Abstract
The structures of infectious human parvovirus B19 and empty wild-type particles were determined by cryoelectron microscopy (cryoEM) to 7.5-Å and 11.3-Å resolution, respectively, assuming icosahedral symmetry. Both of these, DNA filled and empty, wild-type particles contain a few copies of the minor capsid protein VP1. Comparison of wild-type B19 with the crystal structure and cryoEM reconstruction of recombinant B19 particles consisting of only the major capsid protein VP2 showed structural differences in the vicinity of the icosahedral fivefold axes. Although the unique N-terminal region of VP1 could not be visualized in the icosahedrally averaged maps, the N terminus of VP2 was shown to be exposed on the viral surface adjacent to the fivefold β-cylinder. The conserved glycine-rich region is positioned between two neighboring, fivefold-symmetrically related VP subunits and not in the fivefold channel as observed for other parvoviruses.
Human parvovirus B19 (B19) belongs to the genus Erythrovirus within the Parvoviridae family (33, 54) and is highly tropic for erythroid progenitor cells. It is a human pathogen that causes the mild childhood disease erythema infectiosum (4) and has also been associated with a variety of other clinical symptoms, such as arthropathies, hepatitis (34), failure of red cell production (8), hydrops fetalis (18), fetal loss (14), and myocarditis (35).
The single-stranded DNA genome of B19 is encapsidated in a nonenveloped protein shell that has an external diameter of about 260 Å (2). The 60 T=1 icosahedrally related protein subunits of the wild-type B19 virus capsid are comprised of two structural proteins, viral protein 1 (VP1; 83 kDa) and VP2 (58 kDa), that are produced from a single open reading frame by transcriptional regulation. The major structural component of the protein shell is VP2, accounting for about 95% of the total capsid protein (36). VP2 alone is sufficient to form icosahedral virus-like particles (VLPs) with characteristic parvoviral surface features, including protrusions adjacent to the threefold axes, depressions at the icosahedral twofold axes, and canyon-like depressions surrounding the β-cylindrical structure at the fivefold vertices. A pore at the center of the β-cylinder at each pentameric vertex runs between the outer surface of the virion and the interior of the virus. It has been demonstrated for several parvoviruses that VLPs are similar in morphogenicity and antigenicity to native virions (13, 15, 17, 28, 44). However, differences in the neutralizing response against B19 VP2 VLPs and VLPs containing VP2 and VP1 indicated that VP1 modifies VP2 epitopes (17, 41).
The structural protein VP1 differs from VP2 only in an N-terminal extension of 227 amino acids called the unique region (VP1u). A phospholipase A2-like activity that has been linked to VP1u is required during parvoviral infection (49, 53, 56, 60). In contrast to other mammalian parvoviruses and despite its relatively low concentration in the virion, B19 VP1u represents a dominant antigenic target for neutralizing antibodies (17, 25, 43) and has been suggested to be always located outside the B19 virion (5, 22, 41). The enzymatic activity of the surface-exposed B19 VP1u has been linked to autoimmune reactions and inflammatory processes (26, 30). However, some data indicate that critical regions of VP1u, such as the most N-terminal neutralizing epitopes or the phospholipase enzymatic core of infectious virions, are exposed only after receptor binding, heat, or low-pH treatment, whereas these regions seem to be always accessible in VLPs (5, 40). Infectious B19 virions are more sensitive to various methods of inactivation than other parvoviruses (32, 39).
The VP (meaning VP1 or VP2) N termini are not ordered in any parvovirus crystal structure (3, 21, 23, 29, 45, 46, 52, 58, 59), most likely because their positions are different in each particle and because the signal for VP1u is lost during averaging among the low copy number of VP1 molecules. However, a conserved glycine-rich portion close to the VP2 N terminus has been modeled as to electron density observed within the fivefold pores of DNA-containing canine parvovirus and minute virus of mice (MVM) virions (1, 52, 59), indicating the role of the fivefold channel in the externalization of the N-terminal peptides of some VP subunits. Cryoelectron microscopy (cryoEM) data to 10-Å resolution for adeno-associated virus type 2 (AAV-2) suggested that VP1u is located inside the wild-type particle at the icosahedral twofold axes (24). During or after cell entry by receptor-mediated endocytosis, parvoviral particles undergo structural modifications that lead to the exposure of the VP N termini and to genome release (6, 9, 31, 37, 47, 51, 55). Similar structural changes can be induced in vitro by physicochemical treatment of DNA-containing virions (10, 24, 32, 38, 56).
