Abstract
Baculovirus proteins P49 and P35 are potent suppressors of apoptosis in diverse organisms. Although related, P49 and P35 inhibit initiator and effector caspases, respectively, during infection of permissive insect cells. The molecular basis of this novel caspase specificity is unknown. To advance strategies for selective inhibition of the cell death caspases, we investigated biochemical differences between these baculovirus substrate inhibitors. We report here that P49 and P35 use similar mechanisms for stoichiometric inhibition that require caspase cleavage of their reactive site loops (RSL) and chemical contributions of a conserved N-terminal cysteine to stabilize the resulting inhibitory complex. Our data indicated that P49 functions as a homodimer that simultaneously binds two caspases. In contrast, P35 is a monomeric, monovalent inhibitor. P49 and P35 also differ in their RSL caspase recognition sequences. We tested the role of the P4-P1 recognition motif for caspase specificity by monitoring virus-induced proteolytic processing of Sf-caspase-1, the principal effector caspase of the host insect Spodoptera frugiperda. When P49's TVTD recognition motif was replaced with P35's DQMD motif, P49 was impaired for inhibition of the initiator caspase that cleaves and activates pro-Sf-caspase-1 and instead formed a stable inhibitory complex with active Sf-caspase-1. In contrast, the effector caspase specificity of P35 was unaltered when P35's DQMD motif was replaced with TVTD. We concluded that the TVTD recognition motif is required but not sufficient for initiator caspase inhibition by P49. Our findings demonstrate a critical role for the P4-P1 recognition site in caspase specificity by P49 and P35 and indicate that additional determinants are involved in target selection.
The baculoviruses are large double-stranded DNA viruses of insects that trigger widespread apoptosis. To block host cell apoptosis and thereby enhance multiplication, these viruses encode diverse suppressors of apoptosis (reviewed in references 5, 6, and 14). P49 and P35 are two baculovirus-encoded apoptotic suppressors that function by inhibiting a broad range of the cell death proteases known as caspases (2, 3, 19, 30, 40, 45). Although related to one another, P49 and P35 display different caspase specificities in the infected insect cell (21, 22, 45). Because selective inhibition of caspases may be advantageous in therapeutics for apoptosis-associated diseases, the molecular basis of target specificity by P49 and P35 is of considerable interest.
The caspases are a family of cysteinyl aspartate-specific proteases that are critical effectors of apoptosis in metazoans (reviewed in references 15, 23, 32, and 33). Caspase-mediated proteolysis promotes cellular disassembly that includes chromatin condensation, nuclear DNA cleavage, membrane blebbing, and cell fragmentation. Thus, these death proteases are subject to regulation by diverse cellular and viral mechanisms (4, 6, 15, 33, 34, 36). Upon apoptotic signaling, initiator caspases are autoactivated through interactions of their N-terminal prodomain with specific adaptor proteins (1, 31, 35, 42). Subsequently, the effector caspases are activated by initiator caspase-mediated cleavage of their inactive zymogen (procaspase) form. Procaspase cleavage occurs between the large and small subunit domains, allowing for the assembly of the active protease, consisting of a dimer of heterodimers with two active sites (15, 23, 36). The substrate specificity of initiator caspases differs from that of effector caspases, due in part to their unique functions during execution of apoptosis (37). Not surprisingly, because of their central role in cell death, both types of caspases are important therapeutic targets for the treatment of apoptosis-associated diseases (11, 23). The novel selectivity of P49 and P35 for different caspases in insects provides a unique opportunity to define the in vivo determinants of caspase specificity in the experimentally advantageous system provided by the baculovirus-infected cell.
Studying baculovirus caspase inhibitors has led to important insights into the role and regulation of caspases in apoptosis (4, 5, 7, 14). In general, a single baculovirus species encodes only one functional inhibitor, either a substrate inhibitor or an inhibitor of apoptosis protein (IAP). P35 from the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the founding member of the family of substrate inhibitors that includes baculovirus P49 and entomopoxvirus P33 (3, 9, 27). Sequence comparisons have also revealed P35 homologs in Spodoptera littoralis NPV (SlNPV), Spodoptera litura NPV, Leucania separata NPV, and Bombyx mori NPV (reviewed in reference 5). The potency of P35 as a caspase inhibitor is attributed to its novel solvent-exposed reactive-site loop (RSL), which is readily recognized and cleaved at Asp87 by the target caspase (2, 3, 24, 30, 40, 43). Cleavage of the P4-P1 recognition motif DQMD87↓G (see Fig. 1A), located at the apex of the RSL, triggers a conformational change that positions the P35 N terminus in the caspase active site to prevent peptide hydrolysis and thereby form a stable complex (8, 10, 12, 24, 40). The inhibited complex consists of the caspase homodimer with each of the two active sites occupied by a separate monomer of P35.
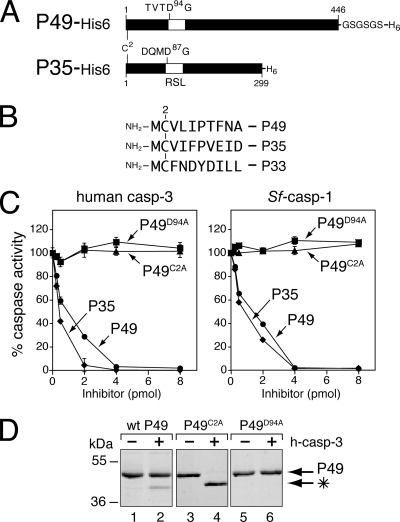
FIG. 1.
Requirement of Cys2 for caspase inhibition by P49. (A) Comparison of P49 and P35. P49 (446 residues) and P35 (299 residues) share significant amino acid sequence identity. Cleavage of P49 and P35 occurs at Asp94 and Asp87, respectively, within the caspase recognition sequences TVTD94↓G and DQMD87↓G, which are located at the apex of an RSL (open box). A Gly-Ser linker (GSGSGS) was inserted between the C terminus and the His6 tag of recombinant P49. (B) N termini of P35 family caspase inhibitors. Residue 2 is a cysteine for SlNPV P49, AcMNPV P35, and Amsacta moorei entomopoxvirus P33. (C) Caspase inhibition assays. Increasing amounts of purified His6-tagged wild-type P49 (circles), P49C2A (triangles), P49D94A (squares), or wild-type P35 (diamonds) were mixed with human caspase-3-His6 or Sf-caspase-1-His6 (1 pmol). After 30 min, residual caspase activity was measured by using DEVD-AMC as the substrate. Plotted values are the averages ± standard deviations of triplicate assays and are expressed as percentages of uninhibited caspase activity for a representative experiment. (D) P49 cleavage. Excess purified His6-tagged wild-type (wt) P49, P49C2A, or P49D94A (40 pmol) was incubated with (+) or without (−) human caspase-3 (h-casp-3)-His6 (5 pmol) for 30 min at 37°C, subjected to SDS-PAGE, and stained with Coomassie blue. The P49 cleavage fragment (*) is indicated. Molecular mass standards are indicated on the left.
P49 is a stoichiometric substrate inhibitor with P35-like properties. First discovered in SlNPV (9), P49 has the capacity to inhibit both effector and initiator caspases (19, 29, 45). Cleavage of P49 at an aspartate residue (Asp94) within the caspase recognition motif TVTD94↓G (see Fig. 1A) is necessary for formation of an inhibitory complex with the target caspase (19, 29, 45). Although the structure of P49 is unknown, sequence alignments suggest that it resembles that of P35, including the presence of a prominent RSL that presents Asp94 for cleavage (29, 45). Thus, we predicted that P49 functions by using a P35-like mechanism for caspase inhibition.
P49 prevents proteolytic processing of effector caspases Sf-caspase-1 and Sf-caspase-2 during baculovirus infection of the moth Spodoptera frugiperda (order Lepidoptera) (45). Caspase cleavage of P49 at TVTD94↓G is required for P49-mediated suppression of virus-induced apoptosis. Thus, P49 is a substrate inhibitor of the initiator caspase designated Sf-caspase-X, which is responsible for the proteolysis and activation of Sf-caspase-1 and -2 (45). In a cellular context, Sf-caspase-X is also inhibited by baculovirus Op-IAP, but not P35, which fails to inactivate other initiator caspases, including those from invertebrates (16, 21, 28, 39, 45). Because P49's TVTD recognition motif resembles the caspase processing sites TETD↓G and AETD↓G of pro-Sf-caspase-1 and -2, respectively, we hypothesized that P49's in vivo specificity is determined by its caspase recognition motif. To test this possibility, we altered the recognition motifs of P49 and P35, delivered the modified caspase inhibitors to Spodoptera cells by using recombinant baculoviruses, and monitored proteolytic processing of Sf-caspase-1 and -2 during infection.
