Abstract
The family of interferon-inducible transmembrane proteins (Ifitm) consists of five highly sequence-related cell surface proteins, which are implicated in diverse cellular processes. Ifitm genes are conserved, widely expressed, and characteristically found in genomic clusters, such as the 67-kb Ifitm family locus on mouse chromosome 7. Recently, Ifitm1 and Ifitm3 have been suggested to mediate migration of early primordial germ cells (PGCs), a process that is little understood. To investigate Ifitm function during germ cell development, we used targeted chromosome engineering to generate mutants which either lack the entire Ifitm locus or carry a disrupted Ifitm3 gene only. Here we show that the mutations have no detectable effects on development of the germ line or on the generation of live young. Hence, contrary to previous reports, Ifitm genes are not essential for PGC migration. The Ifitm family is a striking example of a conserved gene cluster which appears to be functionally redundant during development.
Germ cells in the mouse are derived from proximal epiblast cells of the early embryo in response to instructive signals that ultimately result in the emergence of about 40 specified primordial germ cells (PGCs) at embryonic day 7.5 (E7.5) (13, 17). These nascent PGCs subsequently migrate along the developing hindgut to colonize the genital ridges at E10.5 (4, 8), where they eventually undergo sex-specific differentiation to form mature gametes.
Among the molecules thought to regulate mouse PGC development is the family of interferon-inducible transmembrane proteins (Ifitms). Its five members are Ifitm1 (fragilis2/mil-2), Ifitm2 (fragilis3/mil-3), Ifitm3 (fragilis/mil-1) Ifitm5 (fragilis4), and Ifitm6 (fragilis5) (16, 26, 32). All five Ifitm genes are located within a 67-kb region at the telomeric tip of chromosome 7 (Chr7) which is conserved in humans (16, 18, 32). Ifitms are short, two-transmembrane-domain proteins (5 to 18 kDa) with high core sequence similarity but with more divergent N and C termini (16, 32). Interferon-stimulable response elements within the promoter regions of several of these genes make them potentially responsive to class I interferons (alpha/beta interferons) (see Fig. S1A in the supplemental material) (12, 24, 25).
During development, there is dynamic expression of Ifitm1, -2, and -3, which is tightly associated with germ line competence of epiblast cells and PGC fate (16, 26, 32, 33). Prior to gastrulation, cells in the proximal epiblast express Ifitm3, of which a subset at E6.75 will become germ cell lineage restricted (22). These cells then locate to the posterior extraembryonic mesoderm and initiate expression of the PGC marker stella/PGC7, as well as of Ifitm1 and Ifitm2 (26, 27). The expression of Ifitm1, -2, and -3 is maintained through the entry of PGCs into the genital ridges, whereas expression of Ifitm5 and Ifitm6 is not detected in early embryos (see Fig. S1B in the supplemental material) (16). Generally, Ifitm transcripts are also seen in a variety of other embryonic and adult tissues, and individual family members show partly overlapping yet distinct expression patterns (see Fig. S1B in the supplemental material) (16, 30, 32).
Ifitm genes have been suggested to function in a variety of contexts, including immune cell regulation, cancerogenesis, somitogenesis, and germ cell development. In human leukocyte cell lines, for example, IFITM1 (9-27/Leu-13) is thought to mediate antiproliferative activities and cell-cell adhesion processes (7, 10, 11). IFITM1 and related proteins form complexes in B and T lymphocytes with the tetraspanin proteins CD9 and CD81 (TAPA-1), which are critical cell surface components during B-cell maturation and activation (6, 15, 28, 30). Recently, Ifitm1 has also emerged as a target of Wnt/β-catenin signaling. RNA interference (RNAi) against Ifitm1 during embryogenesis indicated a role in the posterior axis and the epithelialization of somites (19). IFITM genes are also upregulated in colorectal cancer, where hyperactivation of the β-catenin signaling pathway is causally linked to tumor formation (5). A recent study also suggested an active role for the Ifitm family during PGC development (33). Overexpression and knockdown experiments indicated that homo- and/or heterotypic interactions of Ifitm1 and Ifitm3 proteins on PGCs and neighboring somatic cells coordinate the guidance of early germ cells during their migration from the mesoderm into and within the visceral endoderm. These data indicated a possible antagonistic effect between different Ifitm family members (33).
