Abstract
A major challenge in human immunodeficiency virus type 1 (HIV-1) vaccine development is to elicit potent and broadly neutralizing antibodies that are effective against primary viral isolates. Previously, we showed that DNA prime-protein boost vaccination using HIV-1 gp120 antigens was more effective in eliciting neutralizing antibodies against primary HIV-1 isolates than was a recombinant gp120 protein-only vaccination approach. In the current study, we analyzed the difference in antibody specificities in rabbit sera elicited by these two immunization regimens using peptide enzyme-linked immunosorbent assay and a competitive virus capture assay. Our results indicate that a DNA prime-protein boost regimen is more effective than a protein-alone vaccination approach in inducing antibodies that target two key neutralizing domains: the V3 loop and the CD4 binding site. In particular, positive antibodies targeting several peptides that overlap with the known CD4 binding area were detected only in DNA-primed sera. Different profiles of antibody specificities provide insight into the mechanisms behind the elicitation of better neutralizing antibodies with the DNA prime-protein boost approach, and our results support the use of this approach to further optimize Env formulations for HIV vaccine development.
AIDS remains one of the largest human epidemics affecting the world today. More than 33 million people are currently living with human immunodeficiency virus type 1 (HIV-1), the virus that causes AIDS, with an estimated 2.5 million new infections occurring in 2007 alone. Over 20 million deaths have already been attributed to HIV-1 infection (http://www.who.int/hiv/mediacenter/news62/en/index.html). The best hope of controlling this epidemic is the development of a successful prophylactic vaccine. Recent setbacks from the STEP trial (36), which relied purely on the induction of cell-mediated immune responses, further highlight the need to develop vaccines that can elicit neutralizing antibodies (NAbs) against primary HIV-1 isolates. Unfortunately, eliciting a strong NAb response to the virus has proven to be an exceptionally difficult task.
Natural HIV-1 infection usually results in strong antibody responses against the viral envelope protein (Env). However, only weak heterologous neutralization is observed in most HIV-positive patient sera. HIV employs a number of strategies to escape neutralization pressure from the host. One of these is the high variability of Env, which allows the virus to escape from NAbs (31, 45). Epitope masking by variable loops, high levels of glycosylation, and quaternary interactions resulting from the trimerization of Env also play significant roles (28). Despite this, several broadly neutralizing monoclonal antibodies (MAbs) that target conserved regions of Env have been identified (3, 4, 47). Two of these antibodies, 2G12 and immunoglobulin G1b12 (IgG1b12 [b12]), recognize gp120, the surface subunit of Env. 2G12 targets specific carbohydrate epitopes on the heavily glycosylated silent face of gp120, while b12 targets the CD4 binding site (CD4bs) (46). Other broadly neutralizing MAbs, 2F5, Z13, and 4E10, target the membrane-proximal external region of gp41 (25, 49). The V3 region of Env is frequently masked in primary isolate Env trimers; therefore, antibodies targeted to this region usually have only limited neutralization breadth and potency against primary HIV-1 isolates (29). In light of studies showing that broad NAbs, administered as passive immunizations, are able to prevent infection in nonhuman primate challenge models (5, 12, 14, 20, 21), attempts to elicit similar NAbs through active immunization have become a major focus of HIV vaccine development.
We previously reported that DNA prime-protein boost vaccination successfully elicited NAbs against the primary HIV-1 isolate JR-FL, a relatively neutralization-resistant virus (39). Rabbits were first inoculated with a DNA vaccine expressing the JR-FL gp120 antigen, followed by a boost with the homologous recombinant gp120, produced in CHO cells. While the control group, which received the JR-FL gp120 protein vaccine alone, induced NAbs against sensitive HIV-1 isolates, only sera from the group that received the DNA prime-protein boost elicited detectable neutralizing activity against the JR-FL virus in both human peripheral blood mononuclear cell (PBMC)- and cell line-based neutralization assays (39). Subsequently, using the same DNA prime-protein boost approach, we demonstrated that polyvalent Env formulations containing Env antigens from diverse genetic backgrounds elicited low-titer but broad NAbs against a subset of pseudotyped viruses expressing primary HIV-1 Env antigens from clade A, B, C, D, and E isolates (44).
In the current study, we investigated the profiles of antibody specificities from these two different vaccination approaches in an attempt to explain the differences in neutralization activity. We analyzed sera from two studies: one from our published study using the monovalent JR-FL gp120 antigen (39) and another that employed a polyvalent gp120 formulation that was recently evaluated in a phase I clinical trial with human volunteers (43).
MATERIALS AND METHODS
Rabbit immunizations.
Rabbit sera against the monovalent JR-FL gp120 antigen were generated in a previous study (39). We also generated rabbit sera in the current study with the same polyvalent primary gp120 formulation used in our recent clinical study (43). Briefly, female New Zealand White rabbits, 6 to 8 weeks old (∼2 kg body weight), were purchased from Millbrook Farm (Amherst, MA) and housed in the animal facility managed by the Department of Animal Medicine at the University of Massachusetts Medical School. Groups of rabbits were immunized with a pentavalent gp120 DNA vaccine at weeks 0, 4, and 12 and boosted with the homologous pentavalent gp120 proteins at weeks 20 and 28. The control protein-alone group received an empty DNA vector at weeks 0, 4, and 12 and the same pentavalent gp120 proteins at weeks 20 and 28. This formulation includes gp120 antigens from subtype A (92UG037.8), B (92US715.6 and Bal), C (96ZM651), and E (93TH976.17) HIV-1 isolates. The DNA vaccine components were produced by subcloning the five codon-optimized gp120 genes, individually, into the DNA vaccine vector pSW3891 (40). The gp120 DNA vaccine plasmids or the empty pSW3891 vector plasmid were coated onto 1.0-μm gold beads at a ratio of 2 μg of DNA per mg of gold and delivered to the animals via a Bio-Rad Helios gene gun on shaved abdominal skin (42). Each animal received 36 μg of total DNA plasmids at each immunization. The five recombinant gp120 proteins were produced in CHO cell lines (27) and purified with a laboratory-based system using lectin to concentrate the gp120 proteins. Recombinant gp120, in doses of 100 μg, were administered intradermally in incomplete Freund's adjuvant (Sigma). Sera were collected immediately before and 2 weeks after each immunization.
Antibodies.
MAbs IgG1b12, b6, and X5 were obtained from Dennis Burton. MAbs 39F, 7B2, LE311, and 15e were all obtained from James Robinson. 2G12 was obtained from Hermann Katinger.
Viruses.
Pseudotypings of env genes from 92RW020, 92TH021, 92UG046, 93IN905, 94UG103, 94UG114, 98CN006, CMU02, JR-CSF,MN, NL4-3, PVO.4, QH0692.42, SC422661.8, AC10.0.29, and SF162 for the PhenoSense assay were all performed at Monogram Biosciences. The HIV-1 env genes from SF162, SC422661.8, and SS1196 and the pSG3ΔEnv backbone, used for pseudotyping as part of the in-house neutralization assay, were obtained through the NIH AIDS Research and Reference Reagent Program.