We report here that, when P49's TVTD motif was swapped with P35's DQMD motif, P49 was impaired as an initiator caspase inhibitor and instead functioned downstream as an effector caspase inhibitor. In contrast, when P35's DQMD motif was swapped with TVTD, P35's selectivity for effector caspases was unaltered. Thus, P49 requires a TVTD motif for Spodoptera initiator caspase inhibition, but this motif alone is insufficient to confer the same activity upon P35. To search for additional determinants of caspase selectivity, we compared the biochemical properties of P49 and P35. By using recombinant P49, produced and purified from Escherichia coli for the first time, we determined that P49 and P35 use comparable mechanisms for caspase inhibition. In contrast to P35, which acts as a monomer, P49 functions as a homodimer with the capacity to form a stable complex with two individual caspase dimers. Thus, P49 is the first example of a divalent caspase inhibitor in which caspase targeting can be altered by sequence alterations in the caspase recognition motif.
MATERIALS AND METHODS
Cells and virus.
Spodoptera frugiperda IPLB-SF21 (38) cells were propagated at 27°C in TC100 growth medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (HyClone). For infections, SF21 monolayers (106 cells per plate) were overlaid with supplemented TC100 containing virus at a multiplicity of infection (MOI) of 10 PFU per cell and rocked gently at room temperature for 1 h. AcMNPV recombinants vP49 (pIE1prmp49Δ35K/lacZ; p49+, p35−, iap−), vP35 (pIE1prmp35Δ35K/lacZ; p35+, iap−), and vΔP35 (vΔ35K/lacZ; p49−, p35−, iap−) have been described previously (18, 26, 45). Recombinant vP49DQMD (pIE1prmp49DQMDΔ35K/lacZ; p49DQMD+, p35−, iap−) and vP35TVTD (pIE1prmp35TVTDΔ35K/lacZ; p35TVTD+, p35−, iap−) were created by allelic replacement in which the polyhedrin gene of the p35− parent vΔ35K (18) was replaced with genes encoding P4-P1 DQMD94-mutated P49 (P49DQMD) or P4-P1 TVTD87-mutated P35 (P35TVTD) fused to the AcMNPV ie-1 promoter (IE1prm) and linked to a lacZ reporter gene. After plaque purification, all recombinant viruses were verified by PCR and sequence analysis.
Plasmids.
pIE1prm/hr5/PA-based vectors in which the highly active AcMNPV ie-1 promoter directs expression of AcMNPV wild-type P35, TVTD-mutated P35, SlNPV wild-type P49, DQMD-mutated P49, or D94A-mutated P49 have been described previously (45). C2A-mutated P49 was generated from parent plasmid pIE1prm/hr5/p49/PA by overlap extension PCR. P49 containing a Gly-Ser-Gly-Ser-Gly-Ser linker followed by a C-terminal His6 tag [pIE1prm/hr5/p49-(GS)3-His6/PA] was generated by insertion of an oligonucleotide carrying the linker at the NotI site of plasmid pIE1prm/hr5/p49-His6/PA (45). E. coli protein expression plasmid pET22B-p49-GS3-His6 was generated by placement of the XhoI fragment of pIE1prm/hr5/p49-GS3-His6/PA into pET22B-p49-His6 (45). Protein expression plasmids for P35-His6, human caspase-3-His6, and Sf-caspase-1-His6 have been described previously (2, 21, 45). All plasmids and mutations thereof were verified by nucleotide sequencing.
Protein production.
Recombinant wild-type and mutated P49-(GS)3-His6 (designated P49-His6), wild-type and mutated P35-His6, human caspase-3-His6, and Sf-caspase-1-His6 were isolated from E. coli strain BL21(DE3) by Ni2+ affinity chromatography as described previously (2, 21, 45). In brief, cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 or 4 h at 30°C (Sf-caspase-1-His6 or human caspase-3-His6, respectively) or 0.1 mM IPTG overnight at 20°C (wild-type and mutated P49-His6 and P35-His6) and lysed by sonication. Ni2+-nitrilotriacetic acid beads (Qiagen) were mixed with clarified lysate for 2 h at 4°C, washed with 45 mM imidazole in 20 mM Tris, pH 7.9, 500 mM NaCl, and 1 mM dithiothreitol (DTT), and mixed with 500 mM imidazole in 20 mM Tris, pH 7.9, 500 mM NaCl, and 1 mM DTT to elute bound proteins. Protein preparations were judged >95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Protein concentrations were measured by using Bio-Rad protein assay reagent.
Caspase assays.
For in vitro assays, purified recombinant His6-tagged caspases (1 pmol) were incubated at 37°C for 15 min and then mixed with increasing amounts of purified His6-tagged recombinant inhibitor (0.25 to 8 pmol) in caspase activity buffer (25 mM HEPES, pH 7.5, 5 mM EDTA, 10 mM DTT, 10% glucose, and 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}) containing 0.1 mg/ml bovine serum albumin. After 30 min, residual caspase activity was measured using the substrate N-acetyl-DEVD-7-amino-methylcoumarin (Ac-DEVD-7-AMC) (Sigma) as described previously (2). Assays were performed in triplicate, and results are reported as percentages of uninhibited caspase activity. To measure intracellular caspase activity, mock- or virus-infected SF21 cells were harvested from triplicate plates and lysed on ice using CHAPS lysis buffer (10 mM HEPES, pH 7.0, 2 mM EDTA, 0.1% CHAPS, and 5 mM DTT). Cell lysates were clarified by centrifugation, and caspase activity was measured by using substrate Ac-DEVD-7-AMC. The average values determined from triplicate infections are reported as percentages of the caspase activity determined in lysates of vΔP35-infected cells.
DNA transfections and marker rescue assays.
For DNA transfections, SF21 monolayers (106 cells per 60-mm-diameter plate) were overlaid with TC100 medium containing CsCl-purified plasmid (6 μg) that was mixed with cationic liposomes consisting of N-[1-(2,3 dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate-l-α-phosphatidylethanolamine, dioleoyl (C18:1, [cis]-9) (DOTAP-DOPE) (10 μl) as described previously (21). Marker rescue assays were performed as previously described (2). In brief, SF21 cells were transfected with a pIE1prm/hr5/PA-based vector carrying the test gene and infected 24 h later with vΔP35. Extracellular budded virus collected 48 h after infection was quantified by measuring the 50% tissue culture infective dose using SF21 cells and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to determine rescued (nonapoptotic)-virus yields.
Caspase pull-down assays.
Untagged wild-type or mutated P49 or P35 and human caspase-3 were expressed in E. coli strain BL21(DE3) as described above. The bacterial cells were suspended in 20 mM Tris, pH 7.9, 150 mM NaCl, and 1 mM DTT, lysed by sonication, and clarified by centrifugation. Purified recombinant Sf-caspase-1-His6, inhibitor-containing E. coli lysate, and caspase-containing E. coli lysate were mixed and incubated at 27°C for 1 h. After binding to Ni2+-nitrilotriacetic acid beads, protein complexes were washed, eluted as described above, and subjected to immunoblot analysis.
Antisera and immunoblots.
At various times after infection, intact cells and accompanying apoptotic bodies were collected by centrifugation, lysed with 1% SDS-1% β-mercaptoethanol, and subjected to SDS-PAGE. Proteins were transferred to Immobilon-P polyvinyl difluoride membranes (Millipore), which were then incubated with the following antisera diluted as indicated in parentheses: anti-P49 (1:1,000) (45), monoclonal anti-P35 (1:5,000) (gift from Yuri Lazebnik), anti-human caspase-3 (1:1,000) (12), anti-Sf-caspase-1 (1:1,000) (21), anti-Sf-caspase-2 (1:1,000) (20), or antiactin (1:5,000) (BD Transduction Laboratories). The membranes were subsequently treated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G or goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories). The CDP-Star chemiluminescence detection system (Roche) was used for signal detection. Images were scanned at 300 dpi using an Epson Twain Pro scanner and prepared using Adobe Photoshop and Illustrator CS2.