Difficulties in propagating B19 in cell culture systems have forced biochemical and structural studies to resort largely to recombinant VLPs assembled from either VP2 alone or VP2 and VP1 (17). The structure of recombinant B19 VP2 VLPs has been determined to near-atomic resolution by X-ray crystallography (21). Like other parvoviruses, B19 VP2 contains an eight-stranded, antiparallel β-barrel. Large insertions connecting the strands of the β-barrel form most of the intersubunit contacts and define the outer surface of the virus, including receptor attachment sites and immunological determinants. The fivefold channel, suggested to be involved in the externalization of VP N termini in other parvoviruses, seems to be closed at the outer viral surface, gated by five symmetry-related threonines. However, three glycine residues immediately following the gating residues in sequence might provide structural flexibility to switch the channel from closed to open in the presence of VP1 or DNA or during cell entry.
The present study uses cryoEM to examine the structure of intact, DNA-containing, wild-type B19 virions (iB19) and empty wild-type B19 particles (eB19) purified from human sera. Comparison with recombinant B19 VP2 VLPs shows that the presence of VP1 and/or DNA affects the spatial position of the VP2 termini within the virus particle. The VP2 N-terminal peptide is positioned between neighboring VP subunits of wild-type B19 virions and is exposed to the surface adjacent to the fivefold β-cylinder.
MATERIALS AND METHODS
Purification of B19 virus particles from human serum.
Human plasma with high concentrations of B19 genomic DNA (genotype 1; 1 × 1013 genome equivalents/ml) (27) was kindly provided by Volker Schottstedt from the DRK-Blutspendedienst Nordrhein-Westfalen, Hagen, Germany. Plasma samples which tested negative for VP1/VP2-specific immunoglobulin G and immunoglobulin M in an enzyme-linked immunosorbent assay (RecomWell; Mikrogen GmbH, Neuried, Germany) were depleted of remaining cells and large fragments by low-speed centrifugation. The virus was concentrated from the supernatant by ultracentrifugation and subsequently sedimented into a 24% (wt/wt) CsCl cushion in 50 mM Tris-HCl (pH 8.7), 25 mM EDTA, 0.5% Triton X-100 by centrifugation for 4 h at 150,000 × g (10°C). CsCl was removed from the virus fraction by dialysis against 25 mM Tris-HCl buffer, pH 7.5. For further purification, a linear sucrose gradient (0 to 50%) was overlaid with the virus sample and centrifuged for 2.5 h at 125,000 × g (15°C). The virus was harvested from the lower third of the gradient, transferred into 25 mM Tris-HCl buffer, pH 7.5, and concentrated by using Amicon centrifugal filters (Millipore, Billerica, MA). The protein concentration and purity of the final virus preparations were estimated by sodium dodecyl sulfate gel electrophoresis with Coomassie blue staining.
CryoEM and three-dimensional (3D) image reconstructions.
Small aliquots of purified virus were applied to 400-mesh copper grids coated with a holey carbon film and rapidly frozen by being plunged into ethane slush. Micrographs of the frozen-hydrated samples were recorded on Kodak (Rochester, NY) SO-163 films with a CM300 FEG transmission electron microscope (Philips, Eindhoven, The Netherlands). Images were taken at a calibrated magnification of ×47,000 and at a total electron dose of approximately 22 e−/Å2. The cryoEM micrographs were digitized at 7-μm intervals by using a Zeiss SCAI scanner. Sets of four pixels were averaged, resulting in a 2.97-Å separation of pixels on the specimen.
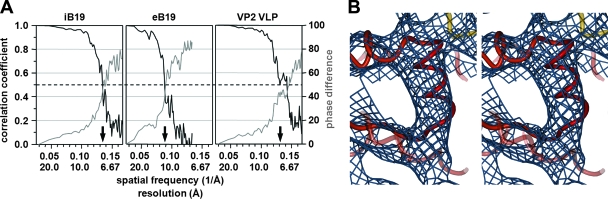
Image analysis and 3D-image reconstructions were performed independently for iB19 and eB19 as previously described (20). The resolution of the resulting maps was estimated by comparing structure factors computed from two independent half data sets (Fig. 1 and Table 1). For the final 3D reconstructions, all data were included to a resolution where the correlation between the half data sets was greater than 0.3 (Fig. 2). The magnification of the cryoEM maps was standardized to a map calculated from the atomic coordinates of the B19 VP2 crystal structure (PDB accession no. 1S58) (21). The icosahedrally averaged cryoEM reconstruction of recombinant B19 VP2 VLPs (19) used to initiate the 3D reconstructions presented here was extended to a 7.7-Å resolution for optimal comparison with wild-type B19 (Fig. 2C). In an attempt to find a putative unique axis formed by VP1, reconstructions assuming only fivefold symmetry were computed from 2,000 particles of each data set by using a modified version of XMIPP (48). This attained an estimated resolution of 17 Å.