Enzyme-linked immunosorbent assay (ELISA).
Microtiter plates (96 wells) were coated with 100 μl of individual overlapping 15-mer peptides (4 μg/ml) derived from the consensus HIV-1 M-group gp120 sequence, obtained from the NIH HIV reference reagent program (catalog number 9487). Plates were incubated at room temperature for 1 h, washed five times with 200 μl wash buffer (0.1% Triton-X in phosphate-buffered saline [PBS]), and blocked overnight in 4% whey dilution buffer containing 1% (by weight) powdered milk. Plates were washed again and incubated with 100 μl rabbit sera normalized to 200 ng/ml gp120-specific IgG for 1 h at room temperature. Plates were washed again and incubated with 100 μl of 1 μg/ml biotinylated anti-rabbit antibody (BA-1000; Vector Labs) for 1 h at room temperature. After additional washes, 100 μl of 500 ng/ml horseradish peroxidase-conjugated streptavidin (SA-5004; Vector Labs) was added for 1 h at room temperature. Plates were washed and developed for 3.5 min in 100 μl of a 3,3′,5,5′-tetramethylbenzidine substrate solution (TMB-E-100; ViroLabs). The reaction was stopped by the addition of 25 μl 2 N H2SO4 to the mixture. Plates were then read using a Dynex OpSys MR plate reader.
The rabbit sera were normalized using a semiquantitative ELISA to determine the concentration of gp120-specific IgG in each sample. The gp120 used to coat plates was matched to the immunogen used for rabbit study, i.e., either JR-FL gp120 alone (5 μg/ml) or a pentavalent mixture (1 μg/ml of each component). The concentration of gp120-specific IgG in each serum sample was determined against a standard curve generated using known rabbit IgG (catalog number 0111-01; Southern Biotechnology Associates), as previously described (41). Rabbit sera were prepared to normalized concentrations of 200 ng/ml of gp120-specific IgG.
Virus capture competition.
Various assays of virus capture by HIV-1 MAbs have been described in literature (24, 26, 30). In the current study, the ability of rabbit sera to inhibit the capture of pseudovirions by various MAbs was investigated using a previously described assay (6, 10). Pseudotyped virions in this assay expressing both HIV-1 Env and vesicular stomatitis virus G protein on the virion surface were produced. The vesicular stomatitis virus G protein will mediate the entry of virions into the target cell line CF2.CD4.CCR5 irrespective of any neutralizing activity against the HIV envelope present in the sera, thus providing a sensitive readout of captured virus as previously described (24). The capture of pseudovirions by HIV-1 MAbs will be blocked if there is a competing antibody in the rabbit sera. Specifically, microwells were coated with individual MAbs (5 μg/ml) overnight and were then washed and blocked with 3% bovine serum albumin in PBS. Graded dilutions of rabbit sera were added to the virus, and the virus-serum mixtures were then added to MAb-coated ELISA wells for 3 h, followed by washing with PBS. CF2.CD4.CCR5 cells were overlaid, and 2 days later, infection was measured by assaying luciferase. The reciprocal serum dilution that inhibited MAb-mediated virus capture by 50% was recorded.
Neutralization assays.
Neutralization was determined by three separate but similar assays. The first assay measured the p24 reduction in PBMC with the monovalent JR-FL Env serum as previously reported (23, 39) The second assay was conducted through a subcontract using the PhenoSense assay system (31). In this system, pseudovirus was harvested 48 h after cotransfecting HEK293 cells with pCXAS-Env libraries plus an HIV genomic vector that contains a firefly luciferase indicator gene. Pseudoviruses were then incubated with heat-inactivated rabbit sera at graded dilutions for 1 h at 37°C. U87 cells expressing CD4 and the CCR5 and CXCR4 coreceptors were inoculated with virus-antibody mixtures in the absence of added cations. Virus infectivity was determined 72 h postinoculation by measuring the amount of luciferase activity expressed in infected cells. Neutralizing activity was calculated as the percent inhibition of viral replication (luciferase activity) at each antibody dilution compared with a rabbit serum-free control as follows: % inhibition = [1 − (luciferase + immune sera)/(luciferase − immune sera)] × 100. Prebleed rabbit sera were also included as negative controls. Murine leukemia virus was also included in each assay to rule out nonspecific neutralizing activities. The third assay, the in-house neutralization assay, was performed as previously described (22). Pseudovirus was generated by the cotransfection of a gp160 envelope with the pSG3ΔEnv backbone. Pseudotyped virus was added at 200 50% tissue culture infective doses/well and incubated with rabbit sera at 37°C for 1 h. TZM-bl cells were then seeded at 10,000 cells/well in a final concentration of 20 μg/ml DEAE-dextran. Plates were incubated at 37°C for 48 h and then developed with luciferase assay reagent according to the manufacturer's instructions (Promega). Neutralization was calculated as the percent reduction in luciferase activity in the presence of rabbit immune sera compared to the luciferase activity induced by the virus in the presence of preimmune sera as follows: [1 − (luciferase + immune sera)/(luciferase + preimmune sera)] × 100 (10).
In peptide adsorption experiments, the same in-house neutralization assay was conducted with one additional step: first incubating rabbit sera with or without 30 μg/ml of peptide for 1 h at 37°C. The percent reduction in neutralization was calculated as follows: [1 − (NAb titer in the presence of peptide)/(NAb titer in the absence of peptide)] × 100.
BN-PAGE.
To analyze antibody binding to Env trimers, we investigated antibody binding to native, virion-derived trimers by a modified blue native polyacrylamide gel electrophoresis (BN-PAGE) protocol as previously described (6, 24). Virus-like particles were mixed with rabbit sera for 5 min. To liberate Env, virus-like particles were incubated in an equal volume of solubilization buffer (0.12% Triton X-100 in 1 mM EDTA-1.5 M aminocaproic acid) and 1 μl of a protease inhibitor cocktail (Sigma). An equal volume of 2× sample buffer containing 100 mM morpholinepropanesulfonic acid (MOPS), 100 mM Tris-HCl (pH 7.7), 40% glycerol, and 0.1% Coomassie blue was then added. Samples were loaded onto a 4% to 12% Bis-Tris NuPAGE gel (Invitrogen). Ferritin (Amersham) was used as a size standard. Samples were electrophoresed at 4°C for 3 h at 100 V with 50 mM MOPS-50 mM Tris (pH 7.7) containing 0.002% Coomassie blue as a cathode buffer and the same buffer without Coomassie blue as the anode buffer. The gel was then blotted onto polyvinylidene difluoride. Excess Coomassie blue dye was removed by washing the membrane with 30% methanol-10% acetic acid, followed by 100% methanol. The blot was then transferred into blocking buffer (4% nonfat milk in PBS) for 30 min and probed using 1 μg/ml each of MAbs 2G12, b12, and 447-52D (anti-gp120 cocktail). Goat anti-human and/or anti-mouse Fc alkaline phosphatase conjugates were used, as appropriate, to detect the primary MAbs at a dilution of 1:3,000 (Jackson Laboratories). The density of native and liganded Env bands was determined using ImageJ software, v. 1.33u (NIH freeware; http://rsb.info.nih.gov/ij/). The antibody dilution was recorded as that in the final sample at the time of loading.