Size exclusion chromatography.
A Superdex-200 10/300 GL column (GE Health Sciences) equilibrated with gel filtration buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT) was used for all experiments. Protein samples were chromatographed at a flow rate of 0.4 ml per min and collected using 400-μl fractions. Size calibration was conducted by using gel filtration standards (Bio-Rad). Cellular extracts were prepared for size exclusion analysis by suspending transfected or infected SF21 cells in gel filtration buffer and subjecting them to three freeze-thaw cycles. The lysates (2 × 106 cell equivalents per column) were clarified by centrifugation (25,000 × g) for 30 min at 4°C, passed through a 0.2-μm polytetrafluoroethylene filter (Millex), and applied directly to the column.
RESULTS
P49 requires residue Cys2 for anticaspase activity.
We postulated that the observed differences in caspase specificity between P49 and P35 are due to variations in each protein's structure or inhibitory mechanism. To rule out the possibility that the mechanism by which P49 mediates caspase inhibition differs from that of P35, we first tested the role of the N-terminal residue cysteine 2 (Cys2) for P49 anticaspase activity. Conserved within P49, P35, and entomopoxvirus caspase inhibitor P33 (Fig. 1B), Cys2 is absolutely required for caspase inhibition by P35 (24, 40, 41). Because of the equilibrium involving the thioester bond between P35's Asp87 and the active-site cysteine and a bond between P35's Cys2 and Asp87, the N terminus of P35 is retained in the caspase active site, thereby inhibiting proteolytic activity. To assess the role of Cys2, we determined the anticaspase activity of C2A-mutated P49. To this end, we produced and purified recombinant His6-tagged P49 from E. coli. Previous efforts to generate functional P49 in E. coli were hindered by problems of insolubility (19, 45). However, by inserting a Gly-Ser-Gly-Ser-Gly-Ser linker between the C terminus of P49 and the His6 tag (Fig. 1A), solubility was increased significantly. The linker-modified P49 and mutations thereof were readily obtained with >95% purity. Moreover, in direct tests of caspase-inhibitory activities (see below), the modified P49 (P49-His6) was as effective or better than that obtained by using baculovirus vectors (45). Thus, for the first time, workable yields of active P49-His6 are available using E. coli.
We tested the activity of C2A-mutated P49-His6 by using dose-dependent caspase inhibition assays. Purified, active His6-tagged effector caspases were mixed with increasing amounts of wild-type, C2A-mutated, or cleavage-resistant D94A-mutated P49-His6. Residual caspase activity was measured by using DEVD-AMC as a substrate. Wild-type P49-His6 inhibited human caspase-3 and Sf-caspase-1 (Fig. 1C) at levels comparable to those for inhibition by P35-His6, which is a stoichiometric inhibitor. Thus, when purified with its C-terminal Gly-Ser-containing linker, P49-His6 was a more effective inhibitor than it was when it lacked the linker (45). In contrast, neither caspase was inhibited by C2A- or D94A-mutated P49-His6 at any concentration. Nonetheless, when mixed in molar excess with human caspase-3, C2A-mutated P49-His6 was cleaved to completion (Fig. 1D, lane 4). The resulting fragments were comparable in size to those generated by cleavage of wild-type P49-His6 (Fig. 1D, lane 2), suggesting that cleavage occurred at Asp94. As expected, wild-type P49-His6 cleavage was incomplete due to caspase inhibition (lane 2). D94A-mutated P49-His6, in which the requisite residue Asp94 was replaced with Ala, was not cleaved (lane 6). It is unlikely that the C2A substitution caused significant misfolding since the same mutation in P35 does not (40). Because C2A-mutated P49-His6 was cleaved normally, we concluded that Cys2 is required for postcleavage caspase inhibition by P49. This loss-of-function phenotype was verified by testing the ability of C2A- or C2S-mutated P49 to block baculovirus-induced apoptosis by using marker rescue assays (data not shown). Consistent with loss of caspase inhibition, C2A- and C2S-mutated P49 failed to inhibit apoptosis. The Cys2 requirement suggested that P49 uses a P35-like mechanism for caspase inhibition that involves Cys2-mediated interactions within the active site.
P49 forms a dimer.
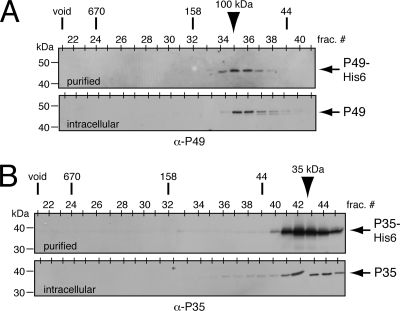
In an effort to define biochemical differences between P49 and P35 and the manner in which they interact with their target caspases, we first assessed the oligomeric state of each inhibitor. P35 is monomeric when purified from E. coli (12, 30), although it has the capacity to form oligomers (44). Moreover, a monomer of P35 interacts with each active site in the P35/caspase complex, as indicated by crystal structure (10, 40). Unexpectedly, when subjected to size exclusion column chromatography, P49-His6 eluted as a complex with a molecular mass of ∼100 kDa (Fig. 2A). Because our E. coli-generated P49-His6 was judged >95% pure, the single complex consisted solely of P49. Thus, recombinant P49-His6 behaves as a dimer. To determine if intracellular P49 exhibits a similar size, we subjected freeze-thaw extracts of Spodoptera frugiperda SF21 cells transfected with a plasmid encoding native P49 to size exclusion chromatography. Immunoblot analysis indicated that P49 eluted as an ∼100-kDa complex (Fig. 2A), suggesting that the principal form of P49 in the nonapoptotic cell is also a dimer. In comparison, purified E. coli-generated P35-His6 eluted with a predicted size of 30 to 40 kDa (Fig. 2B), confirming that the principal form of recombinant P35 is monomeric. When produced in transfected SF21 cells, P35 behaved similarly (Fig. 2B). Thus, whereas P35 is a monomer, P49 prefers to dimerize.
FIG. 2.
Size exclusion chromatography of P49 and P35. (A) P49 profile. Purified E. coli-produced P49-His6 (top) or freeze-thaw extracts (bottom) of SF21 cells transfected with an ie-1 promoter expression plasmid encoding P49 were subjected to size exclusion chromatography and immunoblot analysis with anti (α)-P49. Chromatographic size standards are indicated across the top with fraction numbers. Electrophoretic molecular mass markers are indicated on the left. (B) P35 profile. Purified E. coli-produced P35-His6 (top) or freeze-thaw extracts (bottom) of SF21 cells transfected with a plasmid encoding P35 were analyzed with anti-P35 as described for panel A. Arrowheads denote the peak fractions of P49 and P35.
P49 is a divalent caspase inhibitor.