FIG. 1.
CryoEM analysis of parvovirus B19. (A) Resolution assessment of the 3D reconstructions of iB19, eB19 particles, and recombinant B19 VP2 VLPs (VP2 VLP). Correlation coefficients (black) and phase differences (gray) for different-resolution shells were computed from the structure factors derived from the independent reconstructions of two half data sets. These plots demonstrate that the data are reliable to at least 7.5-Å (iB19), 11.3-Å (eB19), and 7.7-Å (VP2 VLP) resolution based on a correlation coefficient of 0.5 (arrows) or a phase difference of 45°. (B) A stereodiagram of a density region of the iB19 cryoEM map congruent with the α-helical region of the B19 VP2 crystal structure (shown in red) validates the quality of the cryoEM density.
TABLE 1.
Image data for the cryoEM reconstructions
| Specimen | No. of micrographs | Defocus level range (μm) | No. of particles
|
Final resolution in Å | EM databank entryc | |
|---|---|---|---|---|---|---|
| Selected from micrographs | Used for final reconstruction | |||||
| iB19 | 50 | 3.66-1.25 | 10,517 | 8,854 | 7.5a (7.15b) | EMD-1466 |
| eB19 | 31 | 3.66-1.25 | 2,297 | 1,959 | 11.3a (10.5b) | EMD-1467 |
Resolution at a Fourier shell correlation coefficient of 0.5 between half data sets.
Resolution at a Fourier shell correlation coefficient of 0.3 between half data sets.
The cryoEM map of B19 virus VP2 VLPs with a final resolution of 7.7a (6.7b) Å was deposited in the EM databank under accession number EMD-1468.
FIG. 2.
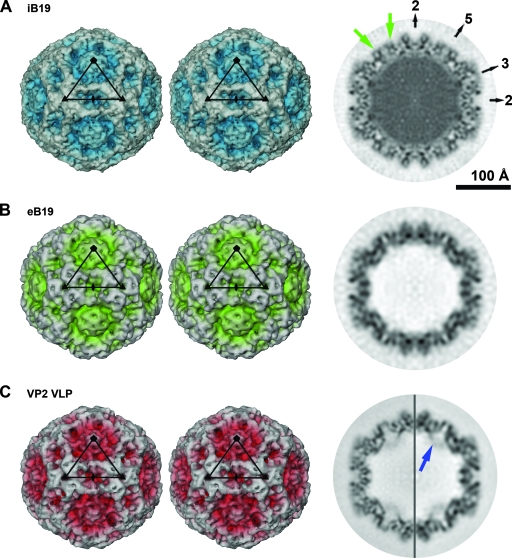
3D image reconstructions of parvovirus B19. (A) Stereoscopic view of a surface rendering of the iB19 particle at 7.5-Å resolution, viewed down an icosahedral twofold axis. The black triangular outline identifies an icosahedral asymmetric unit. The central cross-section is shown on the right, also viewed down an icosahedral twofold axis. Darker coloring in the central cross-section corresponds to higher electron density. The positions of icosahedral two-, three-, and fivefold axes are indicated. Structural differences between wild-type B19 and VP2 VLPs are located around the icosahedral fivefold axes. For one of the fivefold axes, densities near the viral surface, not present in VP2 VLPs, are highlighted with green arrows. (B) Image as described for panel A but illustrating the cryoEM density of eB19 particles at 11.3-Å resolution. (C) Image as described for panel A but showing the cryoEM density of recombinant B19 VP2 VLPs at 7.7-Å resolution. The equatorial slice is depicted at 7.7-Å (left) and 11.3-Å (right) resolution. One of the densities due to internally located VP2 termini pointing toward the center of the VP2 VLP is labeled with a blue arrow. The renderings are at sigma levels of 1.2, 1.5, and 2.0 for the maps in panels A, B, and C, respectively.
