypes). Although some emerging clonotypes came to represent a substantial proportion of the overall repertoire (e.g., 5821 clonotype 24, which accounted for 0, 9.6, and 12% of the clonotypes), the majority of the emerging clonotypes represented only a minor proportion of the repertoire (e.g., 5616 clonotype 3, which accounted for 0 and 1.3% of the clonotypes). Declining clonotypes were elicited by vaccination and lost following viral challenge. Interestingly, some of the dominant clonotypes elicited by vaccination (e.g., 5616 clonotype 9, which accounted for 12 and 0% of the clonotypes) were lost following viral challenge.
FIG. 2.
KP9-specific TCR clonotype modulations over time. TCR clonotypes present in samples from individual animals could be divided into three categories, (i) those that were present at all of the time points studied after vaccination and challenge (relatively stable), (ii) those not present after vaccination that emerged after challenge (emerging), and (iii) those present after vaccination that were subsequently lost after challenge (declining). Examples of clonotypes from each animal that fall into each of these three categories are illustrated here, with clonotype identities indicated by the keys on the right. Other clonotypes are displayed as random shades of gray.
TCR Vβ usage by KP9-specific CD8 T cells.
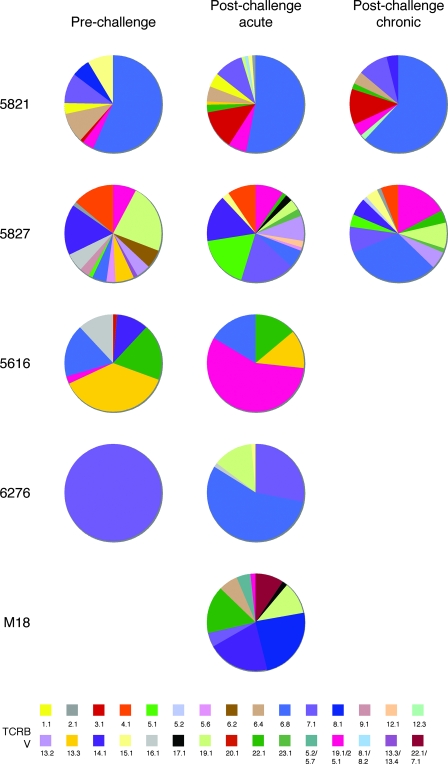
Analysis of Vβ usage by KP9-specific clonotypes again highlighted the remarkably diverse and polyclonal nature of these populations (Fig. 3). The repertoire of VV/FPV-vaccinated animal 5821 was dominated by Vβ 6.8 (blue in Fig. 3); however, patterns of Vβ usage in the other vaccinated animals were not clear-cut. For example, Vβ 6.8 was present in the KP9-specific repertoires from animals 5827, 5616, and 6276 but only dominated following SHIV challenge in animal 6276. Similarly, Vβ 13.3 (gold) was prominent in the 5616 response prechallenge but Vβ 19.1/25.1 (pink) dominated postchallenge. An assessment of which Vβ genes were used across the whole cohort (Table 2) shows that the repertoires in VV/FPV-vaccinated animals 5821 and 5827 comprised a greater range of Vβ genes than those in the other three animals, although all of the responses were highly diverse. Interestingly, no single Vβ gene was used in all five of the animals studied.
FIG. 3.
TCRBV usage in KP9-specific CD8 T-cell populations. The KP9-specific response was characterized by diverse TCRBV gene usage. All of the TCRBV genes detected are listed in accordance with the nomenclature of Arden et al. (1) for the closest matching human sequence after conversion from the TCRBV sequences accessed in IMGT at http://imgt.cines.fr/textes/IMGTrepertoire/Proteins/protein/human/TRB/TRBV/Hu_TRBVallgenes.html.
TABLE 2.