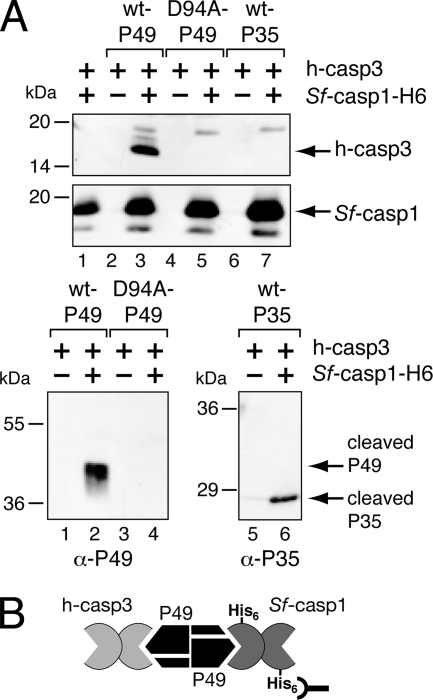
If P49 retains its dimeric structure upon caspase interaction, we predicted that P49 has the capacity to simultaneously interact with more than one caspase. To investigate this possibility and thereby confirm the oligomeric status of P49, we tested the multiplicity of caspase binding by P49. To this end, a bacterial extract containing untagged P49 was mixed with an extract containing untagged human caspase-3 that was processed and active. This reaction mixture was mixed immediately with an excess amount of purified Sf-caspase-1-His6 that was also processed and active. The resulting complexes were subjected to a Ni2+ pull-down. Immunoblot analysis was used to determine the composition of the Sf-caspase-1-His6-containing complexes. When wild-type P49 was present in the reaction mixture, human caspase-3 was detected in the complex (Fig. 3A, top, lane 3). However, in the absence of P49 (lane 1) or in the presence of cleavage-resistant D94A-mutated P49 (lane 5), no caspase-3 was detected. Similarly, human caspase-3 was not pulled down in the absence of Sf-caspase-1 (lanes 2, 4, and 6), verifying that Sf-caspase-1-His6 was required. As confirmation of the requirement of functional P49 for complex formation, only cleaved wild-type P49 was detected by pull-down (Fig. 3A, bottom, lane 2), not cleavage-resistant D94A-mutated P49 (lane 4). These data are most consistent with the formation of a complex of Sf-caspase-1 and human caspase-3 simultaneously bound to a P49 dimer (Fig. 3B). In contrast, when wild-type P35 was mixed with the two caspases and subjected to Ni2+ pull-down, there was no evidence of a tripartite complex. Although cleaved P35 was detected in the Sf-caspase-1-His6-containing complex (Fig. 3A, bottom, lane 6), human caspase-3 was not (Fig. 3A, top, lane 7). Thus, the monovalent nature of P35 was confirmed. We concluded that, unlike P35, P49 is divalent and has the capacity to interact simultaneously with two individual caspases (Fig. 3B).
FIG. 3.
Divalent caspase interactions with P49. (A) Formation of caspase complexes. E. coli extracts containing active human caspase-3 (h-casp3) (+) were mixed with extracts of untagged wild-type (wt) P49 (lanes 2 and 3), P49D94A (lanes 4 and 5), or wild-type P35 (lanes 6 and 7) and incubated with (+) or without (−) purified active Sf-caspase-1-His6 for 1 h at 37°C. Both human caspase-3 and Sf-caspase-1 are spontaneously processed to their mature, active subunits when overproduced in E. coli. The reaction mixtures were subjected to Ni2+ affinity chromatography followed by immunoblot analysis by using large subunit-specific anti (α)-human caspase-3 (top), large subunit-specific anti-Sf-caspase-1 (middle), anti-P49 (bottom left), or anti-P35 (bottom right). Molecular mass standards are indicated to the left of each panel. (B) Model of the P49/caspase complex. By virtue of its interactions with dimeric P49, the active form of untagged human caspase-3 forms a complex with His6-tagged Sf-caspase-1 that can be isolated by Ni2+ pull-down.
P49 forms a stable, higher-order complex with an effector caspase.
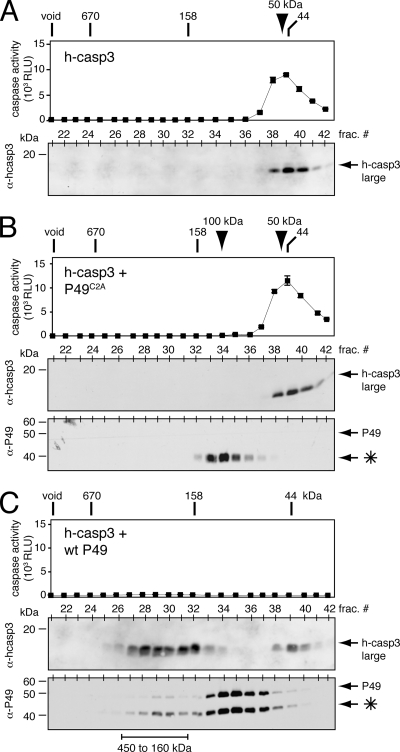
To further understand the stoichiometry of P49 inhibition, we determined the size of a complex between P49 and a well-characterized caspase. To this end, the reaction products of human caspase-3-His6 and excess purified P49-His6 were subjected to size exclusion chromatography. In the absence of P49, human caspase-3-His6 and DEVD-AMC cleavage activity coeluted, with an apparent molecular mass of ∼50 kDa (Fig. 4A), as expected for the caspase's homodimeric structure (30). When caspase-3 was mixed with excess C2A-mutated P49-His6, its elution profile was unchanged (Fig. 4B). Moreover, the fully cleaved C2A-mutated P49-His6 eluted as an ∼100-kDa complex (Fig. 4B), which suggested that P49 remains dimeric even after caspase cleavage and release. When mixed with excess wild-type P49-His6, human caspase-3-His6 eluted as a large 160- to 450-kDa complex and a less abundant 50-kDa complex (Fig. 4C). The absence of caspase activity and the coelution of cleaved P49-His6 identified the larger form as the inhibited P49/caspase complex. The average size of this inhibited complex was 250 kDa (Fig. 4C), which suggested that the most abundant species was a tripartite complex consisting of P49 dimers bound to the two active sites of a single caspase. Nonetheless, the broadness of the chromatographic peak was suggestive of a range of complexes, including higher-order end-to-end assemblies (see below). We concluded that by virtue of its dimeric structure P49 forms postcleavage complexes with various ratios of caspase to inhibitor. The lack of DEVD-ase activity associated with the less abundant 50-kDa caspase-3 complex (Fig. 4C) suggested that this caspase fraction was inactive during purification and before P49 interaction. Consistent with this interpretation was the finding that a comparable peak of inactive caspase-3 was observed among the reaction products of excess P35 and purified human caspase-3-His6 (data not shown). Although unlikely, the possibility that P49 and P35 inactivate caspase-3 and then are released cannot be ruled out.
FIG. 4.
Size of the P49/caspase complex. (A) Human caspase-3 (hcasp3). Purified caspase-3-His6 (100 pmol) was subjected to gel filtration chromatography. Column fractions were analyzed for caspase activity (top) using substrate DEVD-AMC and for protein by immunoblotting with anti (α)-human caspase-3 (bottom). Values for caspase activity are reported as the averages ± standard deviations of relative light units (RLU) from triplicate samples of each fraction. A representative experiment is shown. Chromatographic size standards and electrophoretic molecular mass markers are indicated. (B) Caspase-3 with loss-of-function P49C2A. Purified caspase-3-His6 (100 pmol) was incubated with purified P49C2A-His6 (500 pmol) and analyzed as described for panel A using anti-human caspase-3 (top) and anti-P49 (bottom). The P49 cleavage fragment (*) is indicated. (C) Caspase-3 with wild-type P49. Purified human caspase-3-His6 (100 pmol) was incubated with purified P49-His6 (500 pmol) and analyzed as described for panel B. The portion of P49 cleavage fragments (*) not associated with caspase (fractions 34 to 36) was released by caspase either before or during chromatography.
P35, but not P49, forms inhibitory complexes with effector caspases during infection.