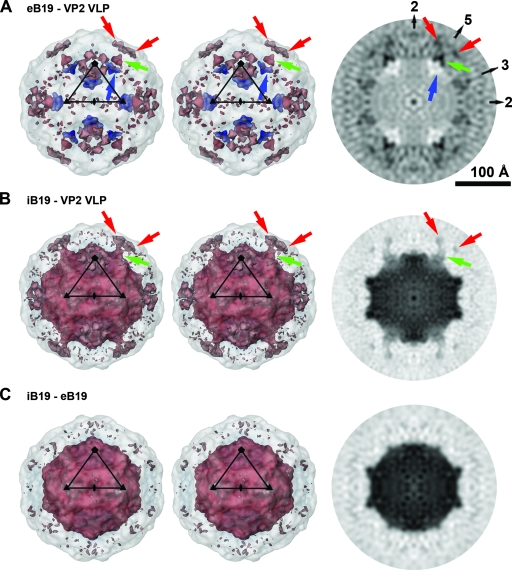
Difference maps between iB19 and eB19, iB19 and VP2 VLPs, and eB19 and VP2 VLPs were calculated after scaling of the densities of the icosahedrally averaged maps to optimize the radial dimensions and average density levels by using the programs EMfit (42) and RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt). Both programs gave essentially the same results. The most significant differences between wild-type B19 (full or empty) and VP2 VLPs were observed at or close to the icosahedral fivefold axes (Fig. 3). These differences were interpreted, based on location and connectivity, as densities representing most of the VP2 termini in wild-type B19. The N-terminal 24 Cα atoms of B19 VP2 were modeled into the difference densities by using the molecular graphics program O (16). The resolution of the maps does not allow the exact placement of the residues. Therefore, the structure database of the program O was used to assure a stereochemically correct secondary structure of the Cα chain.
FIG. 3.
Significant differences between wild-type B19 and recombinant VP2 particles cluster at and around the icosahedral fivefold axes. (A) Stereoview of a surface rendering of a difference map between eB19 and B19 VP2 VLPs at 11.3-Å resolution, viewed down an icosahedral twofold axis. Positive densities are rendered in red, and negative densities in blue. The difference densities are superpositioned onto a semitransparent VP2 VLP. The black triangle marks an icosahedral asymmetric unit. On the right the central cross-section of the difference map is viewed down an icosahedral twofold axis. Black pixels represent positive density. The positions of icosahedral two-, three-, and fivefold axes are indicated. The most-significant positive differences (matter present in eB19 but not VLPs) are located around the fivefold cylindrical structure at the outer viral surface, labeled for one fivefold axis by red arrows, and within the fivefold channel, pointed out by green arrows. Significant negative difference densities (matter present in VLPs but not eB19) can be identified next to the fivefold axes at the inner viral surface (blue arrows). (B) Image as described for panel A but with a difference map between iB19 and B19 VP2 VLPs at 7.7-Å resolution. The largest positive differences besides the central DNA density are located around the fivefold cylinder at the outer viral surface, indicated by red arrows, and at the base of the fivefold channel, highlighted by green arrows. Connecting density diverges off the central fivefold axis halfway up the pore. (C) Image as described for panel A but the difference map is between iB19 and eB19 at 11.3-Å resolution. The most significant difference density is due to the presence of the DNA genome in infectious virions. In panel A, the renderings are at a sigma level of about 0.65 and 0.5 for positive and for negative density, respectively. The maps in panels B and C are rendered at a sigma level of about 0.6. The sigma values are based on the original maps, not on the difference maps.
RESULTS AND DISCUSSION
Parvovirus B19 was purified from human plasma by using a two-step gradient purification procedure. Before the final sucrose gradient, the sample contained not only iB19, but a fraction of eB19. The presence of both empty capsids and intact virions in parvovirus preparations had been observed previously (41, 50). The considerable stability of parvoviral particles (7) suggests that empty capsids are unlikely to be degradation products of complete virions but might be precursors of full particles or remnants of unstable intermediates. Some free nucleic acid was visible in the background of the EM micrographs before sucrose gradient purification but was mostly associated with broken particles. By assuming icosahedral symmetry, 3D cryoEM image reconstructions for both types of particles were calculated to an estimated resolution of 7.5 Å for iB19 virions and 11.3 Å for eB19 particles (Table 1 and Fig. 1 and 2). The β-sheets of the conserved β-barrel, as well as connecting loops, could be clearly recognized in the cryoEM maps, even though the tips of some loops on the viral surface are rather flexible, consistent with high-temperature-factor regions in the VP2 crystal structure. Difference maps were produced between the wild-type B19 maps (iB19 or eB19) and the cryoEM map of B19 VP2 VLPs (Fig. 3). The latter was preferred for calculating the difference map, as opposed to a map calculated from the crystal structure, in order to minimize errors due to differences in data collection.