In the present study, we set out to address the functional importance of the Ifitm gene family during mouse germ cell development, using genetic engineering techniques. First, we targeted the entire Ifitm cluster by creating a chromosomal deletion of 120 kb, encompassing the complete family locus. Second, to examine a dosage-dependent relationship between various Ifitm genes as postulated previously, we created mice with a functional null mutation in only the Ifitm3 gene. Surprisingly, we found that deletion neither of the entire cluster nor of Ifitm3 alone had a detectable phenotypic effect specifically on germ cells or generally during development to adulthood.
MATERIALS AND METHODS
Generation of IfitmDel and Ifitm3EGFP mutants.
For IfitmDel mutants, E14tg2a cells were targeted using MICER vector (1) MHPN60K05 (5′Hprt; containing a neomycin selection cassette and Tyr marker) and selected clones were retargeted using a replacement vector (3′Hprt; containing a puromycin selection marker) constructed by recombineering (9). Double-targeted clones were electroporated with CAGGs-Cre (34) and selected in hypoxanthine-aminopterin-thymidine medium to identify clones carrying the IfitmDel deletion (36). Correct targeting was confirmed by screening for puromycin sensitivity and fluorescent in situ hybridization (FISH) on metaphase spreads using a fluorescein isothiocyanate-labeled genomic probe inside and a Texas red-labeled bacterial artificial chromosome (BAC) clone outside the deletion (performed as described previously [36]). For Ifitm3EGFP mutants, E14tg2a cells were targeted using a pBluescript-based vector, constructed by BAC modification and conventional restriction digest cloning, containing a puromycin selection cassette and enhanced green fluorescent protein (EGFP) inserted into exon 1 of Ifitm3. Targeted clones were selected, and targeting was confirmed by Southern blot analysis. Generation of chimeras and germ line transmission of mutant alleles were done as described previously (20).
Southern/Northern blot analysis and reverse transcription-PCR.
DNA for Southern blot analysis or PCR was extracted using proteinase K digestion and ethanol precipitation. RNA was extracted using Trizol reagent. DNA/RNA was blotted onto Hybond N+ membranes and probed with [α-32P]dCTP-labeled DNA probes. For reverse transcription-PCR, RNA was DNase I treated, reverse transcribed, and PCR amplified (Ifitm1 to -6, 30 cycles; Ifitm7, 35 cycles). See the supplemental material for oligonucleotide sequences.
Histology.
For immunohistochemistry and in situ hybridization, whole-mount embryos were processed as described previously (16, 23). For cryosectioning, tissues were fixed in 4% paraformaldehyde in phosphate buffered saline and embedded in OCT compound. Cryosections (12-μm thickness) were postfixed prior to incubation with primary and secondary antibodies. Antibodies were obtained from Upstate (Oct3/4 and H3K9me2) and Abcam (Ifitm3); Stella/PGC7, H3K27me3, and MetCyt antibodies were kind gifts from T. Nakano, T. Jenuwein, and A. Niveleau, respectively. Samples were mounted in Vectashield-DAPI (4′,6′-diamidino-2-phenylindole) and imaged using an Olympus fluorescence or Zeiss confocal microscope. For paraffin sections, testes and ovaries were fixed in Bouin's fixative, dehydrated, and embedded in paraffin wax. Sections (10-μm thickness) were stained with hematoxylin-eosin and imaged with an Olympus microscope.
RESULTS
Deletion of the Ifitm gene locus.