RESULTS
Detection of antibodies recognizing conformational V3 epitopes and the CD4bs in monovalent DNA prime-protein boost HIV vaccine sera.
We previously observed that both the DNA prime-protein boost and protein-alone vaccination regimens elicit high titers of anti-Env antibodies in New Zealand White rabbits; however, only sera from the DNA prime-protein boost group effectively neutralized isolate JR-FL (39). This observation is supported by studies from several groups who provided evidence that DNA prime-protein boosting more effectively elicits NAbs against JR-FL and other primary HIV-1 viruses than does protein alone (1, 16, 44).
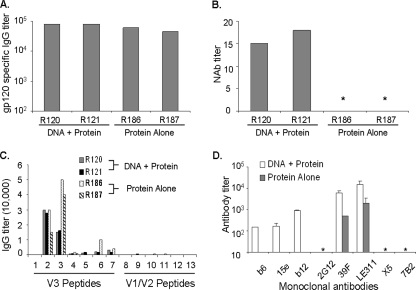
Two rabbit sera (rabbit number 120 [R#120] and R#121) from the DNA prime-protein boost group and two from the protein-alone group (R#186 and R#187), reported in a previous study (39), were included in the current study. The anti-gp120 IgG titers were very similar between the DNA prime-protein boost and protein-alone sera (Fig. 1A). However, positive NAb titers against JR-FL were found only in the group that received the DNA prime (Fig. 1B) (39). Mapping using a panel of overlapping peptides derived from the V3 region of JR-FL Env antigen, 15 amino acids (aa) in length with 11 aa of overlap (see Table S1 in the supplemental material), revealed that every V3 peptide recognized by the DNA prime-protein boost sera was also recognized by the protein-only sera (Fig. 1C). A more than twofold greater IgG titer was found in the protein-alone sera than in the DNA prime-protein boost sera against peptide 3, suggesting that linear V3 epitopes are well preserved in recombinant gp120 antigens. Overall, there was no significant reactivity to the V1/V2 region peptides for either type of rabbit sera (Fig. 1C).
FIG. 1.
Antibody responses elicited from immunization with JR-FL in a DNA prime-protein boost or protein-alone immunization regimen. (A) Anti-gp120 IgG endpoint binding titers as measured by ELISA. (B) NAb titers against JR-FL at 50% neutralization of the virus as seen in a PBMC-based neutralization assay. An * indicates that less than 50% neutralization was seen at a starting dilution of 1:10. (C) Serum recognition of linear peptides derived from the V1/V2 and V3 regions of JR-FL among DNA prime-protein boost and protein-alone rabbit sera. The lowest serum dilution tested was 1:100. (D) Virus capture competition assay using seven MAbs specific for domains on HIV-1 Env and sera from both DNA prime-protein-boosted and protein-immunization-alone animals. Antibody levels are reported as titers necessary to prevent 50% of virus binding. Data shown are average titers, with bars indicating standard deviations of data from rabbits in each group. An * indicates that no antibody competition was detected.
We next performed virus capture competition experiments using various MAbs including three MAbs (b6, 15e, and b12) against the CD4bs, two (39F and LE311) against the V3 loop, one (2G12) specific for carbohydrate, one (X5) recognizing a CD4-inducible epitope (CD4i), and one (7B2) specific for the gp41 cluster I region of Env, which also served as a negative control since the rabbits were immunized with gp120 (Fig. 1D). Similar to the peptide ELISAs, high titers of antibodies competing with MAbs LE311 and 39F for binding to the V3 loop were found in both immunization groups. However, different from the peptide ELISAs, the titers were two- to fivefold higher in sera from DNA-primed animals than in sera from the protein-alone group (Fig. 1D). This difference may reflect the fact that the viral particle-based competition assay preserves Env conformation. Therefore, the DNA prime-protein boost approach appears to induce more antibodies against conformation-dependent V3 epitopes.
More importantly, high levels of antibodies that compete against three CD4bs-specific MAbs were found in DNA prime-protein boost sera (Fig. 1D). DNA prime-protein boost sera were positive for b6- and 15e-like antibodies; however, these same antibodies were undetectable in protein-only immune sera. The DNA-primed sera also competed for binding to b12 at titers approaching 1:1,000, while sera from the group immunized with protein only could not compete for binding at detectable levels (Fig. 1D). Competition assays using MAbs 2G12 and 7B2 revealed no antibodies against either epitope (Fig. 1D).
Eliciting NAbs in rabbit sera using a polyvalent DNA prime-protein boost Env vaccine formulation, DP6-001.
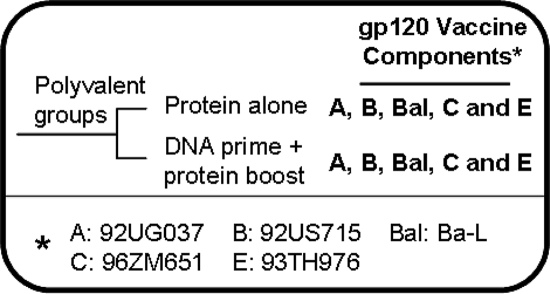
In our preclinical and clinical immunogenicity studies, we reported that polyvalent gp120 formulations, delivered by the DNA prime-protein boost approach, elicited NAbs that were effective against viruses belonging to several different major subtypes of HIV-1 (43, 44). To further investigate the antibody specificities induced by the DNA prime-protein boost regimens, we immunized rabbits with the same pentavalent gp120 vaccine formulation (DP6-001) that induced low-titer but positive NAbs against a wide range of primary Env antigens in a recently completed phase I clinical study (43). New Zealand White rabbits were immunized with either a DNA prime-protein boost or protein-alone regimen with the pentavalent gp120 formulation as shown in Fig. 2. Sera collected from rabbits 2 weeks after the second protein boost in both groups were used to evaluate peak-level gp120-specific IgG and NAb responses.
FIG. 2.
Immunization groups and pentavalent gp120 vaccine formulation. Rabbits received either DNA prime-protein boost or protein-alone immunizations. An * indicates the HIV-1 primary isolates from which the gp120 vaccine components were derived.