The absence of genome in eB19 simplified the initial comparison with VP2 VLPs, although the resolution of eB19 is less than that of iB19. The biggest differences between the structure of eB19 and VP2 VLPs were found around the icosahedral fivefold axes (Fig. 2 and 3A). Positive differences, corresponding to densities present in eB19 but not VLPs, are located at the base of the fivefold channel and next to the fivefold β-cylinder on the outer viral surface. No significant differences between wild-type particles and VLPs of AAV-2 or MVM have been observed on the viral outer surface (23, 24), making the differences on the outer surface of B19 found here unique among parvoviruses. Negative differences between eB19 and VP2 VLPs, corresponding to densities present in the VLPs but not eB19, were found inside the viral particle close to the icosahedral fivefold axes. This negative difference density overlaps partially with the last ordered N-terminal residues of the B19 VP2 crystal structure (Fig. 4A). In the crystal structure of B19 VP2 VLPs, well-ordered electron density starts at amino acid 18 (21). In contrast to the ordered VP2 termini in other mammalian parvoviruses, residues 18 to 23 of B19 VP2 point away from the fivefold channel toward the interior of the virus particle (Fig. 2 and 4A). The absence of such inward-pointing density in eB19, as well as the presence of positive density extending toward and into the fivefold pore, suggests that the VP2 terminal sequence, stretching from residues 23 back to 18, enters into the basal opening of the fivefold channel in a way that is similar to the equivalent residues of parvoviruses such as MVM (Fig. 3A and 4A). Accordingly, the Cα atoms of these B19 VP2 residues were placed in a position similar to that of the equivalent residues of MVM. Such differences in the positions of the VP2 N termini in wild-type B19 virus in comparison to their positions in recombinant particles are most likely the result of the presence of VP1 and/or DNA in the former (1, 59). The positive difference density does not continue through the fivefold channel to the viral surface, but at low contour levels, connections diverge off the fivefold-symmetry axis and connect to the outside difference density adjacent to the fivefold β-cylinder. No globular density was observed inside the capsids at the twofold axes, as had been described for AAV-2 (24).
FIG. 4.
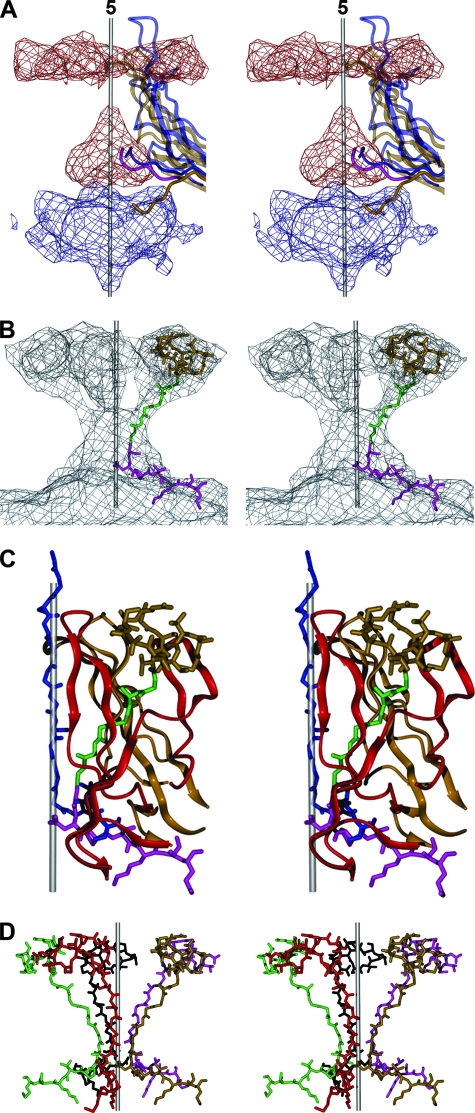
Interpretation of difference densities between wild-type B19 and B19 VP2 VLPs. (A) Stereoview of the difference densities between eB19 and VP2 VLPs close to an icosahedral fivefold axis. Positive densities are rendered in red and negative densities in blue. The icosahedral axis is indicated by a gray rod. The fivefold proximal portions of one of five symmetry-related B19 VP2 molecules (PDB ID code 1S58) are shown as a ribbon diagram in half-transparent gold, while its N terminus is rendered nontransparent. Similarly, one MVM VP2 molecule (PDB ID code 1MVM) is shown in blue. Significant negative density (matter present in VLPs but not in eB19) is located at the inside of the virus particle and overlaps with the positions of the last ordered N-terminal residues in the crystal structure of B19 VP2 VLPs that point toward the viral center. The position of positive density at the base of the fivefold channel suggests that in eB19, the VP2 terminal sequence (residues 23 through 18, shown in pink) enters into the inner basal opening of the fivefold channel in a manner similar to that of the N-terminal residues of MVM. (B) Stereoview of the positive difference densities between iB19 and B19 VP2 VLPs close to an icosahedral fivefold axis. The difference map shows tubular density connecting the inside and outside of the viral particle. The Cα chain of B19 VP2 upstream of residue 18 was modeled into the difference density. The glycine-rich region (residues 17 through 12), shown in green, diverges from the central axis of the channel and connects with the difference density on the outside of the viral particle. The latter difference density could accommodate the remaining outermost N-terminal 11 residues of VP2, rendered in gold. (C) The VP2 N terminus (residues 1 through 24) of one VP, shown as described for panel B, is located between two symmetry-related VP2 molecules (gold and red). In contrast, the N-terminal residues 28 through 41 of MVM VP2, shown in blue, are positioned in the center of the fivefold channel (1). (D) Organization of symmetry-related B19 VP2 termini (residues 1 through 24) at an icosahedral fivefold axis. The position of the B19 VP2 terminus makes it spatially possible for all VP termini to be externalized at the same time and closes the base of the fivefold channel.