TCRBV gene usage across pigtail macaque KP9-specific CD8 T cells
| TCRBV gene | Presence in animal:
|
||||
|---|---|---|---|---|---|
| M18 | 6276 | 5616 | 5821 | 5827 | |
| 1.1 | No | No | No | Yes | No |
| 2.1 | No | No | No | Yes | Yes |
| 3.1 | No | No | No | Yes | No |
| 4.1 | No | No | No | No | Yes |
| 5.1 | No | No | No | No | Yes |
| 5.2 | No | No | No | No | Yes |
| 5.6 | No | No | No | No | Yes |
| 6.2 | No | No | No | No | Yes |
| 6.4 | Yes | No | No | Yes | No |
| 6.8 | No | Yes | Yes | Yes | Yes |
| 7.1 | Yes | Yes | No | Yes | Yes |
| 8.1 | Yes | No | No | Yes | No |
| 9.1 | No | No | No | No | Yes |
| 12.1 | No | No | No | No | Yes |
| 12.3 | No | No | No | Yes | No |
| 13.2 | No | No | No | No | Yes |
| 13.3 | No | No | Yes | Yes | No |
| 14.1 | Yes | No | Yes | Yes | Yes |
| 15.1 | No | Yes | No | Yes | Yes |
| 16.1 | No | Yes | Yes | No | Yes |
| 17.1 | Yes | No | No | No | Yes |
| 19.1 | Yes | Yes | No | Yes | Yes |
| 20.1 | No | No | Yes | No | No |
| 22.1 | Yes | No | Yes | Yes | Yes |
| 23.1 | No | No | No | No | Yes |
| 5.2/5.7 | Yes | No | No | No | No |
| 19.1/25.1 | Yes | No | Yes | Yes | Yes |
| 8.1/8.2 | No | No | No | Yes | No |
| 13.3/13.4 | No | No | No | No | Yes |
| 22.1/7.1 | Yes | No | No | No | No |
Although the Vβ/Jβ pairing was diverse, with the same Vβ being paired with multiple Jβ segments even within the same animal, some pairing patterns were apparent. For example, Vβ 19.1/25.1 was found in the repertoires of four animals, M18, 5616, 5821, and 5827. While macaque M18 had only one Vβ 19.1/25.1 clonotype and paired it with Jβ 1.4, the other three animals preferentially paired Vβ 19.1/25.1 with Jβ 1.2 (5616, one clonotype; 5821, three of five clonotypes; 5827, five of six clonotypes; see Table S1 in the supplemental material). Similarly, Vβ 6.8 was also detected in the repertoires of four animals, 5616, 6276, 5821, and 5827. In three of the four animals (5616, 5821, and 5827), the dominant Vβ 6.8 clonotypes paired with Jβ 2.3. In macaque 6276, however, the Vβ 6.8 clonotypes, which all emerged after challenge, used four different Jβs, 1.1, 1.2, 1.6, and 2.2.
TCR diversity analyses of KP9-specific T-cell populations.
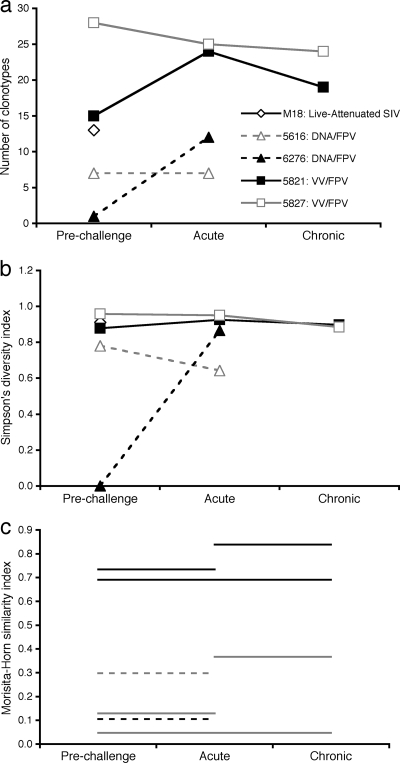
The overall diversity of the KP9-specific TCR repertoires can be approximated by comparing the number of clonotypes detected at each time point for each animal, estimated as if all samples were of the same size (Fig. 4a). The estimated number of clonotypes detected at each time point was relatively similar for animals 5821 and 5827, and clonotype numbers were greater than those detected for DNA/FPV-vaccinated animals 5616 and 6276.
FIG. 4.
Analyses of TCR repertoire similarity and diversity. The diversity of the TCR repertoire was assessed in three different ways, (a) comparison of the number of clonotypes found in each animal at each time point; (b) calculation of the Simpson diversity index for each sample, a measurement of diversity that is sensitive to the presence of dominant clonotypes (0 = minimum diversity, 1 = maximum diversity); and (c) calculation of the Morisita-Horn similarity index, comparing the clonotypes present in paired samples. In panel c, each line represents the similarity between the samples indicated on the x axis; for example, the grey solid line at the bottom compares the prechallenge and postchallenge chronic samples from macaque 5827 (0 = minimum similarity, 1 = maximum similarity).
A more meaningful measurement of diversity, however, is one that accounts for the different clone frequencies of the clonotypes in each sample (see Table S1 in the supplemental material for clonotype frequencies). Simpson's diversity index is an appropriate measurement of diversity for analyzing TCR repertoires, with 0 representing minimal diversity and 1 representing maximal diversity (39). Using this index, following sample size compensation, it is clear that VV/FPV-vaccinated animals 5821 and 5827 exhibited a more diverse repertoire than the DNA/FPV-vaccinated animals (Fig. 4b). The analysis of the prechallenge sample from DNA/FPV-vaccinated animal 5616 is skewed, however, because only a single clonotype was detected from a very small cell sample. Apart from this sample, however, the repertoire diversities were relatively stable across the time points studied. In particular, there was no clear narrowing of TCR repertoires after challenge.