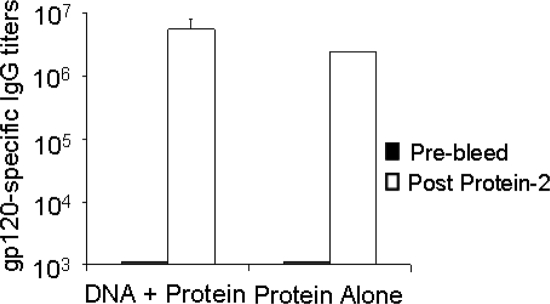
The peak-level gp120-specific binding titers were very similar between the two immunization groups (Fig. 3). However, sera from rabbits that received the polyvalent DNA prime-protein boost showed higher neutralizing activities against a panel of pseudotyped viruses in the PhenoSense assay (Table 1). Here, we examined three sensitive viruses (MN, NL4-3, and SF162) and 12 viruses that are relatively resistant to neutralization with Env antigens derived from primary isolates of clades A, B, C, D, and E. Serum dilution was started at 1:10, but positive neutralizing activities (defined as >50% inhibition) at a serum dilution of 1:30 are summarized to reflect a more stringent cutoff for scoring positive responses and to exclude nonspecific inhibitions in this assay. All sera, regardless of the immunization regimen, neutralized the tier 1 sensitive isolates MN, NL4-3, and SF162 at this serum dilution. Sera from the DNA prime-protein boost group also neutralized 8 out of the 12 more resistant viruses. Neutralization of primary isolates from clades A, C, and D was observed in almost every case in the DNA-primed rabbit sera. Three viruses (AC10.0.29, PVO.4, and QH0692.42) from the NIH tier 2 clade B standardized panel (17) and JR-CSF (another neutralization-resistant isolate) were not neutralized by any of the rabbit sera (data not shown). Two rabbit sera had neutralizing activity against SC422661.8 from the NIH tier 2 clade B standardized panel. Clade E isolate 92TH021 was neutralized by only two out of four DNA-primed sera. In contrast, sera from rabbits immunized with protein only did not neutralize most of the primary viruses. At a 1:30 serum dilution, no inhibition of murine leukemia virus infection was observed (data not shown), demonstrating that the neutralization was HIV-1 specific. Additionally, a broadly neutralizing human serum, N16, was used as a positive control. This serum neutralized all primary isolates with titers between 1:30 and 1:90 (data not shown). Preimmune rabbit sera were also included as a negative control for the PhenoSense assay. No neutralization was observed with the preimmune sera at a 1:30 serum dilution.
FIG. 3.
gp120-specific IgG titers in rabbits immunized with DNA prime-protein boost and protein-alone formulations against autologous antigens as measured by ELISA. Data are shown as geometric mean titers, with bars indicating standard deviations of data from two or four rabbits in the group immunized with protein alone or the group immunized with DNA plus protein, respectively. “Prebleed” and “Post Protein-2” indicate the sera collected at preimmunization and at 2 weeks after the second protein immunization, respectively.
TABLE 1.
NAb responses elicited by polyvalent Env formulationsa
| HIV-1 isolate (subtype) | % Neutralization for vaccine group:
|
|||||
|---|---|---|---|---|---|---|
| DNA prime + protein boost
|
Protein alone
|
|||||
| 29-1 | 29-2 | 30-1 | 30-2 | 33-1 | 33-2 | |
| MN (B) | 99.9 | 99.9 | 99.9 | 99.9 | 99.1 | 92.7 |
| NL4-3 (B) | 97.6 | 98.6 | 99.7 | 96 | 94.3 | 93.9 |
| SF162 (B) | 99.9 | 99.9 | 99.9 | 99.9 | 99.8 | 98.4 |
| 92RW020 (A) | 54 | 57.5 | 55.6 | 50.4 | — | — |
| 94UG103 (A) | — | 51 | 51.6 | 55.4 | — | — |
| SC422661.8 (B) | — | 58.6 | — | 56.4 | — | — |
| 93IN905 (C) | 62.1 | 66.6 | 66.7 | 63.3 | — | 59.2 |
| 98CN006 (C) | 53 | 56.1 | — | 50 | — | — |
| 92UG046 (D) | 53.4 | 56.1 | 54.7 | 54.1 | — | — |
| 94UG114 (D) | 57.2 | 57.7 | 55.2 | 60.3 | — | — |
| 92TH021 (E) | — | — | 51.9 | 53.9 | — | 50.2 |
The data are shown as percent neutralization at a 1:30 serum dilution as determined by the PhenoSense neutralization assay. —, less than 50% inhibition.
Epitope mapping of the polyvalent sera using linear overlapping peptides.
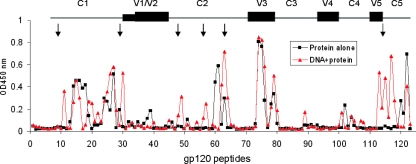
To determine any difference in antibody specificities between DNA-primed versus protein-alone polyvalent rabbit sera, we first measured serum binding to linear 15-mer peptides with 11-residue overlaps derived from the group M consensus gp120 sequence (catalog number 9487; NIH AIDS Reference and Reagent Program). To test this, rabbit sera was normalized to equivalent total gp120-specific IgG titers. Sera from both groups had strong reactivity to peptides derived from the C1, C2, V3, and C5 segments of gp120 (Fig. 4), including two prominent regions within the N terminus of the V3 loop, which is similar to the peptide binding results, as measured by ELISA using monovalent JR-FL-immunized rabbit sera (Fig. 1C). Very little reactivity was observed against peptides derived from the V1/V2 and V4 loops of gp120 for any of the sera tested.
FIG. 4.
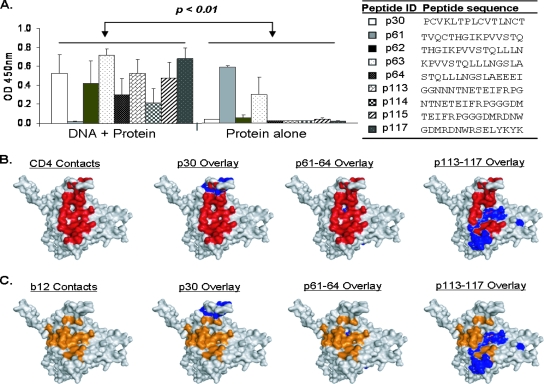
gp120 peptide-specific IgG responses as measured by ELISA. The red and black curves indicate rabbit sera from DNA prime-protein boost or protein-alone immunization with polyvalent vaccine, respectively. Rabbit sera were normalized to equivalent gp120 binding titers. The gp120 peptides were linear overlapping peptides derived from the group M consensus gp120 sequence of HIV-1. Data shown are the average optical density (OD) values for two rabbits in the protein-alone group or four rabbits in the group immunized with DNA plus protein. Arrows indicate positive binding responses unique to the DNA-primed group against six different regions (i.e., no positive responses were observed in the protein-alone group). These six regions include the regions at p11, p30, p48 to p49, p56 to p57, p61 to p64, and p113 to p117.