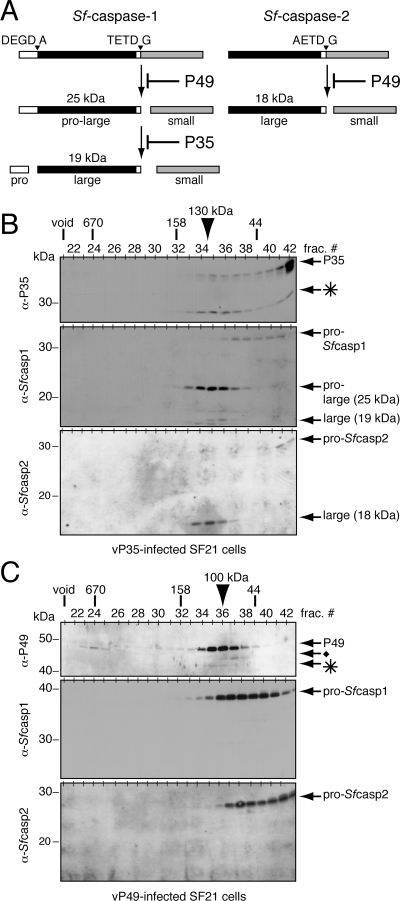
To further understand P35 and P49 specificity, we investigated their in vivo targets during baculovirus infection. In Spodoptera frugiperda cells, P35 functions after pro-Sf-caspase-1 is processed at TETD↓G, whereas P49 functions upstream to prevent this cleavage event by inhibiting the responsible Spodoptera initiator caspase, Sf-caspase-X (Fig. 5A). Processing of pro-Sf-caspase-1 generates the pro-large/small subunit complex and then the large/small complex; pro-Sf-caspase-2 is cleaved once at AETD↓G to a large/small subunit complex (20, 21, 45). We examined the inhibitory complexes formed with the Spodoptera caspases by using AcMNPV recombinants vP35 and vP49, which encode P35 and P49, respectively. Synthesized early in infection, P35 and P49 block apoptosis that is triggered in every cell (21, 45). When freeze-thaw extracts of vP35-infected SF21 cells were subjected to size exclusion chromatography, cleaved P35 eluted as an ∼130-kDa complex in association with the pro-large/small subunit form of Sf-caspase-1 and the large/small subunit form of Sf-caspase-2 (Fig. 5B). The proforms of both caspases eluted as smaller complexes. As expected, full-length, uncleaved P35 eluted as a 35-kDa monomer (Fig. 5B). The P35/Sf-caspase-1 and -2 complexes were similar in size (∼130 kDa) to the complex formed between purified P35 and human caspase-3 (30), suggesting that no other proteins were present. Because the pro-large/small intermediate was the predominant form of Sf-caspase-1 to complex with P35, we concluded that this subunit form is preferentially targeted by P35. Less-abundant quantities of Sf-caspase-1 consisting of the mature large subunit or the large subunit lacking the linker domain also coeluted with cleaved P35 (Fig. 5B). Because the formation of a stable P35/caspase complex requires cleavage of P35 by the targeted caspase, the association of P35 with Sf-caspase-2 comprising its large and small subunits indicated that this effector caspase is also active during infection. This finding provides the first evidence that Sf-caspase-2 is a functional enzyme involved in apoptosis.
FIG. 5.
Sf-caspase-1 and -2 form a complex with P35, but not P49, during infection. (A) Activation of Sf-caspase-1 and -2. Pro-Sf-caspase-1 is proteolytically cleaved by P49-sensitive, P35-resistant Sf-caspase-X at TETD↓G to yield the pro-large and small subunits. The pro-large subunit is cleaved by a P35-sensitive caspase at DEGD↓G to yield the mature large subunit. The linker region (open box) at the C terminus of the large subunit can also be processed. P49-sensitive Sf-caspase-X cleaves Sf-caspase-2 at AETD↓G to yield the large and small subunits. Because our polyclonal antisera detect the large subunit of each respective caspase, the molecular mass of each large subunit is indicated. (B) vP35-infected cells. Gel filtration chromatography was conducted on extracts of SF21 cells infected 24 h earlier with vP35 (MOI, 10). Column fractions were subjected to immunoblot analysis using anti (α)-P35 (top), anti-Sf-caspase-1 (middle), and anti-Sf-caspase-2 (bottom). The P35 cleavage fragment (*) is indicated. Chromatographic size standards and electrophoretic molecular mass markers are indicated. (C) vP49-infected cells. Extracts of SF21 cells infected 24 h earlier with vP49 (MOI, 10) were analyzed as described for panel B by using anti-P49 (top), anti-Sf-caspase-1 (middle), and anti-Sf-caspase-2 (bottom). The P49 cleavage fragment (*) and an unidentified P49-related protein (⧫) are indicated.
In vP49-infected cells, pro-Sf-caspase-1 and pro-Sf-caspase-2 remained in their intact ∼60-kDa complexes (Fig. 5C). Thus, as expected, P49 blocked proteolytic processing of both effector caspases. A majority of the intracellular P49 eluted in its full-length 100-kDa homodimeric form (Fig. 5C). Although a low-abundance protein with an electrophoretic mobility distinct from that of P49 was detected in the 670-kDa fractions, it most likely represents a nonspecific viral protein because it was not detected in plasmid transfected cells (Fig. 2A). Minor quantities of another P49-related protein, probably an internal initiation product (45), were also detected. We routinely observed limited cleavage of P49 in vP49-infected cell extracts. Because P49 functions as a stoichiometric inhibitor in vitro, the limited cleavage of P49 in vivo is likely due to a low abundance of Sf-caspase-X. Thus, under these conditions, we were unable to assess the size of the P49/Sf-caspase-X complex. Efforts are under way to isolate and characterize this P49-inhibitable, P35-insensitive initiator caspase from Spodoptera frugiperda.
The P49 TVTD recognition motif is required for initiator caspase inhibition.
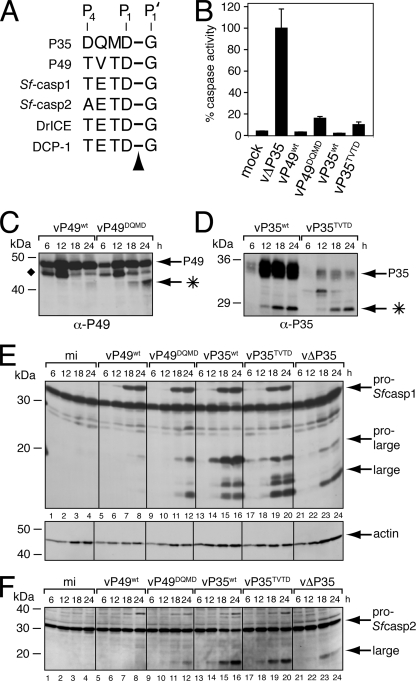
When the P35 cleavage site residues (P4 to P1) reside in the active site, they form the principal contacts between P35 and its target caspase (10, 40). Importantly, the P49 recognition motif TVTD94↓G resembles the P4 to P1 residues at the caspase cleavage site (TETD↓G) located at the junction of the large/small subunit of Sf-caspase-1 (Fig. 6A). Similarly, the P35 recognition motif DQMD87↓G most closely resembles sequences (DXXD↓G/A) cleaved by insect effector caspases. These striking similarities suggested that the P49 and P35 P4 to P1 residues function as a caspase recognition motif that determines target specificity. To test this hypothesis, we determined the effect of swapping the P4 to P1 residues of one inhibitor for those of the other on caspase targeting during infection. To this end, we generated AcMNPV recombinants vP35TVTD and vP49DQMD, which encoded P49DQMD and P35TVTD, respectively, as the sole apoptotic suppressor. The principal advantage of using these recombinant viruses is the capacity to monitor caspase processing in a context where all cells receive the same virus-induced apoptotic signal in the presence of physiologically relevant levels of apoptotic inhibitors.
FIG. 6.
P49DQMD and P35TVTD fail to inhibit the Spodoptera initiator caspase during infection. (A) Caspase cleavage sites. The P4 to P1 caspase recognition residues of P35 and P49 are compared with those at the large/small subunit junctions of Spodoptera and Drosophila effector caspases. The arrowhead denotes the cleaved P1-P1′ peptide bond. (B) Intracellular caspase activity. Cytoplasmic extracts of SF21 cells prepared 24 h after infection with recombinant baculoviruses encoding the indicated proteins as their sole apoptotic inhibitor were assayed for caspase activity by using substrate DEVD-AMC. Plotted values are the averages ± standard deviations of samples taken from triplicate infections and are reported as percentages of the 24-h caspase activity of vΔP35-infected cells. α, anti. (C to F) Immunoblots. Total lysates of cells harvested at the indicated hours after infection with the viruses described for panel B were subjected to immunoblot analysis by using (C) anti-P49, (D) anti-P35, (E) anti-Sf-caspase-1 (top) and antiactin (bottom), or (F) anti-Sf-caspase-2. Actin levels indicate comparable protein loading. The P49 and P35 cleavage fragments (*) and the unidentified P49-related protein (⧫) are indicated. Molecular mass standards are included on the left of each panel.
Upon infection of SF21 cells with vP35TVTD or vP49DQMD, intracellular caspase activity was dramatically reduced compared to that in cells infected by apoptosis-inducing vΔP35, which lacks p35 and p49 (Fig. 6B). DEVD-AMC cleavage activity in vP35TVTD- and vP49DQMD-infected cells was comparable to that in vP35wt- or vP49wt-infected cells, which produced wild-type P35 or P49, respectively. P49DQMD and P35TVTD successfully blocked virus-induced apoptosis (data not shown), a finding consistent with caspase inhibition. Intracellular cleavage of P49DQMD, unlike that of wild-type P49, was readily detected and yielded a fragment indicative of cleavage at Asp94 (Fig. 6C). The increased yield of cleaved P49DQMD compared to that of wild-type P49 suggested that a different caspase was targeted (see below). P35TVTD was cleaved to produce a fragment consistent with caspase-mediated proteolysis at Asp87 (Fig. 6D). Collectively, these data indicated that both modified proteins were functional and produced in quantities sufficient to inhibit virus-activated host caspases.