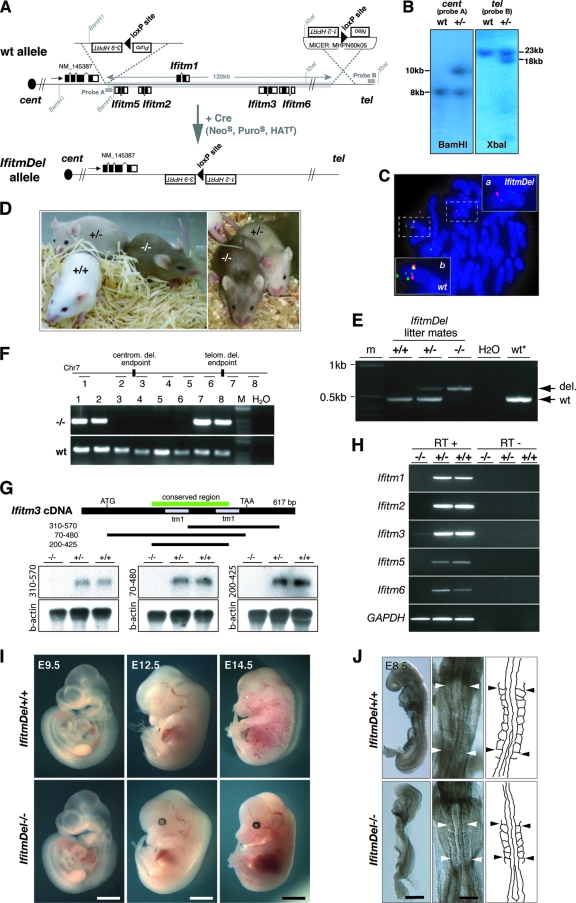
We engineered a deletion of a 120-kb region on Chr7, harboring the entire Ifitm family locus but no other known gene. The mutant allele was designated IfitmDel. In brief, two loxP selection cassettes were sequentially inserted via homologous recombination into the centromeric and telomeric regions flanking Ifitm5 and Ifitm6, respectively (Fig. 1A). Transient transfection of double-targeted embryonic stem (ES) cell clones with a Cre-expressing plasmid resulted in the generation of IfitmDel+/− ES cells, which were validated by PCR and Southern blot analysis (Fig. 1B and data not shown). IfitmDel+/− ES cells were used to generate chimeric animals, and the mutation was transmitted through the germ line onto the C57BL/6Jc/c background. The resulting F1 animals were backcrossed for another generation to C57BL/6Jc/c prior to phenotyping. FISH analysis confirmed the deletion of the entire Ifitm gene cluster (Fig. 1C).
FIG. 1.
Targeted deletion of the mouse Ifitm gene cluster. (A) Schematic of the targeting strategy used to create the IfitmDel allele. (B) Southern blot analysis of targeted ES cell clones prior to Cre-mediated excision. (C) FISH analysis of IfitmDel+/− ES cell metaphase spreads using a fluorescein isothiocyanate-labeled genomic probe in the deletion (green) and a Texas red-labeled BAC-derived probe outside the deletion (red). Inset a, IfitmDel allele; inset b, wt allele. (D) Fourteen-day-old IfitmDel−/−, IfitmDel+/−, and wt littermates. Coat color marking of the targeted allele enables genotyping by eye. (E) Confirmation of genotyping by PCR on tail extracts. (F) PCR on genomic DNA of IfitmDel−/− and wt tissues using primers amplifying fragments flanking (lanes 1, 2, 7, and 8) and within (lanes 3 to 6) the deletion confirms deletion of the Ifitm locus from the targeted genome. (G) Northern blot analysis using E17.5 whole-embryo RNA from IfitmDel−/−, IfitmDel+/−, and wt littermates. Three probes corresponding to different fragments of Ifitm3 cDNA were used for hybridization: 310 bp to 570 bp, Ifitm3-specific probe; 70 bp to 480 bp, Ifitm3 coding region probe, expected to cross-hybridize with all known mouse Ifitm transcripts; 200 bp to 425 bp, Ifitm gene highly conserved region probe, expected to cross-hybridize with any Ifitm-like transcript. (H) Reverse transcription-PCR analysis on E17.5 whole embryo cDNA from IfitmDel−/−, IfitmDel+/−, and wt littermates using Ifitm gene-specific primer sets. (I) IfitmDel−/− and wt littermates at different stages of development. (J) E8.5 IfitmDel−/− embryos show no defects in somite organization. Left panels, lateral view; middle panels, dorsal view; right panels, outline of somites and neural tube in dorsal view. Bars, 400 μm (I, left panels), 2.25 mm (I, middle panels), 3.25 mm (I, right panels), 200 μm (J, left panels), and 100 μm (J, middle panels).
IfitmDel mutants show normal development and are viable.
Intercrosses of heterozygous IfitmDel animals gave rise to offspring of all expected genotypes, including 17% IfitmDel−/−, 52% IfitmDel+/−, and 30% IfitmDel+/+ (n = 164). The insertion of the tyrosinase marker Tyr at the telomeric end of the deletion enabled genotyping by coat color, which was confirmed by genomic PCR (Fig. 1D and E). To confirm the absence of all five Ifitm genes in homozygous IfitmDel animals, we performed PCR analysis (Fig. 1F) as well as Northern blot analysis and reverse transcription-PCR analysis on whole embryo material at E17.5 (Fig. 1G and H).