Interestingly, rabbit sera from the DNA-primed group exhibited unique positive binding against six regions that were poorly recognized by the protein-only sera (see arrows in Fig. 4). The sizes of these six regions vary: peptides 11 (p11) and 30 (p30) were recognized as single peptides, p48/p49 and p56/p67 were recognized as two adjacent peptides, and p61 to p64 and p113 to p117 were recognized as clusters of several neighboring peptides. Further analysis of these six highly reactive regions indicated that three of them, p30, p61 to p64, and p113 to p117, contain amino acid residues that either are part of the CD4bs or are involved in binding with the neutralizing MAb b12 based on a previously published Env structure (15, 46). The sequences of these peptides are listed in Fig. 5A. The average levels of binding of rabbit sera from the two immunized groups to these peptides were compared. Sera from the DNA prime-protein boost group had significantly greater recognition of these three regions than did sera from the protein-alone group (P < 0.01) (Fig. 5A). The overlay of these sequences onto the crystal structure of gp120 demonstrates that the locations of these peptides (in blue) are in the known CD4 binding region on gp120, overlapping with either the above-described CD4bs (Fig. 5B, red) or b12 binding residues (Fig. 5C, orange).
FIG. 5.
Additional analysis of three highly reactive regions of gp120. (A) Recognition of linear peptides that contain CD4 or b12 contact residues by DNA-primed or protein-alone rabbit sera as measured by ELISA. Bars represent the average optical density (OD) values with standard deviations for two rabbits in the protein-alone group or four rabbits in the group immunized with DNA plus protein against each peptide listed on the right. (B) Locations of group M consensus linear peptides (blue) overlaid onto the crystal structure of JR-FL gp120 liganded with CD4 and MAb X5. CD4 contact residues are highlighted in red. (C) Locations of group M consensus linear peptides (blue) overlaid onto the crystal structure of JR-FL gp120 liganded with CD4 and MAb X5. b12 contact residues are highlighted in orange.
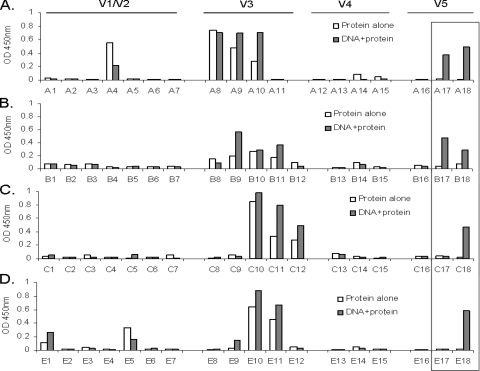
Because a polyvalent Env formulation was used in this rabbit immunization study and because the sequence differences of these Env proteins, particularly in the variable loops, may have precluded the recognition of the consensus M peptides, we generated overlapping peptides (20-mer peptides with 10 aa overlapping) for each of the variable loops from four primary gp120 antigens that were used in our study: 92UG037 (clade A), 92US715 (clade B), 96ZM651 (clade C), and 93TH976 (clade E) (see Table S2 in the supplemental material). These peptides were then tested for antibody recognition with the polyvalent rabbit sera by ELISA. Some sequence-specific variable-loop recognition was observed in sera from immunized rabbits against peptides derived from the V1/V2 loop of clade A and E Env antigens, in contrast to peptides derived from the V1/V2 loop of clade B and C Env antigens. However, there was no differential recognition in this region between the DNA-primed and protein-alone groups. Reactivity was also found in the V3 loop for both the group immunized with the DNA prime-protein boost and the group immunized with protein alone; however, the locations of these recognized epitopes differed between Env antigens. The recognition of the V3 loop was more focused on the N-terminal side and crown of the V3 loop for the clade A Env, on the crown of the V3 loop for the clade E Env, and on the crown and C-terminal strand for the clade C Env and was evenly distributed against the V3 loop of the clade B Env (Fig. 6). Once again, very little reactivity against peptides in V4 region was generated (Fig. 6). For the V5 region, it is striking to find that there are 1 to 2 peptides, located at the junction of the V5 and C5 regions, that were recognized by the sera elicited with the DNA prime-protein boost approach but not the protein-alone sera (Fig. 6). Peptides 17 and 18 in Fig. 6 correspond to the amino acid sequences included in peptides 115 to 117 as shown in Fig. 4 and are therefore in the region involved in CD4 binding (Fig. 5) (32).
FIG. 6.
Polyvalent rabbit serum recognition of peptide sequences derived from the variable loops V1/V2, V3, V4, and V5 from individual gp120 proteins of the polyvalent vaccine components as measured by ELISA. Rabbits were immunized with DNA prime-protein boost (gray bars) or protein only (white bars). Peptides from gp120 A, B, C, and E were used in graphs A, B, C, and D, respectively. Bars represent the average optical density (OD) values for two rabbits in the protein-alone group or four rabbits receiving DNA plus protein against each peptide. Peptide sequences are shown in Table S2 in the supplemental material.
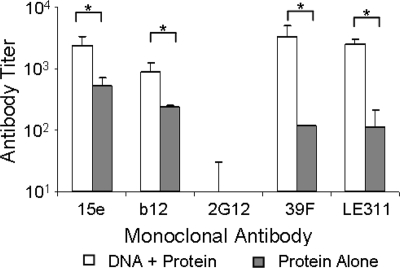
Mapping polyvalent sera by virus capture competition.
Competitive virus capture was used to further map antibody specificities. Titers of 39F- and LE311-like antibodies (V3 loop specific) were much higher in animals that received a DNA prime (Fig. 7); this mirrors the pattern observed with the monovalent sera (Fig. 1C and D) but contradicts data from the peptide ELISAs (Fig. 4 and 5). Additionally, the competitive virus capture assay revealed higher titers of CD4bs-specific antibodies (15e and b12) in animals that received a DNA prime compared to those who received protein-only immunizations.
FIG. 7.
Detection of antibody specificities in DNA-primed or protein-alone polyvalent rabbit sera. This competitive virus capture assay used MAbs to study the specificities and titers of antibodies present in the polyvalent sera. Bars indicate the average dilutions of sera for two rabbits in the protein-alone group or four rabbits in the group receiving DNA plus protein, resulting in a 50% reduction in virus binding. Error bars indicate standard deviations within the group. Significance was calculated using a one-tailed Student's t test. *, P < 0.05.
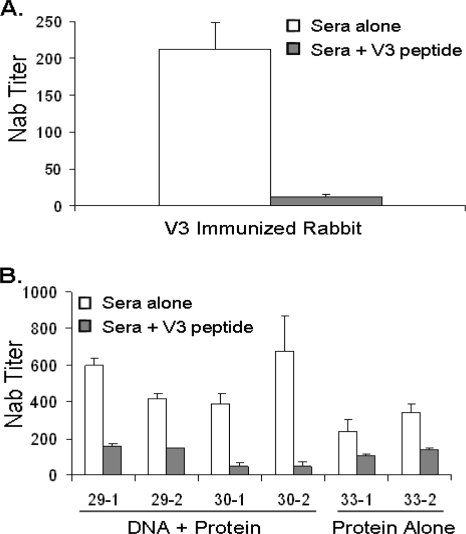
The data described here demonstrate that the DNA prime-protein boost approach is more effective in eliciting conformational V3 and CD4bs antibodies than the protein-alone approach. However, these data do not prove that these antibodies are responsible for the improved NAb activities observed in the DNA-primed rabbit sera (Table 1). To elucidate the roles of the elevated V3 antibodies in the neutralization of HIV isolates, we attempted to adsorb the V3-specific antibodies by incubating the sera with 15-mer clade B consensus peptides spanning the N-terminal strand of the V3 loop (TRPNNNTRKSIHIGPGRAF). This region was chosen because several recent studies indicated that this area is the target of several neutralizing V3 MAbs (2, 33-35). A pilot experiment was conducted to confirm that these peptides were capable of adsorbing V3-mediated neutralizing activity in rabbit sera by using a control rabbit serum that was immunized with only a V3 peptide fused with a carrier protein (48). V3 peptide adsorption of this serum resulted in a greater than 95% decrease in the neutralizing activity against pseudotyped virus expressing Env from HIV-1 isolate SF162, an isolate very sensitive to V3-mediated neutralization (Fig. 8A).