Another way of analyzing TCR repertoires is to ask how much the repertoires change, or remain the same, over time. The Morisita-Horn similarity index can be used to compare paired samples once the sample sizes have been normalized. In this index, 0 represents minimal similarity (and therefore maximal divergence) and 1 represents maximal similarity (and therefore minimal divergence). This index was applied to the KP9-specific repertoire samples from each animal, using paired time points (Fig. 4c). The repertoires for the two VV prime/FPV boost-vaccinated animals showed differing patterns, with all of the 5821 samples being quite similar to each other and the 5827 samples being quite dissimilar between time points. In particular, the prechallenge repertoire in 5827 was quite different from the repertoires at either of the postchallenge time points. The two postchallenge samples (acute and chronic) were more similar, although not as similar as the two postchallenge time points from 5821. The similarity between the prechallenge and postchallenge (acute) samples for the two SHIV A/E animals was also quite low.
While only a small number of animals was studied, it appears that the KP9-specific CD8 T-cell repertoire is highly diverse, with, to date, very little sharing of identical TCRs between individuals. It also appears that different vaccination regimens and challenge viruses are likely to influence the repertoires that subsequently develop.
Phenotype of KP9-specific CD8 T cells.
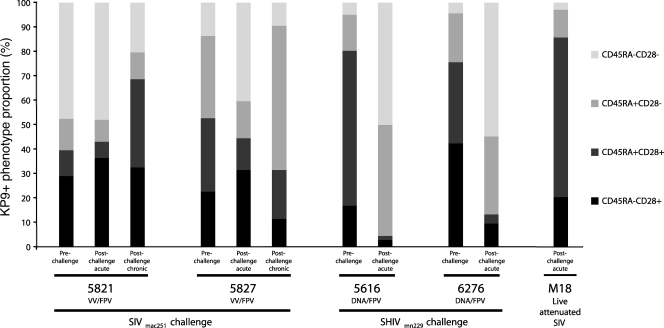
Our findings of a diverse TCR repertoire expressed by most KP9-specific CD8 T cells suggested that there were likely phenotypic differences within the KP9-specific CD8 T-cell population. The costimulatory and memory molecule CD28/CD45RA phenotypes for each sample were therefore assessed in parallel in the tetramer+ populations (Fig. 5). The phenotypic profiles of KP9-specific CD8 T cells from the postchallenge VV/FPV trial samples were consistent with those observed in fresh whole blood, suggesting that future phenotypic studies could be performed with sequential frozen samples. Notably, the phenotypic profiles of the prechallenge samples (1 week postboost) from these two VV/FPV-vaccinated animals were quite different, with a larger CD28-CD45RA− (purple) population in 5821 and a larger CD28-CD45RA+ (red) population in 5827. The DNA/FPV-vaccinated animals had a larger proportion of CD28+ cells prechallenge (green and yellow) than the VV/FPV vaccinees, potentially owing to priming via DNA rather than VV. Following an acute SHIVmn229 challenge, the phenotypic profiles of the KP9-specific CD8 T cells in both of the DNA/FPV-vaccinated animals were strongly dominated by CD28− populations (purple and red), contrasting with the profiles seen during acute SIV infection of the VV/FPV animals. The ×4-tropic SHIVmn229 challenge virus resulted in a much more dramatic early loss of CD4 T cells than the R5-tropic SIVmac251 challenge (5), potentially explaining the dramatic difference in phenotype early after infection. The KP9-specific CD8 T cells from the live attenuated SIV vaccinee, M18, showed a phenotypic profile remarkably similar to those at the prechallenge time point in the DNA/FPV-vaccinated animals.
FIG. 5.
CD45RA/CD28 phenotypes of sorted KP9-specific CD8 T cells. The CD45RA/CD28 phenotypes are shown for the KP9-specific CD8 T-cell populations sorted for TCR analysis from each of the 11 pigtail macaque PBMC samples. The two VV/FPV-vaccinated animals (5821, 5827) were challenged with SIVmac251, and the two DNA/FPV-vaccinated animals (5616, 6276) were challenged with SHIVmn229. M18 was administered live attenuated SIV (proviral SIVmac239 DNA with nef and long terminal repeat deletions) (20).
Broad recognition of mutants by KP9-specific CD8 T cells.