To identify the caspase targets of the modified inhibitors, we monitored the processing of pro-Sf-caspase-1 during the course of infection. As expected, Sf-caspase-1 was not proteolytically cleaved in vP49wt-infected cells (Fig. 6E, lanes 5 to 8), confirming that wild-type P49 inhibited Sf-caspase-X, which mediates pro-Sf-caspase-1 processing. In contrast, vP49DQMD allowed significant processing of Sf-caspase-1 to its pro-large and large subunits (lanes 9 to 12). The steady-state levels of wild-type P49 and P49DQMD were comparable (Fig. 6C), which indicates that viruses vP49DQMD and vP49wt were equally infectious. The level of Sf-caspase-1 processing in vP49DQMD-infected cells was similar to that in p35-deficient vΔP35-infected cells (lanes 21 to 24). P49DQMD also allowed processing of pro-Sf-caspase-2 (Fig. 6F, lanes 9 to 12), which was blocked in vP49wt-infected cells (lanes 5 to 8). We concluded that P49DQMD is less effective than wild-type P49 at inhibiting initiator Sf-caspase-X. Because caspase activity was low and apoptosis was blocked in the presence of proteolytically processed Sf-caspase-1, we also concluded that P49DQMD inhibited Sf-caspase-1. In vitro assays confirmed this finding by demonstrating that Sf-caspase-1 inhibition by P49DQMD-His6 was comparable to that by wild-type P49-His6 (data not shown). Moreover, P49DQMD was cleaved more abundantly than wild-type P49 during infection (Fig. 6C), which was expected if an effector caspase rather than an initiator caspase was targeted by P49DQMD. We concluded that substitution of recognition sequence TVTD for DQMD diminished P49's anti-initiator caspase activity but had no obvious effect on its anti-effector caspase activity.
Upon infection with vP35wt, processing of pro-Sf-caspase-1 was vigorous (Fig. 6E, lanes 13 to 16). Levels of Sf-caspase-1 processing were similar in vP35TVTD-infected cells (lanes 17 to 20), with the exception that accumulation of the pro-large subunit of Sf-caspase-1 was reduced compared to that in vP35wt-infected cells. Levels of Sf-caspase-2 processing in vP35wt- and vP35TVTD-infected cells were also comparable (Fig. 6F, lanes 13 to 16 and 17 to 20). Thus, the substitution of recognition sequence DQMD for TVTD failed to convert P35 to a P49-like inhibitor of initiator Sf-caspase-X. Purified P35TVTD and wild-type P35-His6 were equally potent as in vitro inhibitors of Sf-caspase-1 (data not shown). Collectively, these data indicated that the P4-P1 TVTD caspase recognition sequence of P49 is required but not sufficient for initiator caspase targeting by the baculovirus caspase inhibitors.
P49DQMD forms an inhibitory complex with intracellular Sf-caspase-1.
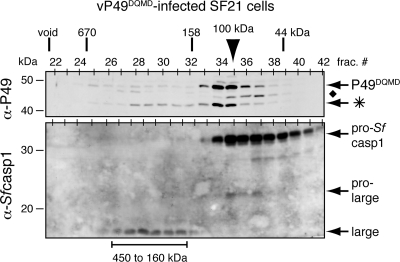
To verify that P49DQMD functioned by inhibiting endogenous effector caspases activated during infection, we determined whether P49DQMD formed a complex with Sf-caspase-1. To this end, extracts from vP49DQMD-infected cells were subjected to size exclusion chromatography. As expected, full-length P49DQMD was in excess and eluted as an ∼100-kDa dimeric complex (Fig. 7, top). Cleavage of P49DQMD was more abundant than that of wild-type P49 (Fig. 5C). A portion of the cleaved P49 coeluted with full-length P49 (Fig. 7, top), suggesting substrate dissociation. Importantly, a significant fraction of cleaved P49DQMD was found in a larger complex that coeluted with the mature large subunit of Sf-caspase-1 (Fig. 7, bottom). The elution profile of this larger complex was indistinguishable from that for the complex formed between purified P49 and human caspase-3 (Fig. 4C), suggesting similar stoichiometries of P49 to caspase in the inhibitory complex. In independent in vitro assays, purified P49DQMD-His6 was as potent a substrate inhibitor of Sf-caspase-1-His6 as P35-His6 and wild-type P49-His6 (data not shown). We therefore concluded that upon cleavage P49DQMD formed a stable inhibitory complex with processed, active Sf-caspase-1 during infection. These results suggested that, unlike wild-type P49, P49DQMD failed to completely block initiator caspase-mediated processing of Sf-caspase-1 and instead inhibited downstream effector caspases. It is noteworthy that a significant level of pro-Sf-caspase-1 remained unprocessed in P49DQMD-infected cells (Fig. 7) compared to vP35wt-infected cells (Fig. 5B). This observation suggested that Sf-caspase-X was partially inhibited in vP49DQMD-infected cells. We concluded that P49DQMD was impaired but not fully defective for initiator caspase inhibition.
FIG. 7.
P49DQMD forms an inhibitory complex with Sf-caspase-1 during infection. Extracts prepared from SF21 cells 24 h after infection with vP49DQMD (MOI, 10) were subjected to gel filtration chromatography and immunoblot analysis with anti (α)-P49 (top) and anti-Sf-caspase-1 (bottom). The P49 cleavage fragment (*) and an unidentified P49-related protein (⧫) are indicated. Chromatographic size standards and electrophoretic molecular mass markers are included. The arrowhead denotes the peak fraction of P49DQMD.
DISCUSSION
By virtue of their effectiveness as caspase inhibitors that prevent virus-induced apoptosis, P49 and P35 provide a powerful replication advantage to the baculoviruses. These apoptotic suppressors have also provided valuable insight into the role and regulation of caspases in diverse organisms (6, 14, 15). Here we investigated the novel difference in target specificity exhibited in vivo by P49 and P35 in permissive insect cells, the host system in which these inhibitors evolved to function. Our studies indicated that the P4-P1 caspase recognition and cleavage motif TVTD is required for P49's capacity to inhibit the Spodoptera initiator caspase, Sf-caspase-X. However, this cleavage motif is not sufficient to confer analogous apical caspase inhibitor activity upon P35. Thus, P49 possesses another molecular determinant necessary for initiator caspase inhibition that is lacking in P35. The biochemical differences between P49 and P35 defined herein, including oligomerization and stoichiometry of caspase interaction, argue that multiple determinants may contribute to target specificity for each molecule.
Role of the caspase recognition motif for P49 and P35 specificity.
By using recombinant viruses to test the in vivo target specificity of P49 or P35 during infection, we have shown that the P4-P1 caspase recognition motif is critical but not sufficient to confer caspase specificity. Substitution of P49's TVTD recognition motif for P35's DQMD diminished initiator caspase inhibition by P49 (Fig. 6) without affecting inhibition of effector caspases, which were subsequently targeted by P49DQMD (Fig. 7). In our previous study in which caspase inhibitors were delivered by plasmid transfection rather than virus vectors (45), the loss of initiator caspase inhibition by P49DQMD was less pronounced. It is likely that the lower efficiency of transfection and resulting inconsistencies in inhibitor production accounted for this difference. Here, by using recombinant vectors vP49DQMD and vP35TVTD, we gained the important advantage of monitoring in vivo processing of endogenous caspases in a context wherein all cells received comparable inhibitors and a synchronized apoptotic signal.
Because the only known anticaspase activity of P49 is by substrate inhibition, it is likely that Sf-caspase-X failed to cleave P49DQMD in vP49DQMD-infected cells. The S4 pocket, second only to the S1 pocket in a caspase's substrate binding cleft, which is occupied by the requisite P1 Asp residue of the substrate, is most influential in discriminating caspase substrates (15). Thus, Sf-caspase-X, which is targeted by P49, probably prefers a Thr in the P4 position since the cleavage sites of wild-type P49 and pro-Sf-caspase-1 share a P4 Thr residue. Use of cell-permeable tetrapeptide inhibitors has already demonstrated that Sf-caspase-X has a substrate preference distinct from that of the Spodoptera effector caspases (25). Analysis of the cleavage site preference of Sf-caspase-X will benefit from the isolation and purification of this low-abundance initiator caspase, which are under way.