FIG. 8.
Effect of V3 peptide adsorption on neutralization of HIV-1 isolate SF162. Clade B consensus overlapping peptides corresponding to the N-terminal strand and crown of the V3 loop were pooled and incubated with sera at 30 μg/ml prior to exposure to pseudotyped virus expressing Env from HIV-1 isolate SF162. Neutralization titers indicate the serum dilution that can achieve a 50% inhibition of virus infection. (A) Effect of peptide adsorption on the neutralizing activity of sera from a rabbit immunized with only a V3 fusion protein as a control. Error bars indicate standard deviations of data from a one-time multiwell neutralization assay. (B) Effect of V3 peptide adsorption on the neutralizing activity of individual rabbit sera from polyvalent DNA prime-protein boost (29-1, 29-2, 30-1, and 30-2) and protein-alone immunizations (33-1 and 33-2). Error bars indicate standard deviations from replicate experiments.
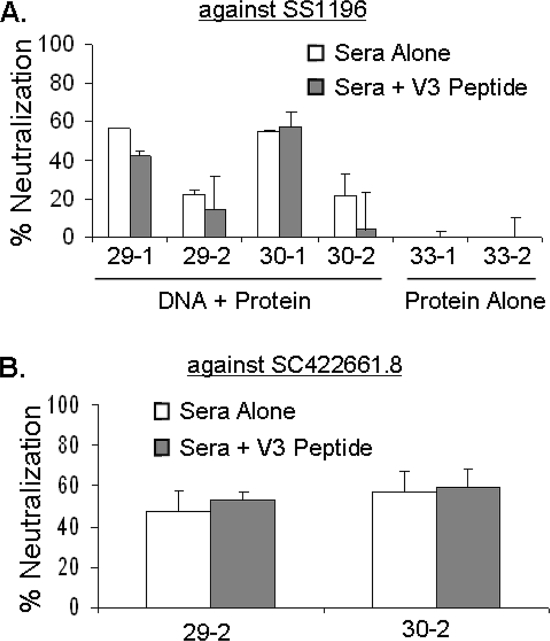
When sera from the polyvalent DNA prime-protein boost rabbits were first incubated with these V3 peptides and then tested for their neutralizing activities against pseudotyped virus expressing Env from HIV-1 isolate SF162, we observed that the sera isolated from DNA prime-protein-boosted animals were more sensitive to a depletion in neutralization (Fig. 8B). Sera from rabbits that received a DNA prime had an average of 79% reduction in NAb titers, compared to the 50% reduction in animals that received protein-only-based immunizations. The greater sensitivity of the DNA-primed sera to V3 peptide adsorptions supports the above-described data that V3-directed antibodies are present in higher levels in DNA-primed animals. We next addressed the role of V3-mediated antibodies in the neutralization of more resistant primary isolates. We repeated the V3 peptide adsorptions in an attempt to neutralize two clade B primary isolates, SS1196 (Fig. 9A) and SC422661.8 (Fig. 9B). V3 peptide adsorption had very little effect on the neutralization of SS1196: only an average of 9% reduction in neutralization was observed. Additionally, no reduction in neutralization was observed in the presence of competing V3 peptide with the other primary isolate, SC422661.8. This indicates that while V3 antibody levels may be elevated in the DNA-primed animals, they play a minimal role in the neutralization of more resistant primary isolates. This leaves the increased levels of CD4bs antibodies or other unknown conformational antibodies as possible candidates responsible for the enhanced neutralizing activities of the DNA-primed sera against more resistant primary viral isolates (Table 1).
FIG. 9.
Effect of V3 peptide adsorption on neutralization of primary HIV-1 isolates. Clade B consensus overlapping peptides corresponding to the N-terminal strand and crown of the V3 loop were pooled and incubated with sera at 30 μg/ml prior to exposure to pseudotyped virus. Percent neutralization at a 1:10 dilution of rabbit sera is reported. “Protein Alone” and “DNA + Protein” indicate rabbits receiving polyvalent protein-alone (33-1 and 33-2) or DNA prime-protein boost (29-1, 29-2, 30-1, and 30-2) immunization, respectively. (A) Effect of V3 adsorption on neutralization of HIV-1 clade B virus SS1196. (B) Effect of V3 adsorption on neutralization of HIV-1 clade B virus SC422661.8. Error bars indicate standard deviations from replicate experiments.
DNA-primed rabbit sera have higher gp120 trimer binding abilities.
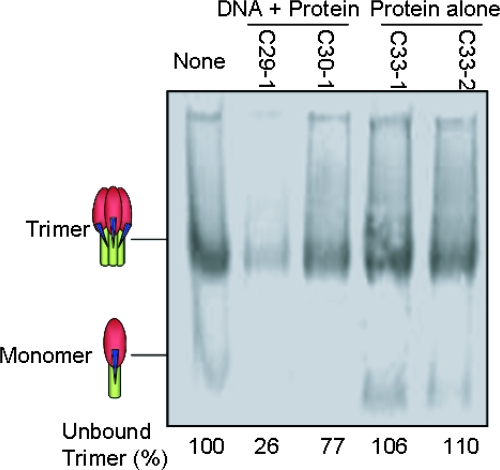
Finally, we measured serum binding to native Env trimers thought to be crucial for neutralization (24). Trimer binding was measured by the depletion of unliganded trimers in a BN-PAGE assay. Only sera from rabbits that received the DNA prime-protein boost immunizations, particularly C29-1, effectively depleted unliganded trimers (Fig. 10). These data support neutralization results showing that the DNA prime-protein boost immunization strategy elicits more effective NAb responses.
FIG. 10.
Binding of rabbit immune sera to HIV-1 JR-FL Env trimers by BN-PAGE analysis. Sera from either DNA prime-protein boost immunization (29-1 and 30-1) or protein-only immunization (33-1 and 33-2) were incubated with solubilized trimers prior to being run on a BN-PAGE gel. Numbers below each lane indicate the percentage of unliganded trimer present in the gel based upon densitometry analysis.