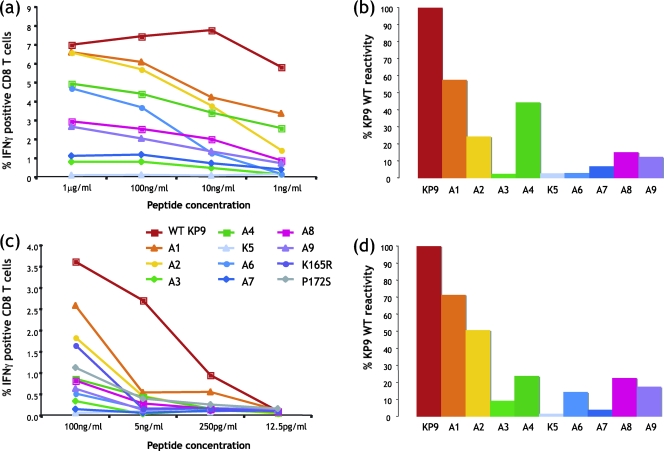
We have previously shown that viral escape at the KP9 epitope involves a lysine-to-arginine change at position 2 (K165R) or, less commonly, a proline-to-serine change at position 9 (P172S) as the exclusive escape mutations selected (14). Given that the TCR repertoire directed toward the KP9 epitope is so diverse and the dominant escape mutations are so uniform, we studied the KP9/Mane-A*10/TCR interaction by evaluating which residues within the KP9 epitope are critical for T-cell recognition. Peptides containing sequential alanine substitutions across the KP9 epitope (except the position 5 alanine, which was replaced with a lysine) were used at various concentrations in an IFN-γ intracellular cytokine assay and compared to the wild-type KP9 epitope. Two separate assays with different peptide concentrations were conducted with fresh blood from two separate Mane-A*10+ animals (Fig. 6).
FIG. 6.
Recognition of KP9 mutants. Peptides containing sequential alanine mutations across the KP9 epitope were studied in IFN-γ intracellular cytokine assays of whole blood from Mane-A*10+ animals with KP9-specific CD8 T-cell responses. (a) Alanine scan titration of blood from VV/FPV-immunized animal 5827. (b) Comparative reactivity of each alanine mutant with wild-type (WT) KP9 peptide at 100 ng/ml (from the titration shown in panel a). (c) Additional alanine scan titration, including the common K165R and P172S mutations, of a separate blood sample. (d) Comparative reactivity of each alanine mutant with the WT KP9 peptide at 100 ng/ml (from the titration shown in panel c). A1 = AKFGAEVVP, A2 = KAFGAEVVP, A3 = KKAGAEVVP, A4 = KKFAAEVVP, K5 = KKFGKEVVP, A6 = KKFGAAVVP, A7 = KKFGAEAVP, A8 = KKFGAEVAP, A9 = KKFGAEVVA, K165R = KRFGAEVVP, and P172S = KKFGAEVVS (mutations are in bold).
We observed almost identical patterns of partial recognition of KP9 mutants in the two separate titrations covering a large dynamic range of peptide concentrations (Fig. 6). Alanine substitutions at positions 1, 2, and 4 are tolerated reasonably well. Alanine mutations at positions 3, 6, and 7 of the KP9 epitope abrogate most CD8+ T-cell reactivity, as does the lysine mutation at position 5. Alanine mutations at positions 8 and 9 reduce reactivity compared to the wild-type epitope. Common escape mutant peptides K165R and P172S show limited reactivity to both peptides at 100 ng/ml, although this drops to background levels at 5 ng/ml. Since the TCR repertoires are diverse, it is likely that this reactivity is restricted to a subset of clonotypes within the overall repertoire.
DISCUSSION
In this study, we characterized the TCR repertoire of CD8 T-cell populations specific for the immunodominant Mane-A*10-restricted SIV Gag164-172 KP9 epitope in pigtail macaques. TCR repertoires have not previously been investigated in pigtail macaques, and it was necessary to sequence the TRB constant region to ensure unbiased amplification (29). Several single nucleotide substitutions were detected within the sequenced constant region, although only two of these substitutions were present exclusively in pigtail macaque sequences. Our pigtail macaque TRB amplifications therefore included novel primers based on these sequences.
Several intriguing insights into the molecular characteristics of KP9-specific CD8 T-cell populations emerged. Remarkably, only 1 out of 161 different KP9-specific clonotypes was found in more than one animal, suggesting that the KP9-specific response is mediated by a diverse repertoire of clonotypes that are highly “private” to individual animals in this outbred populations. This contrasts with TCR repertoires dominated by “public” or shared repertoires between individuals which occur either when a particular TCR has a selection advantage over others (2) or when unusual structural features limit the number of reactive clonotypes (24). The selection of highly “private” TCR repertoires suggests that the bound KP9 epitope presents sufficient physical features to elicit recognition by multiple clonotypes but does not have any unusual features that might restrict recognition or give some clonotypes a selection advantage over others (36). Our findings that there was significant recognition of mutated KP9 variants and a diverse phenotype of KP9-specific CD8 T cells are also consistent with the broad TCR repertoire we identified. Future analyses of the TCR repertoires of subpopulations of KP9-specific CD8 T cells with different avidities (as identified by using lower concentrations of the tetramer) are also suggested by these studies (30).