The most-significant differences between iB19 and VP2 VLPs (Fig. 3B), other than the genome density, were concentrated around the fivefold axes and coincide with the differences observed between eB19 and VP2 VLPs (Fig. 3A). Consequently, there are only minor differences between the capsids of iB19 and eB19 (Fig. 3C). The higher-resolution difference map between iB19 and VP2 VLPs confirmed that the difference density between wild-type B19 virus and VP2 VLPs does not connect through the fivefold pore to the viral surface but diverges away from the fivefold symmetry axis and extends to the surface adjacent to the fivefold β-cylinder (Fig. 3B and 4B). The Cα atoms of the glycine-rich region (residues 12 through 17 of VP2) were built into the difference density connecting the outside difference volume with the difference density at the base of the fivefold channel (Fig. 4B). The difference density on the virus surface could accommodate all the remaining N-terminal 11 amino acids of VP2 (Fig. 4B). The position of VP1u could not be identified in the icosahedrally averaged maps of either iB19 or eB19, possibly due to VP1u not obeying icosahedral symmetry or being disordered or the signal being lost during the averaging because of the low copy number of molecules per capsid.
The glycine-rich region in DNA-containing particles of MVM and canine parvovirus is in the fivefold channel, suggesting that only one of five VP termini can be extruded through the pore at any given time (1, 52, 59). Proteolytic cleavage of the externalized N terminus converts VP2 into the amino-terminal-shortened VP3 (9, 12, 37, 51, 57), a maturation step that has been suggested to prime the virions of the genus Parvovirus for VP1u exposure during cellular trafficking (11). Such maturation cleavage does not occur in B19, possibly because VP1u is always external. A priming event for externalization is therefore not necessary. The position of the B19 VP2 N terminus is shown here to differ from its position in other parvoviruses, adding to the many unique characteristics observed for B19. The glycine-rich region of B19 VP2 is not located in the fivefold pore but is positioned between two neighboring symmetry-related VP subunits (Fig. 4C), allowing all VP termini to be external at one time. As the density due to the VP2 N termini in the wild-type B19 maps is almost as high as the density of the capsid, most of them must be exposed. The position of the polypeptide chain does not sterically interfere with its environment. This structural organization of the VP2 N termini closes the pore on the inside of the particle (Fig. 4D), making the extrusion of VP1u through the fivefold pore unlikely. Hence, B19 VP1u is probably always external to the virus, consistent with other observations (5, 17, 22, 25). The search for a unique fivefold axis, as in tailed bacteriophages, resulted in a structure exhibiting no significant deviation from icosahedral symmetry (data not shown). Further investigation using either EM tomography or an asymmetric reconstruction might allow the visualization of the B19 VP1u and provide further structural insight.
Acknowledgments
We are grateful to Sheryl Kelly and Cheryl A. Towell for help in the preparation of the manuscript and to Karin Beckenlehner for technical assistance during virus purification.