DISCUSSION
Most of our understanding on the specificities of NAbs against HIV-1 has been learned through the study of a few broad NAbs (4) and, more recently, through the mapping of neutralizing epitope specificities in HIV-infected people (7, 8, 11, 13, 18). One key neutralizing epitope that has been identified is the CD4bs of gp120. Antibodies specific for this epitope have been shown to be responsible for the activities of broadly neutralizing human sera (18, 32). One MAb, b12, specific for the CD4bs, is known to be capable of neutralizing primary patient isolates. Therefore, the CD4bs is a vulnerable site that could serve as a target for NAbs in HIV vaccine design.
Because the CD4bs is a conformational site formed by residues that are far apart from each other based on the gp120 primary sequence, it is exciting to observe increased levels of CD4bs antibodies in immune sera from the DNA prime-protein boost gp120-vaccinated rabbits in the current study. This was seen in both the monovalent and polyvalent gp120 formulations, indicating a consistent advantage of the DNA priming approach. This finding implies that a DNA priming step may be effective in preserving conformationally sensitive epitopes and/or that this step is important for priming a subset of B cells specific for CD4bs epitopes, which are responsive to a subsequent gp120 protein boost. Due to the relatively low neutralizing titers in rabbit immune sera, we could not definitively prove that CD4bs antibodies are in fact responsible for the neutralization of primary isolates by using the gp120 protein absorption method reported in a recent study (18). However, using peptide absorption experiments, we ruled out a possible contribution by V3-specific antibodies, leaving anti-CD4bs antibodies, among other conformationally sensitive epitopes, as a likely explanation for the neutralizing activity.
Our study identified one particular highly reactive region, involving multiple adjacent peptides, at the junction of the V5 and C5 regions. This region of gp120 includes both the CD4 and b12 binding epitopes as determined by sequence alignment against previously reported HIV-1 Env structures (15). This reactivity to the V5-C5 region is highly conserved, as it can be observed in different peptides derived from four gp120 proteins of different clades included in the polyvalent gp120 formulation. This was seen despite the fact that the exact amino acid sequences of these peptides were not identical. This result highlights the uniqueness of this site and excludes the possibility that one dominant peptide is responsible for a high level of recognition against the M-group consensus peptides. Future studies with additional primary Env antigens should prove whether this region is important for other primary HIV-1 isolates.
We also observed that the DNA prime-protein boost regimen induced higher anti-V3 antibodies in rabbit immune sera. High-level V3-specific antibodies were responsible for the neutralizing activities against the sensitive isolate SF162 but generally not for other primary isolates. It is also interesting that polyvalent rabbit immune sera showed different binding preferences for subdomains of the V3 loops from gp120 proteins of different clades of HIV-1. Our results suggest that the V3 loops of different clades are oriented differently and therefore elicit different specificities. Most of the previously reported data on V3 antibodies are based on clade B viruses; however, recently, several studies indicated that different V3 sequences bind differently to various V3-specific MAbs (2, 33-35). The DNA prime-protein boost vaccination approach, which induces higher levels of anti-V3 antibodies, provides a practical method to understand the fine specificity of V3 antibodies that recognize Env antigens from different clades.
A trimer gel shift assay demonstrated the presence of antibodies that are capable of recognizing the trimeric form of HIV-1 Env in our DNA-primed sera. Sera from DNA-primed animals bound native trimers more efficiently than did sera collected from animals that were immunized with protein only. Since the binding of native trimers predicts neutralization, trimer recognition by DNA-primed sera correlates with increased neutralization activity. Collectively, with improved CD4bs and V3 antibodies, the trimer gel shift results further confirm that a DNA priming step may increase the levels of neutralization for a vaccine formulation by focusing the serum antibody responses to a more favorable specificity profile.
Previous studies demonstrated that antibody induced by one subtype E antigen, rgp120CM235, preferentially binds natively folded gp120 and retains strong cross-reactivity against multiple gp120 strains within subtype E as well as against strains from subtype B (38). In contrast, in that same study, antibody responses to another gp120 from isolate SF2 were shown to be directed predominantly toward linear epitopes that are poorly exposed on native gp120, and it was also shown that these antibodies have a more limited ability to cross-recognize divergent gp120. Similarly, an antibody response preferentially reactive with natively folded gp120/gp160 was dependent on immunization conditions that preserved the structure of the HIV-1 envelope immunogen (37). Several primary HIV-1 isolates were neutralized in that study, which used Env antigens purified directly from cultured HIV-1 viruses, but recombinant protein-based Env antigens used in another study could induce antibodies that could neutralize only T-cell-line-adapted HIV-1 but not primary isolates (38). Analysis of the MAbs elicited by trimeric protein-based HIV-1 gp140 immunogens with modified Env sequences did not reveal neutralizing activity against heterologous primary viruses, suggesting that a particular immunogen was unable to elicit antibodies that can bind to native forms of Env (9). Previously, the same virus capture competition used in the current study was performed to analyze monkey sera from either modified SF162 gp140 immunizations or SHIVSF162P4 infection. Minimal CD4bs- or V3-directed antibodies were seen using the modified gp140 immunogens. However, very high levels of CD4bs- and V3-directed antibodies were observed in the simian/human immunodeficiency virus-infected animals (10), a pattern that is very similar to what we saw in the current study using the natural gp120 immunogens in a DNA prime-protein boost approach. Therefore, an immunization approach that can preserve natural Env epitopes is critical to elicit NAbs.
In order to compensate for the weak immunogenicity of primary Env proteins, strong adjuvants are frequently required in order for recombinant Env proteins to first elicit high-titer antibody responses in experimental animals before any neutralizing activities can be studied (19, 37). In our rabbit studies, a conventional incomplete Freund's adjuvant was used at the protein boost step with both the monovalent and polyvalent Env formulations. This implies another advantage of the DNA prime-protein boost approach in that DNA immunization can effectively prime the antigen-specific B cells, which can in turn respond with a high-level antibody response upon administration of the protein boost without the need for a particularly powerful adjuvant. Furthermore, the variation of antibody responses within a study group is minimal with DNA priming compared to that with immunization with protein alone (1).
Although these results originated from a small-animal study, it is again interesting that similar patterns of antibody recognition were seen in two entirely independent studies. Given this observation, additional work with increased sample sizes and in higher-primate models would be useful in confirming that the observed pattern remains consistent. In summary, we analyzed the antibody profiles induced by a DNA prime-protein boost vaccination approach and discovered that antibody specificities against key neutralizing epitopes are improved with this approach. Given the increasing evidence that CD4bs-specific antibodies are responsible for broad neutralization, as discovered in HIV-infected patients with potent and broad NAb responses, the DNA prime-protein boost vaccine approach offers an attractive platform for the development of next-generation HIV vaccines that focus on broad NAbs.
Supplementary Material
Acknowledgments
This work was supported in part by grants R01 AI06250 (S.L.) and R01 AI58763 (J.B.) from the National Institutes of Allergy and Infectious Diseases, NIH; Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery Vaccine Immune Monitoring Consortium grant 38619 (J.B.); and the AIDS and Infectious Disease Science Center at the Torrey Pines Institute for Molecular Studies (J.B.).