Despite the overall diversity of the KP9-specific repertoire, some animals did demonstrate a bias toward the use of a particular Vβ gene within this response (for example, Vβ 6.8 in animal 5821). These patterns were not consistent across all of the animals studied, however. Notably, the Vβ bias was not reflected in the CDR3 sequences, with multiple CDR3 sequences occurring in conjunction with individual Vβ segments. The CDR3 loops are likely to be more important in the specificity of antigen recognition than TRBV-encoded CDR1 and CDR2 (17). In this study, the CDR3 sequences did not show any common features or motifs. CDR3 lengths were highly variable, with an average of 14 amino acids, but the dominant clonotypes, even within an individual macaque, contained CDR3 loops of various lengths. Long TRB CDR3 sequences have been associated with improved recognition of emerging escape mutants and long-term viral control in HLA-B8+ HIV-infected humans (10). In the latter study, however, the CDR3 sequences were highly conserved between individuals and were paired with common Vβ and Jβ segments; this is in marked contrast to the KP9-specific repertoire.
In mice, study of a single TCR binding to the same MHC molecule presenting two different peptides has shown that the CDR3 loop is crucial for TCR cross-reactivity (31). This is consistent with studies with SIV-infected Mamu-A*01+ rhesus macaques, where a restricted repertoire directed toward the Tat TL8 epitope was associated with consistent patterns of viral escape (29). It is therefore conceivable that the wide repertoire of KP9-specific CD8 T cells in general, and the diverse CDR3 lengths and sequences in particular, contributes to the recognition of potential escape variants, therefore limiting successful viral escape to mutations such as the dominant K165R mutation (14). Such a mechanism appears to account for the development of escape at the Gag CM9 epitope in rhesus macaques (7, 29), although escape at CM9 generally occurs much later than escape at the KP9 epitope (3, 4, 14). Additionally, repertoire breadth resulting from multiple reactive clonotypes within the naïve T-cell repertoire appears to contribute significantly to the immunodominance of the Gag CM9 epitope (18) and may also account, at least in part, for the immunodominance of the KP9 epitope.
The mechanistic basis for the availability of a large repertoire of potentially reactive clonotypes specific for the KP9 epitope may lie in the structural nature of the antigen. Manipulations of a murine influenza virus epitope have elegantly demonstrated the impact of epitope structural features on TCR repertoire diversity (37). A diverse TCR repertoire directed toward the wild-type H-2Db-restricted PA224 epitope with a prominent central arginine residue was transformed into a more uniform and less diverse repertoire when the arginine residue was mutated to alanine (37). Thus, the highly diverse KP9-specific repertoire suggests that the KP9 epitope complexed to Mane-A*10 may have prominent structural features that enable the selection of broad TCR specificities. Structural studies of the KP9 epitope bound to Mane-A*10 would help to profile the features that contribute to TCR repertoire diversity in this case.
The initial studies described herein focused on serial samples from DNA/poxvirus-vaccinated macaques subsequently challenge with SIV or SHIV. A common observation, regardless of the vaccination type or challenge virus, was the retention of only a small subset of vaccine-induced clonotypes after challenge (stable TCRs in Fig. 2). Many clonotypes induced by vaccination were not detectable after viral challenge, and many new clonotypes, not detected prechallenge, appeared after challenge. These observations could suggest that only a limited number of KP9-specific clonotypes induced by vaccination can expand and respond effectively to a viral challenge.
Our studies, using an unbiased TCR sequencing approach, complement a recently reported study that used TRBV-specific primers to examine the SIV-specific repertoire in vaccinated rhesus macaques (33). In the latter study, diverse Vβ usage after prime/boost vaccination was also observed, although a transient narrowing of Vβ usage was detected early, but not late, after SHIV89.6 challenge. In contrast to our studies, they found common CDR3 sequences within particular Vβ PCR products from SIV-specific cytotoxic T lymphocytes present in several rhesus macaques. The differences may reflect, in part, the different methodologies used for repertoire analysis but perhaps more likely reflect biological differences in the nature of the antigenic epitopes targeted and the SIV/macaque models used.
Understanding how to induce effective CD8 T cells that can respond to challenge and limit immune escape is a key goal of future T-cell-based vaccination strategies. A recent study has shown that therapeutic vaccination can modulate CD8 T-cell repertoires in HIV-infected individuals (40), although the long-term impact of such manipulations on viral control has not been evaluated. Investigating the preferred clonotypic characteristics of effective CD8 T cells and how to elicit them through prophylactic or therapeutic vaccination remains an important field of enquiry. Single-cell cloning of CD8 T cells may contribute to our understanding through in vitro studies of clonal T-cell efficacy (25, 41). However, our data raise theoretical concerns about the representative nature of such approaches because individual antigen-specific clonotypes clearly exhibit differential abilities to expand upon viral exposure and hence presumably to assist in the control of viremia. Furthermore, such procedures can skew the biological properties of individual CD8 T-cell clones, regardless of TCR expression. Thus, detailed characterization of antigen-specific clonotypes directly ex vivo will perhaps prove to be more informative.