Interestingly, when tested in baculovirus-infected Drosophila melanogaster cells, wild-type P49 fails to inhibit the Drosophila initiator caspase DRONC (22), which processes Drosophila effector caspases at a site TETD↓G (16, 28) identical to that in pro-Sf-caspase-1 cleaved by Sf-caspase-X. Instead, P49 inhibits the active Drosophila effector caspases, including DrICE. Recently, we have generated substitutions of the P6-P3 recognition motif residues within P49 that increase the anti-DRONC activity of P49 (M. Guy and P. Friesen, unpublished data). These findings suggest that the substrate specificity of DRONC is determined by a larger recognition motif that extends from residue P6 to P1.
Role of a P49 exosite in target specificity.
Our studies here also confirm and extend our earlier conclusions that the caspase recognition sites of P49 and P35 are not sufficient to confer target specificity (45). Because substitution of P35's DQMD recognition motif for P49's TVTD motif did not convert P35 to an inhibitor of Sf-caspase-X (Fig. 6), P35 must lack another P49 determinant that confers anti-initiator caspase activity. Nonetheless, P35TVTD still functioned to inhibit downstream effector caspases. Thus, the S4 pocket of the Spodoptera effector caspases, including Sf-caspase-1 (13), appears less stringent than that of the initiator caspase for substrate recognition. Our data argue that P49 possesses a domain, referred to as an exosite, which interacts noncovalently with the Spodoptera initiator caspase outside of the active site and thereby contributes to target specificity. The P35/human caspase-8 crystal structure defines potential interactions between the L4 loop/α4 helix of the inhibited enzyme and the large protruding K-L loop of P35 (40). Thus, the P49 exosite may make analogous interactions with insect initiator caspases that do not interfere with P49's anti-effector caspase activity. Importantly, P49 contains an ∼120-residue insert that is absent in P35 (29, 45). Due to the hydrophilic nature of the domain, it probably resides on the surface of P49 and therefore could contribute to caspase specificity or P49 oligomerization (see below). Alternatively, P49's in vivo initiator caspase specificity may be the result of interactions with other factors that are required for initiator activation or activity. Such factors include the unidentified Spodoptera homolog of Drosophila Apaf1-related killer (Dark), which by virtue of its required caspase association could juxtapose P49 and Sf-caspase-X. We continue to investigate these possibilities by taking advantage of the in vivo setting provided by baculovirus-infected cells.
P49 as a homodimer.
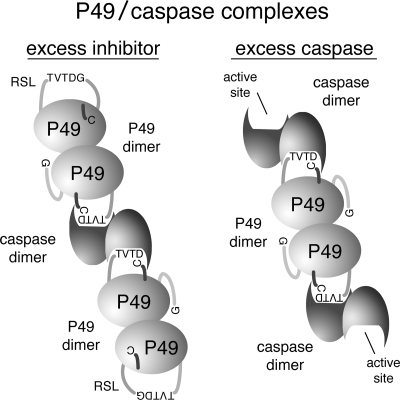
The predominant form of intracellular and purified P49 is a homodimer (Fig. 2A and 5C). Thus, P49 is the first oligomeric example of a P35-related caspase inhibitor. Although P35 has the capacity to form dimers (44), it functions as a monomer (Fig. 2B, 3A, and 5B) when interacting with a single caspase active site (12, 30). In contrast, the P49 dimer possesses the ability to interact simultaneously with two distinct caspases (Fig. 3), indicating that each P49 molecule in the dimer is functional for binding and inhibiting an active site (Fig. 8). The capacity of P49 to interact with separate caspases argues that the P49 dimer is arranged such that each RSL has unobstructed access to each caspase active site. Thus, the P49 domain contributing to homophilic interactions probably lies opposite the RSL. Because the 120-residue domain unique to P49 is also predicted to lie opposite the RSL, it may function as a dimerization domain. Our study also suggests that it is unlikely that a P49 homodimer interacts with a single caspase homodimer by binding to both active sites simultaneously. The structure of the P35/caspase-8 complex (40), in which both P35 molecules point in opposite directions from each occupied active site, suggests that P49 would behave similarly, even as a homodimer (Fig. 8).
FIG. 8.
Model for P49 inactivation of caspases. In the presence of excess P49 (left), each active site of the caspase dimer is occupied by a P49 dimer in which cleavage of the P49 RSL at TVTD↓G promotes placement of the N-terminal Cys2 into the caspase active site and generates a stable inhibitory complex. The RSL of the linked P49 subunit is available for caspase interaction and inhibition. In the presence of excess caspase (right), both RSLs of a P49 dimer are bound to separate caspases. The unoccupied active site remains available for substrates. Thus, the model predicts the formation of higher-order end-to-end assemblies of P49-inhibited caspases when the ratio of P49 to caspase is appropriate.
What is the biological relevance of P49 dimerization? One possibility is to increase the local concentration of P49 at the apoptosome, which serves as an activation platform with a high density of initiator and effector caspases (17, 31, 33). As one P49 subunit binds a caspase, the sister subunit is immediately available for subsequent inhibition of adjacent caspases (Fig. 8), presumably present in the same apoptosome complex. To address this possibility, it will be of interest to compare the intracellular distributions of P49 and P35 and to assess association with the Spodoptera apoptosome. Another potential function for dimerization is to promote P49 stability and resistance to proteosome degradation. Similarly, dimerization may contribute to intracellular solubility of P49. The solubility of P49-His6 in bacteria increased markedly when we extended the hexahistidine tag away from the C terminus by insertion of a linker peptide. Thus, the C terminus of P49 may lie near its dimerization interface.
P49 as a pan-caspase inhibitor of the Cys2-dependent P35 family.
Site-directed mutagenesis demonstrated that residue Cys2 is required for P49 anticaspase activity (Fig. 1). C2A-mutated P49 was readily cleaved by caspases but failed to form an inhibitory complex (Fig. 4B). Thus, P49 and P35 appear to use comparable inhibitory mechanisms that depend on chemical interactions with Cys2 that anchor the inhibitor's N terminus in the caspase active site through formation of a thioester bond between caspase and inhibitor (24). The conservation of Cys2 in Amsacta moorei entomopoxvirus P33, the most distantly related P35 homolog (27), suggests that this mechanism is shared among family members.
Among the P35-related caspase inhibitors, P49 is unique in its capacity to inactivate both initiator and effector caspases in vivo. We have demonstrated here that, upon inactivation of its anti-initiator caspase activity, P49 functions as an effector caspase inhibitor (Fig. 6 and 7). This finding implies that, if an apical caspase escapes inactivation by P49, then the downstream anti-effector caspase activity of P49 will ensure suppression of apoptosis. Indeed, P49 fails to block initiator caspase DRONC upon baculovirus infection of cultured Drosophila cells but still blocks apoptosis through inhibition of the downstream effector caspases, including DrICE (22). This type of dual-action anticaspase activity likely provides significant multiplicative advantages to P49-encoding baculoviruses that encounter a wide variety of insect (lepidopteran) species in nature. P49's capacity to irreversibly bind and inhibit a variety of caspases also suggests that it will be a useful tool for identification of novel initiator and effector caspases. Recombinant P49 molecules engineered for enhanced caspase selectivity will be of special interest in continued investigations of the role of specific caspases in apoptosis as well as in the design of anticaspase strategies for the treatment of apoptosis-associated pathologies.
Acknowledgments
We thank Yuri Lazebnik (Cold Spring Harbor Laboratory) for the gift of P35 monoclonal antibody. We also thank Diccon Fiore and Rianna Vandergaast for assistance in virus and plasmid generation.
This work was supported in part by Public Health Service grant AI40482 from the National Institute of Allergy and Infectious Diseases (P.D.F.) and NIH Predoctoral Traineeship GM07215 (M.P.G.).