We thank Phillip Markham and Ranajit Pal (Advanced BioScience Laboratories, Inc.) for providing some of the gp120 proteins used in the current study and Jill M. Grimes-Serrano for critical reading of the manuscript.
Footnotes
Published ahead of print on 21 May 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Beddows, S., N. Schulke, M. Kirschner, K. Barnes, M. Franti, E. Michael, T. Ketas, R. W. Sanders, P. J. Maddon, W. C. Olson, and J. P. Moore. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 798812-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, C. H., R. Pantophlet, A. Schiefner, L. A. Cavacini, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2008. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J. Mol. Biol. 375969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 10214943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conley, A. J., J. A. Kessler II, L. J. Boots, P. M. McKenna, W. A. Schleif, E. A. Emini, G. E. Mark III, H. Katinger, E. K. Cobb, S. M. Lunceford, S. R. Rouse, and K. K. Murthy. 1996. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 706751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crooks, E. T., P. L. Moore, M. Franti, C. S. Cayanan, P. Zhu, P. Jiang, R. P. de Vries, C. Wiley, I. Zharkikh, N. Schulke, K. H. Roux, D. C. Montefiori, D. R. Burton, and J. M. Binley. 2007. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 366245-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crooks, E. T., P. L. Moore, D. Richman, J. Robinson, J. A. Crooks, M. Franti, N. Schulke, and J. M. Binley. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14101-113. [PMC free article] [PubMed] [Google Scholar]
- 8.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derby, N. R., S. Gray, E. Wayner, D. Campogan, G. Vlahogiannis, Z. Kraft, S. W. Barnett, I. K. Srivastava, and L. Stamatatos. 2007. Isolation and characterization of monoclonal antibodies elicited by trimeric HIV-1 Env gp140 protein immunogens. Virology 366433-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derby, N. R., Z. Kraft, E. Kan, E. T. Crooks, S. W. Barnett, I. K. Srivastava, J. M. Binley, and L. Stamatatos. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 808745-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, A. K., H. Donners, R. Pantophlet, W. E. Johnson, J. M. Decker, G. M. Shaw, F. H. Lee, D. D. Richman, R. W. Doms, G. Vanham, and D. R. Burton. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 816548-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emini, E. A., W. A. Schleif, J. H. Nunberg, A. J. Conley, Y. Eda, S. Tokiyoshi, S. D. Putney, S. Matsushita, K. E. Cobb, C. M. Jett, et al. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355728-730. [DOI] [PubMed] [Google Scholar]
- 13.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, S. Jiang, P. L. Li, T. W. Baba, D. C. Montefiori, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, and R. M. Ruprecht. 2002. Postnatal pre- and postexposure passive immunization strategies: protection of neonatal macaques against oral simian-human immunodeficiency virus challenge. J. Med. Primatol. 31109-119. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 3101025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law, M., R. M. Cardoso, I. A. Wilson, and D. R. Burton. 2007. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J. Virol. 814272-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 131032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 801414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 734009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6207-210. [DOI] [PubMed] [Google Scholar]
- 22.Montefiori, D. C. 2004. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays, p. 1-15. In J. E. Coligan, A. M. Kruisbeek, D. H. Margullies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology, vol. 12. John Wiley, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- 23.Montefiori, D. C., G. Pantaleo, L. M. Fink, J. T. Zhou, J. Y. Zhou, M. Bilska, G. D. Miralles, and A. S. Fauci. 1996. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J. Infect. Dis. 17360-67. [DOI] [PubMed] [Google Scholar]
- 24.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 802515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 676642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 729384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal, R., Q. Yu, S. Wang, V. S. Kalyanaraman, B. C. Nair, L. Hudacik, S. Whitney, T. Keen, C. L. Hung, L. Hocker, J. S. Kennedy, P. Markham, and S. Lu. 2006. Definitive toxicology and biodistribution study of a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 (HIV-1) vaccine in rabbits. Vaccine 241225-1234. [DOI] [PubMed] [Google Scholar]
- 28.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24739-769. [DOI] [PubMed] [Google Scholar]
- 29.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 1004144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selvarajah, S., B. Puffer, R. Pantophlet, M. Law, R. W. Doms, and D. R. Burton. 2005. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 7912148-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12193-204. [DOI] [PubMed] [Google Scholar]
- 34.Stanfield, R. L., M. K. Gorny, S. Zolla-Pazner, and I. A. Wilson. 2006. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J. Virol. 806093-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanfield, R. L., and I. A. Wilson. 2005. Structural studies of human HIV-1 V3 antibodies. Hum. Antibodies 1473-80. [PubMed] [Google Scholar]
- 36.Steinbrook, R. 2007. One step forward, two steps back—will there ever be an AIDS vaccine? N. Engl. J. Med. 3572653-2655. [DOI] [PubMed] [Google Scholar]
- 37.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 714319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.VanCott, T. C., J. R. Mascola, L. D. Loomis-Price, F. Sinangil, N. Zitomersky, J. McNeil, M. L. Robb, D. L. Birx, and S. Barnett. 1999. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J. Virol. 734640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, S., J. Arthos, J. M. Lawrence, D. Van Ryk, I. Mboudjeka, S. Shen, T. H. Chou, D. C. Montefiori, and S. Lu. 2005. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J. Virol. 797933-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S., T. H. Chou, P. V. Sakhatskyy, S. Huang, J. M. Lawrence, H. Cao, X. Huang, and S. Lu. 2005. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 791906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, S., D. Heilman, F. Liu, T. Giehl, S. Joshi, X. Huang, T. H. Chou, J. Goguen, and S. Lu. 2004. A DNA vaccine producing LcrV antigen in oligomers is effective in protecting mice from lethal mucosal challenge of plague. Vaccine 223348-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, S., S. Joshi, and S. Lu. 2004. Delivery of DNA to skin by particle bombardment. Methods Mol. Biol. 245185-196. [DOI] [PubMed] [Google Scholar]
- 43.Wang, S., J. Kennedy, K. West, D. C. Montefiori, A. Bansal, P. Goepfert, S. Coley, J. Lawrence, S. Shen, S. Green, A. Rothman, F. Ennis, R. Pal, P. Markham, and S. Lu. 2008. Balanced cellular and antibody responses induced by the polyvalent DNA prime-protein boost HIV-1 vaccine formulation DP6-001 in healthy human volunteers. Vaccine 261098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S., R. Pal, J. R. Mascola, T. H. Chou, I. Mboudjeka, S. Shen, Q. Liu, S. Whitney, T. Keen, B. C. Nair, V. S. Kalyanaraman, P. Markham, and S. Lu. 2006. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 35034-47. [DOI] [PubMed] [Google Scholar]
- 45.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolla-Pazner, S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolla-Pazner, S., S. S. Cohen, C. Krachmarov, S. Wang, A. Pinter, and S. Lu. 2008. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology 372233-246. [DOI] [PubMed] [Google Scholar]
- 49.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 7510892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.