The study presented here was limited by the modest number of pigtail macaques studied at serial time points. Additionally, although we attempted to clone nearly 100 separate TCRs at each time point, we cannot be sure that we captured the entire population of CD8 T-cell clonotypes, particularly in cases where low numbers of KP9-specific CD8 T cells were present in the samples (e.g., prechallenge in animal 6267). These sampling issues are especially relevant to highly polyclonal populations, such as those specific for KP9 in animals 5821 and 5827. Further, although we observed a great degree of clonotypic diversity, even within individual pigtail macaques on the same vaccination/challenge protocol, studies of larger numbers of animals receiving identical vaccines might reveal subtle patterns that are not yet apparent. For example, the VV/FPV-vaccinated animals had moderately higher numbers of clonotypes and exhibited greater clonotypic diversity compared to DNA/FPV vaccinees (Fig. 4). This observation, which could have several underlying mechanistic explanations, suggests that different vaccine formulations can elicit different repertoires specific for the same antigen. Thus, while the nature of the antigen is likely the primary determinant of which clonotypes can be recruited, the mode of delivery might dictate which clonotypes are actually recruited; importantly, such differences might translate into differential outcomes after challenge. However, first and foremost, confirmation of these findings in larger comparative studies is warranted. Another limitation of this study is that we restricted our analysis to TCR β chains. To confirm the overall diversity of the KP9-specific TCR repertoire, studies should be extended to the TCR α chains that pair with the TCR β chains described here (18). Such studies could ultimately lead to a structural analysis of TCR engagement with the KP9/Mane-A*10 complex and a more detailed understanding of the generation of TCR repertoire diversity and the emergence of viral escape.
It is clear that the immunodominant Gag KP9-specific CD8 T-cell response is a key component of adaptive immunity during vaccination and SIV or SHIV challenge in pigtail macaques. In this study, we have begun to characterize the diverse nature of this useful CD8 T-cell response at the clonotypic level. These first insights into the substantial TCR repertoire that can be mobilized in response to KP9 reveal the diversity of CD8 T-cell populations that recognize this epitope and suggest that, within this complexity, there might be multiple features that contribute to biological outcome. Further detailed phenotypic and functional studies of antigen-specific clonotypes in different vaccine/challenge models will, we hope, clarify the central determinants of successful CD8 T-cell-mediated immunity and potentially guide vaccine development.
Supplementary Material
Acknowledgments
We thank Steve Turner, Andrew Brooks, and Rob De Rose for helpful advice.
This study was supported by Australian NHMRC award 299907. D.A.P. is a Medical Research Council (United Kingdom) Senior Clinical Fellow. This study was supported in part by the intramural program of the NIAID.
Footnotes
Published ahead of print on 28 May 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arden, B., S. P. Clark, D. Kabelitz, and T. W. Mak. 1995. Human T-cell receptor variable gene segment families. Immunogenetics 42455-500. [DOI] [PubMed] [Google Scholar]
- 2.Argaet, V. P., C. W. Schmidt, S. R. Burrows, S. L. Silins, M. G. Kurilla, D. L. Doolan, A. Suhrbier, D. J. Moss, E. Kieff, T. B. Sculley, and I. S. Misko. 1994. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 1802335-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 777367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415335-339. [DOI] [PubMed] [Google Scholar]
- 5.Batten, C. J., R. D. Rose, K. M. Wilson, M. B. Agy, S. Chea, I. Stratov, D. C. Montefiori, and S. J. Kent. 2006. Comparative evaluation of simian, simian-human, and human immunodeficiency virus infections in the pigtail macaque (Macaca nemestrina) model. AIDS Res. Hum. Retrovir. 22580-588. [DOI] [PubMed] [Google Scholar]
- 6.Charini, W. A., M. J. Kuroda, J. E. Schmitz, K. R. Beaudry, W. Lin, M. A. Lifton, G. R. Krivulka, A. Necker, and N. L. Letvin. 2001. Clonally diverse CTL response to a dominant viral epitope recognizes potential epitope variants. J. Immunol. 1674996-5003. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z. W., A. Craiu, L. Shen, M. J. Kuroda, U. C. Iroku, D. I. Watkins, G. Voss, and N. L. Letvin. 2000. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J. Immunol. 1646474-6479. [DOI] [PubMed] [Google Scholar]