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Bao, Q., and Y. Shi. 2007. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 1456-65. [DOI] [PubMed] [Google Scholar]
- 2.Bertin, J., S. M. Mendrysa, D. J. LaCount, S. Gaur, J. F. Krebs, R. C. Armstrong, K. J. Tomaselli, and P. D. Friesen. 1996. Apoptotic suppression by baculovirus P35 involves cleavage by and inhibition of a virus-induced CED-3/ICE-like protease. J. Virol. 706251-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bump, N. J., M. Hackett, M. Hugunin, S. Seshagiri, K. Brady, P. Chen, C. Ferenz, S. Franklin, T. Ghayur, P. Li, et al. 1995. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 2691885-1888. [DOI] [PubMed] [Google Scholar]
- 4.Callus, B. A., and D. L. Vaux. 2007. Caspase inhibitors: viral, cellular and chemical. Cell Death Differ. 1473-78. [DOI] [PubMed] [Google Scholar]
- 5.Clem, R. J. 2007. Baculoviruses and apoptosis: a diversity of genes and responses. Curr. Drug Targets. 81069-1074. [DOI] [PubMed] [Google Scholar]
- 6.Clem, R. J. 2001. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 8137-143. [DOI] [PubMed] [Google Scholar]
- 7.Clem, R. J. 2005. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top. Microbiol. Immunol. 289113-129. [DOI] [PubMed] [Google Scholar]
- 8.dela Cruz, W. P., P. D. Friesen, and A. J. Fisher. 2001. Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage. J. Biol. Chem. 27632933-32939. [DOI] [PubMed] [Google Scholar]
- 9.Du, Q., D. Lehavi, O. Faktor, Y. Qi, and N. Chejanovsky. 1999. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J. Virol. 731278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddins, M. J., D. Lemongello, P. D. Friesen, and A. J. Fisher. 2002. Crystallization and low-resolution structure of an effector-caspase/P35 complex: similarities and differences to an initiator-caspase/P35 complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 58299-302. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, U., and K. Schulze-Osthoff. 2005. Apoptosis-based therapies and drug targets. Cell Death Differ. 12(Suppl. 1)942-961. [DOI] [PubMed] [Google Scholar]
- 12.Fisher, A. J., W. Cruz, S. J. Zoog, C. L. Schneider, and P. D. Friesen. 1999. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 182031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsyth, C. M., D. Lemongello, D. J. LaCount, P. D. Friesen, and A. J. Fisher. 2004. Crystal structure of an invertebrate caspase. J. Biol. Chem. 2797001-7008. [DOI] [PubMed] [Google Scholar]
- 14.Friesen, P. D. 2007. Insect viruses, p. 707-736. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 15.Fuentes-Prior, P., and G. S. Salvesen. 2004. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384201-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins, C. J., S. J. Yoo, E. P. Peterson, S. L. Wang, S. Y. Vernooy, and B. A. Hay. 2000. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 27527084-27093. [DOI] [PubMed] [Google Scholar]
- 17.Hay, B. A., and M. Guo. 2006. Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 22623-650. [DOI] [PubMed] [Google Scholar]
- 18.Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 665525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jabbour, A. M., P. G. Ekert, E. J. Coulson, M. J. Knight, D. M. Ashley, and C. J. Hawkins. 2002. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ. 91311-1320. [DOI] [PubMed] [Google Scholar]
- 20.LaCount, D. J. 1998. Virus replication events and host factors involved in baculovirus-induced apoptosis. Ph.D. thesis. University of Wisconsin-Madison, Madison.
- 21.LaCount, D. J., S. F. Hanson, C. L. Schneider, and P. D. Friesen. 2000. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem. 27515657-15664. [DOI] [PubMed] [Google Scholar]
- 22.Lannan, E., R. Vandergaast, and P. D. Friesen. 2007. Baculovirus caspase inhibitors P49 and P35 block virus-induced apoptosis downstream of effector caspase DrICE activation in Drosophila melanogaster cells. J. Virol. 819319-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavrik, I. N., A. Golks, and P. H. Krammer. 2005. Caspases: pharmacological manipulation of cell death. J. Clin. Investig. 1152665-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M., T. Min, D. Eliezer, and H. Wu. 2006. Native chemical ligation in covalent caspase inhibition by p35. Chem. Biol. 13117-122. [DOI] [PubMed] [Google Scholar]
- 25.Manji, G. A., and P. D. Friesen. 2001. Apoptosis in motion. An apical, P35-insensitive caspase mediates programmed cell death in insect cells. J. Biol. Chem. 27616704-16710. [DOI] [PubMed] [Google Scholar]
- 26.Manji, G. A., R. R. Hozak, D. J. LaCount, and P. D. Friesen. 1997. Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J. Virol. 714509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Means, J. C., T. Penabaz, and R. J. Clem. 2007. Identification and functional characterization of AMVp33, a novel homolog of the baculovirus caspase inhibitor p35 found in Amsacta moorei entomopoxvirus. Virology 358436-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier, P., J. Silke, S. J. Leevers, and G. I. Evan. 2000. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 19598-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei, Z., G. Reske, Q. Huang, B. D. Hammock, Y. Qi, and N. Chejanovsky. 2002. Characterization of the apoptosis suppressor protein P49 from the Spodoptera littoralis nucleopolyhedrovirus. J. Biol. Chem. 27748677-48684. [DOI] [PubMed] [Google Scholar]
- 30.Riedl, S. J., M. Renatus, S. J. Snipas, and G. S. Salvesen. 2001. Mechanism-based inactivation of caspases by the apoptotic suppressor p35. Biochemistry 4013274-13280. [DOI] [PubMed] [Google Scholar]
- 31.Riedl, S. J., and G. S. Salvesen. 2007. The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8405-413. [DOI] [PubMed] [Google Scholar]
- 32.Riedl, S. J., and Y. Shi. 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5897-907. [DOI] [PubMed] [Google Scholar]
- 33.Salvesen, G. S., and J. M. Abrams. 2004. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 232774-2784. [DOI] [PubMed] [Google Scholar]
- 34.Salvesen, G. S., and C. S. Duckett. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3401-410. [DOI] [PubMed] [Google Scholar]
- 35.Schafer, Z. T., and S. Kornbluth. 2006. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10549-561. [DOI] [PubMed] [Google Scholar]
- 36.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9459-470. [DOI] [PubMed] [Google Scholar]
- 37.Timmer, J. C., and G. S. Salvesen. 2007. Caspase substrates. Cell Death Differ. 1466-72. [DOI] [PubMed] [Google Scholar]
- 38.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13213-217. [DOI] [PubMed] [Google Scholar]
- 39.Vier, J., C. Furmann, and G. Hacker. 2000. Baculovirus P35 protein does not inhibit caspase-9 in a cell-free system of apoptosis. Biochem. Biophys. Res. Commun. 276855-861. [DOI] [PubMed] [Google Scholar]
- 40.Xu, G., M. Cirilli, Y. Huang, R. L. Rich, D. G. Myszka, and H. Wu. 2001. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature 410494-497. [DOI] [PubMed] [Google Scholar]
- 41.Xu, G., R. L. Rich, C. Steegborn, T. Min, Y. Huang, D. G. Myszka, and H. Wu. 2003. Mutational analyses of the p35-caspase interaction. A bowstring kinetic model of caspase inhibition by p35. J. Biol. Chem. 2785455-5461. [DOI] [PubMed] [Google Scholar]
- 42.Yan, N., and Y. Shi. 2005. Mechanisms of apoptosis through structural biology. Annu. Rev. Cell Dev. Biol. 2135-56. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, Q., J. F. Krebs, S. J. Snipas, A. Price, E. S. Alnemri, K. J. Tomaselli, and G. S. Salvesen. 1998. Interaction of the baculovirus anti-apoptotic protein p35 with caspases. Specificity, kinetics, and characterization of the caspase/p35 complex. Biochemistry 3710757-10765. [DOI] [PubMed] [Google Scholar]
- 44.Zoog, S. J., J. Bertin, and P. D. Friesen. 1999. Caspase inhibition by baculovirus P35 requires interaction between the reactive site loop and the beta-sheet core. J. Biol. Chem. 27425995-26002. [DOI] [PubMed] [Google Scholar]
- 45.Zoog, S. J., J. J. Schiller, J. A. Wetter, N. Chejanovsky, and P. D. Friesen. 2002. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. EMBO J. 215130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]