The work was supported by NIH grants AI11219 to M.G.R. and AI33468 to Colin R. Parrish (Cornell University, Ithaca, NY) and M.G.R. and a grant from the Deutsche Forschungsgemeinschaft (Mo620/7-1) to S.M. We are grateful for the Keck Foundation grant for the purchase of the CM300 field emission gun electron microscope used in this study.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 61369-1381. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203106-115. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16155-171. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. J., S. E. Jones, H. S. P. Fisher, E. Lewis, S. M. Hall, C. L. Bartlett, B. J. Cohen, P. P. Mortimer, and M. S. Pereira. 1983. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Lancet 3211378. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, S., M. Momoeda, M. Kawase, S. Kajigaya, and N. S. Young. 1995. Peptides derived from the unique region of B19 parvovirus minor capsid protein elicit neutralizing antibodies in rabbits. Virology 206626-632. [DOI] [PubMed] [Google Scholar]
- 6.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 792528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boschetti, N., K. Wyss, A. Mischler, T. Hostettler, and C. Kempf. 2003. Stability of minute virus of mice against temperature and sodium hydroxide. Biologicals 31181-185. [DOI] [PubMed] [Google Scholar]
- 8.Brown, K. E. 2000. Haematological consequences of parvovirus B19 infection. Baillieres Best. Pract. Res. Clin. Haematol. 13245-259. [DOI] [PubMed] [Google Scholar]
- 9.Clinton, G. M., and M. Hayashi. 1976. The parvovirus MVM: a comparison of heavy and light particle infectivity and their density conversion in vitro. Virology 7457-63. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., A. M. D'Abramo, Jr., C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254169-181. [DOI] [PubMed] [Google Scholar]
- 11.Farr, G. A., S. F. Cotmore, and P. Tattersall. 2006. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 80161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farr, G. A., and P. Tattersall. 2004. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 323243-256. [DOI] [PubMed] [Google Scholar]
- 13.Gall-Recule, G., V. Jestin, P. Chagnaud, P. Blanchard, and A. Jestin. 1996. Expression of muscovy duck parvovirus capsid proteins (VP2 and VP3) in a baculovirus expression system and demonstration of immunity induced by the recombinant proteins. J. Gen. Virol. 772159-2163. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, G. L. 2000. Parvovirus B19 infection and its significance in pregnancy. Commun. Dis. Intell. 2469-71. [DOI] [PubMed] [Google Scholar]
- 15.Hernando, E., A. L. Llamas-Saiz, C. Foces-Foces, R. McKenna, I. Portman, M. Agbandje-McKenna, and J. M. Almendral. 2000. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 267299-309. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 17.Kajigaya, S., H. Fujii, A. Field, S. Anderson, S. Rosenfeld, L. J. Anderson, T. Shimada, and N. S. Young. 1991. Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad. Sci. USA 884646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz, V. L., N. C. Chescheir, and M. Bethea. 1990. Hydrops fetalis from B19 parvovirus infection. J. Perinatol. 10366-368. [PubMed] [Google Scholar]
- 19.Kaufmann, B., U. Baxa, P. R. Chipman, M. G. Rossmann, S. Modrow, and R. Seckler. 2005. Parvovirus B19 does not bind to membrane-associated globoside in vitro. Virology 332189-198. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann, B., A. Lopez-Bueno, M. G. Mateu, P. R. Chipman, C. D. Nelson, C. R. Parrish, J. M. Almendral, and M. G. Rossmann. 2007. Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody. J. Virol. 819851-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. USA 10111628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawase, M., M. Momoeda, N. S. Young, and S. Kajigaya. 1995. Most of the VP1 unique region of B19 parvovirus is on the capsid surface. Virology 211359-366. [DOI] [PubMed] [Google Scholar]
- 23.Kontou, M., L. Govindasamy, H. J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 7910931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronenberg, S., B. Bottcher, C. W. der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 795296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurtzman, G., N. Frickhofen, J. Kimball, D. W. Jenkins, A. W. Nienhuis, and N. S. Young. 1989. Pure red-cell aplasia of 10 years' duration due to persistent parvovirus B19 infection and its cure with immunoglobulin therapy. N. Engl. J. Med. 321519-523. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann, H. W., P. von Landenberg, and S. Modrow. 2003. Parvovirus B19 infection and autoimmune disease. Autoimmun. Rev. 2218-223. [DOI] [PubMed] [Google Scholar]
- 27.Liefeldt, L., A. Plentz, B. Klempa, O. Kershaw, A. S. Endres, U. Raab, H. H. Neumayer, H. Meisel, and S. Modrow. 2005. Recurrent high level parvovirus B19/genotype 2 viremia in a renal transplant recipient analyzed by real-time PCR for simultaneous detection of genotypes 1 to 3. J. Med. Virol. 75161-169. [DOI] [PubMed] [Google Scholar]
- 28.Livingston, R. S., D. G. Besselsen, E. K. Steffen, C. L. Besch-Williford, C. L. Franklin, and L. K. Riley. 2002. Serodiagnosis of mice minute virus and mouse parvovirus infections in mice by enzyme-linked immunosorbent assay with baculovirus-expressed recombinant VP2 proteins. Clin. Diagn. Lab. Immunol. 91025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llamas-Saiz, A. L., M. Agbandje-McKenna, W. R. Wikoff, J. Bratton, P. Tattersall, and M. G. Rossmann. 1997. Structure determination of minute virus of mice. Acta Crystallogr. D 5393-102. [DOI] [PubMed] [Google Scholar]