- 8.Dale, C. J., R. De Rose, I. Stratov, S. Chea, D. Montefiori, S. A. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, M. Law, and S. J. Kent. 2004. Efficacy of DNA and fowlpox virus priming/boosting vaccines for simian/human immunodeficiency virus. J. Virol. 7813819-13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rose, R., C. J. Batten, M. Z. Smith, C. S. Fernandez, V. Peut, S. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, V. Venturi, M. P. Davenport, and S. J. Kent. 2007. Comparative efficacy of subtype AE simian-human immunodeficiency virus priming and boosting vaccines in pigtail macaques. J. Virol. 81292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, T., G. Stewart-Jones, N. Chen, P. Easterbrook, X. Xu, L. Papagno, V. Appay, M. Weekes, C. Conlon, C. Spina, S. Little, G. Screaton, A. van der Merwe, D. D. Richman, A. J. McMichael, E. Y. Jones, and S. L. Rowland-Jones. 2004. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. J. Exp. Med. 2001547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douek, D. C., M. R. Betts, J. M. Brenchley, B. J. Hill, D. R. Ambrozak, K. L. Ngai, N. J. Karandikar, J. P. Casazza, and R. A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 1683099-3104. [DOI] [PubMed] [Google Scholar]
- 12.Douek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 41795-98. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, C. S., M. Z. Smith, C. J. Batten, R. De Rose, J. C. Reece, E. Rollman, V. Venturi, M. P. Davenport, and S. J. Kent. 2007. Vaccine-induced T cells control reversion of AIDS virus immune escape mutants. J. Virol. 814137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez, C. S., I. Stratov, R. De Rose, K. Walsh, C. J. Dale, M. Z. Smith, M. B. Agy, S. L. Hu, K. Krebs, D. I. Watkins, D. H. O'Connor, M. P. Davenport, and S. J. Kent. 2005. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J. Virol. 795721-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. St. John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7173-178. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. Capuano III, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 813465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, K. C., and E. J. Adams. 2005. How the T cell receptor sees antigen—a structural view. Cell 122333-336. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa, A., C. Moriya, H. Liu, W. A. Charini, H. C. Vinet, R. A. Subbramanian, P. Sen, N. L. Letvin, and M. J. Kuroda. 2007. Analysis of TCRαβ combinations used by simian immunodeficiency virus-specific CD8+ T cells in rhesus monkeys: implications for CTL immunodominance. J. Immunol. 1783409-3417. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, C. S., Y. Zheng, Y. Liang, J. D. Fontenot, and A. Y. Rudensky. 2006. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 7401-410. [DOI] [PubMed] [Google Scholar]
- 20.Kent, S. J., C. J. Dale, S. Preiss, J. Mills, D. Campagna, and D. F. Purcell. 2001. Vaccination with attenuated simian immunodeficiency virus by DNA inoculation. J. Virol. 7511930-11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent, S. J., R. De Rose, V. V. Mokhonov, E. A. Mohkonova, C. S. Fernandez, S. Alcantara, E. Rollman, R. D. Mason, L. Loh, V. Peut, J. Reece, X. J. Wang, K. M. Wilson, A. Suhrbier, and A. A. Khromykh. 2008. Evaluation of recombinant Kunjin replicon SIV vaccines for protective efficacy in macaques. Virology 374528-534. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. K., G. Stewart-Jones, T. Dong, K. Harlos, K. Di Gleria, L. Dorrell, D. C. Douek, P. A. van der Merwe, E. Y. Jones, and A. J. McMichael. 2004. T cell cross-reactivity and conformational changes during TCR engagement. J. Exp. Med. 2001455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefranc, M. P. 2005. IMGT, the international ImMunoGeneTics information system: a standardized approach for immunogenetics and immunoinformatics. Immunome Res. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehner, P. J., E. C. Wang, P. A. Moss, S. Williams, K. Platt, S. M. Friedman, J. I. Bell, and L. K. Borysiewicz. 1995. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the Vβ17 gene segment. J. Exp. Med. 18179-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffredo, J. T., E. G. Rakasz, J. P. Giraldo, S. P. Spencer, K. K. Grafton, S. R. Martin, G. Napoé, L. J. Yant, N. A. Wilson, and D. I. Watkins. 2005. Tat28-35SL8-specific CD8+ T lymphocytes are more effective than Gag181-189CM9-specific CD8+ T lymphocytes at suppressing simian immunodeficiency virus replication in a functional in vitro assay. J. Virol. 7914986-14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahajan, V. S., I. B. Leskov, and J. Z. Chen. 2005. Homeostasis of T cell diversity. Cell. Mol. Immunol. 21-10. [PubMed] [Google Scholar]
- 27.Manuel, E. R., W. A. Charini, P. Sen, F. W. Peyerl, M. J. Kuroda, J. E. Schmitz, P. Autissier, D. A. Sheeter, B. E. Torbett, and N. L. Letvin. 2006. Contribution of T-cell receptor repertoire breadth to the dominance of epitope-specific CD8+ T-lymphocyte responses. J. Virol. 8012032-12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perfetto, S. P., P. K. Chattopadhyay, L. Lamoreaux, R. Nguyen, D. Ambrozak, R. A. Koup, and M. Roederer. 2006. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J. Immunol. Methods 313199-208. [DOI] [PubMed] [Google Scholar]
- 29.Price, D. A., S. M. West, M. R. Betts, L. E. Ruff, J. M. Brenchley, D. R. Ambrozak, Y. Edghill-Smith, M. J. Kuroda, D. Bogdan, K. Kunstman, N. L. Letvin, G. Franchini, S. M. Wolinsky, R. A. Koup, and D. C. Douek. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21793-803. [DOI] [PubMed] [Google Scholar]
- 30.Ranasinghe, C., S. J. Turner, C. McArthur, D. B. Sutherland, J.-H. Kim, P. C. Doherty, and I. A. Ramshaw. 2007. Mucosal HIV-1 pox virus prime-boost immunization induces high-avidity CD8+ T cells with regime-dependent cytokine/granzyme B profiles. J. Immunol. 1782370-2379. [DOI] [PubMed] [Google Scholar]
- 31.Reiser, J. B., C. Darnault, C. Gregoire, T. Mosser, G. Mazza, A. Kearney, P. A. van der Merwe, J. C. Fontecilla-Camps, D. Housset, and B. Malissen. 2003. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 4241-247. [DOI] [PubMed] [Google Scholar]
- 32.Rollman, E., M. Z. Smith, A. G. Brooks, D. F. Purcell, B. Zuber, I. A. Ramshaw, and S. J. Kent. 2007. Killing kinetics of simian immunodeficiency virus-specific CD8+ T cells: implications for HIV vaccine strategies. J. Immunol. 1794571-4579. [DOI] [PubMed] [Google Scholar]
- 33.Sen, P., W. A. Charini, R. A. Subbramanian, E. R. Manuel, M. J. Kuroda, P. A. Autissier, and N. L. Letvin. 2008. Clonal focusing of epitope-specific CD8+ T lymphocytes in rhesus monkeys following vaccination and simian-human immunodeficiency virus challenge. J. Virol. 82805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, M. Z., C. J. Dale, R. De Rose, I. Stratov, C. S. Fernandez, A. G. Brooks, J. T. Weinfurter, K. Krebs, C. Riek, D. I. Watkins, D. H. O'Connor, and S. J. Kent. 2005. Analysis of pigtail macaque major histocompatibility complex class I molecules presenting immunodominant simian immunodeficiency virus epitopes. J. Virol. 79684-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, M. Z., C. S. Fernandez, A. Chung, C. J. Dale, R. De Rose, J. Lin, A. G. Brooks, K. C. Krebs, D. I. Watkins, D. H. O'Connor, M. P. Davenport, and S. J. Kent. 2005. The pigtail macaque MHC class I allele Mane-A*10 presents an immunodominant SIV Gag epitope: identification, tetramer development and implications of immune escape and reversion. J. Med. Primatol. 34282-293. [DOI] [PubMed] [Google Scholar]
- 36.Turner, S. J., P. C. Doherty, J. McCluskey, and J. Rossjohn. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6883-894. [DOI] [PubMed] [Google Scholar]
- 37.Turner, S. J., K. Kedzierska, H. Komodromou, N. L. La Gruta, M. A. Dunstone, A. I. Webb, R. Webby, H. Walden, W. Xie, J. McCluskey, A. W. Purcell, J. Rossjohn, and P. C. Doherty. 2005. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat. Immunol. 6382-389. [DOI] [PubMed] [Google Scholar]
- 38.Venturi, V., K. Kedzierska, M. M. Tanaka, S. J. Turner, P. C. Doherty, and M. P. Davenport. 2008. Method for assessing the similarity between subsets of the T cell receptor repertoire. J. Immunol. Methods 32967-80. [DOI] [PubMed] [Google Scholar]
- 39.Venturi, V., K. Kedzierska, S. J. Turner, P. C. Doherty, and M. P. Davenport. 2007. Methods for comparing the diversity of samples of the T cell receptor repertoire. J. Immunol. Methods 321182-195. [DOI] [PubMed] [Google Scholar]
- 40.Yang, H., T. Dong, E. Turnbull, S. Ranasinghe, B. Ondondo, N. Goonetilleke, N. Winstone, K. di Gleria, P. Bowness, C. Conlon, P. Borrow, T. Hanke, A. McMichael, and L. Dorrell. 2007. Broad TCR usage in functional HIV-1-specific CD8+ T cell expansions driven by vaccination during highly active antiretroviral therapy. J. Immunol. 179597-606. [DOI] [PubMed] [Google Scholar]
- 41.Yang, O. O., P. T. Sarkis, A. Ali, J. D. Harlow, C. Brander, S. A. Kalams, and B. D. Walker. 2003. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 1971365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, X. G., M. Lichterfeld, S. Chetty, K. L. Williams, S. K. Mui, T. Miura, N. Frahm, M. E. Feeney, Y. Tang, F. Pereyra, M. X. Labute, K. Pfafferott, A. Leslie, H. Crawford, R. Allgaier, W. Hildebrand, R. Kaslow, C. Brander, T. M. Allen, E. S. Rosenberg, P. Kiepiela, M. Vajpayee, P. A. Goepfert, M. Altfeld, P. J. Goulder, and B. D. Walker. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol. 811619-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, X. G., M. Lichterfeld, K. L. Williams, J. Martinez-Picado, and B. D. Walker. 2007. Random T-cell receptor recruitment in human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells from genetically identical twins infected with the same HIV-1 strain. J. Virol. 8112666-12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.