- 30.Lu, J., N. Zhi, S. Wong, and K. E. Brown. 2006. Activation of synoviocytes by the secreted phospholipase A2 motif in the VP1-unique region of parvovirus B19 minor capsid protein. J. Infect. Dis. 193582-590. [DOI] [PubMed] [Google Scholar]
- 31.Mani, B., C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, and C. Ros. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 801015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani, B., M. Gerber, P. Lieby, N. Boschetti, C. Kempf, and C. Ros. 2007. Molecular mechanism underlying B19 virus inactivation and comparison to other parvoviruses. Transfusion 471765-1774. [DOI] [PubMed] [Google Scholar]
- 33.Mortimer, P. P., R. K. Humphries, J. G. Moore, R. H. Purcell, and N. S. Young. 1983. A human parvovirus-like virus inhibits haematopoietic colony formation in vitro. Nature 302426-429. [DOI] [PubMed] [Google Scholar]
- 34.Naides, S. J. 1999. Infection with parvovirus B19. Curr. Infect. Dis. Rep. 1273-278. [DOI] [PubMed] [Google Scholar]
- 35.Orth, T., W. Herr, T. Spahn, T. Voigtlander, D. Michel, T. Mertens, W. J. Mayet, W. Dippold, and B. K. H. Meyer-zum. 1997. Human parvovirus B19 infection associated with severe acute perimyocarditis in a 34-year-old man. Eur. Heart J. 18524-525. [DOI] [PubMed] [Google Scholar]
- 36.Ozawa, K., and N. Young. 1987. Characterization of capsid and noncapsid proteins of B19 parvovirus propagated in human erythroid bone marrow cell cultures. J. Virol. 612627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paradiso, P. R. 1981. Infectious process of the parvovirus H-1: correlation of protein content, particle density, and viral infectivity. J. Virol. 39800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reguera, J., A. Carreira, L. Riolobos, J. M. Almendral, and M. G. Mateu. 2004. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA 1012724-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ros, C., C. Baltzer, B. Mani, and C. Kempf. 2006. Parvovirus uncoating in vitro reveals a mechanism of DNA release without capsid disassembly and striking differences in encapsidated DNA stability. Virology 345137-147. [DOI] [PubMed] [Google Scholar]
- 40.Ros, C., M. Gerber, and C. Kempf. 2006. Conformational changes in the VP1-unique region of native human parvovirus B19 lead to exposure of internal sequences that play a role in virus neutralization and infectivity. J. Virol. 8012017-12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld, S. J., K. Yoshimoto, S. Kajigaya, S. Anderson, N. S. Young, A. Field, P. Warrener, G. Bansal, and M. S. Collett. 1992. Unique region of the minor capsid protein of human parvovirus B19 is exposed on the virion surface. J. Clin. Investig. 892023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136190-200. [DOI] [PubMed] [Google Scholar]
- 43.Saikawa, T., S. Anderson, M. Momoeda, S. Kajigaya, and N. S. Young. 1993. Neutralizing linear epitopes of B19 parvovirus cluster in the VP1 unique and VP1-VP2 junction regions. J. Virol. 673004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saliki, J. T., B. Mizak, H. P. Flore, R. R. Gettig, J. P. Burand, L. E. Carmichael, H. A. Wood, and C. R. Parrish. 1992. Canine parvovirus empty capsids produced by expression in a baculovirus vector: use in analysis of viral properties and immunization of dogs. J. Gen. Virol. 73369-374. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 61355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 3151189-1198. [DOI] [PubMed] [Google Scholar]
- 47.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 8011040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorzano, C. O. S., R. Marabini, J. Velázquez-Muriel, J. R. Bilbao-Castro, S. H. W. Scheres, J. M. Carazo, and A. Pascual-Montano. 2004. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148194-204. [DOI] [PubMed] [Google Scholar]
- 49.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316267-280. [DOI] [PubMed] [Google Scholar]
- 50.Tattersall, P., P. J. Cawte, A. J. Shatkin, and D. C. Ward. 1976. Three structural polypeptides coded for by minute virus of mice, a parvovirus. J. Virol. 20273-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tattersall, P., A. J. Shatkin, and D. C. Ward. 1977. Sequence homology between the structural polypeptides of minute virus of mice. J. Mol. Biol. 111375-394. [DOI] [PubMed] [Google Scholar]
- 52.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 2511456-1464. [DOI] [PubMed] [Google Scholar]
- 53.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 55.Vihinen-Ranta, M., S. Suikkanen, and C. R. Parrish. 2004. Pathways of cell infection by parvoviruses and adeno-associated viruses. J. Virol. 786709-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 761884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weichert, W. S., J. S. Parker, A. T. Wahid, S. F. Chang, E. Meier, and C. R. Parrish. 1998. Assaying for structural variation in the parvovirus capsid and its role in infection. Virology 250106-117. [DOI] [PubMed] [Google Scholar]
- 58.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 9910405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264497-520. [DOI] [PubMed] [Google Scholar]
- 60.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1291-302. [DOI] [PubMed] [Google Scholar]