Throughout development in utero, homozygous IfitmDel embryos appeared morphologically normal (Fig. 1I) and without any somitic or posterior axis defects, contrary to previous conclusions, drawn from Ifitm1 RNAi-treated embryos (Fig. 1J) (19). Furthermore, there were no overt differences in size, weight, or behavior of homozygous IfitmDel animals (Fig. 1D and data not shown). Nevertheless, it should be noted that the number of homozygous IfitmDel animals born from heterozygous IfitmDel intercrosses was slightly reduced from the expected Mendelian ratio at weaning age (χ22df = 6.23; P < 0.05 [χ2 test]). Notably, no increased lethality in IfitmDel−/− mutants was observed during embryonic stages E7.5 to E17.5 (n = 120; χ21df = 0.178 [χ2 test]) or after birth. Therefore, the reduction in frequency at 21 days postpartum is most likely due to a slight increase in prenatal death (fetal period after E17.5) or perinatal lethality, both of which would lead to a stillbirth and are hence not readily detectable.
IfitmDel mutants had normal unchallenged B- and T-cell counts as assessed by fluorescence-activated cell sorting (Student's two-tailed t test) (see Fig. S2 in the supplemental material). Prompted by previous studies using anti-hIFITM1 blocking antibody (7, 10, 11), we in addition tested the mobility and adhesion properties of IfitmDel−/− fibroblasts using an in-culture “wound healing” assay (see Fig. S3 in the supplemental material). Compared to wild-type (wt) cells, mutant embryonic fibroblasts were not impaired in immigrating into or closing the monolayer “lesion.”
While a sixth Ifitm gene on Chr16, Ifitm7, has been previously reported (32), it shows no expression in early postimplantation embryos (32), and its later expression appears confined to the adult testis, as suggested by expressed sequence tag expression profiling and confirmed by whole-mount in situ hybridization and reverse transcription-PCR analysis (see Fig. S4A in the supplemental material). Notably, the Northern blot probe spanning the region of high sequence conservation between Ifitm genes (bp 200 to 425 of Ifitm3 cDNA) also did not recognize any transcripts in homozygous IfitmDel embryos (Fig. 1G) (16). This strongly suggests that apart from the five genes within the targeted genomic cluster on Chr7, no other Ifitm-like genes are embryonically expressed. Hence, it is unlikely that the absence of an apparent phenotype in homozygous IfitmDel mice is due to redundancy and compensation by a sequence-related protein during development.
Germ cell specification, migration, and gonadal development are unperturbed in IfitmDel mutants.
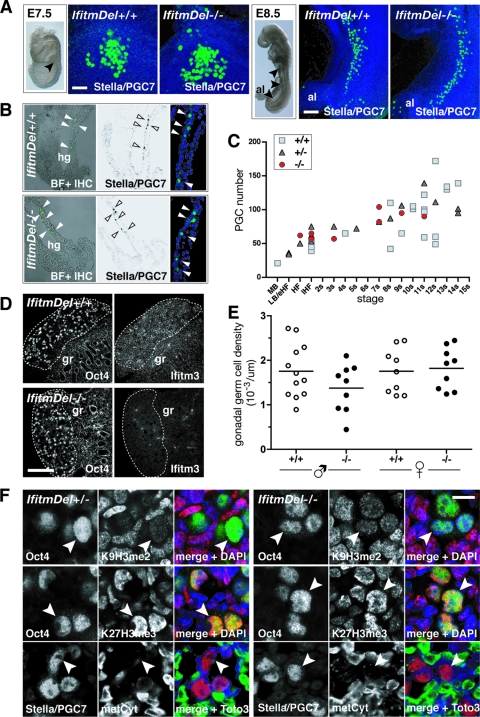
Since several Ifitm genes show pronounced expression during germ cell development and since Ifitm1 and Ifitm3 have been postulated to have a role in PGC migration (26, 32, 33), we next focused on investigating the developing germ cell lineage in the mutants. Whole-mount immunohistochemistry against Stella/PGC7 revealed a cluster of founder PGCs at the base of the allantoic bud in IfitmDel embryos at E7.5 (Fig. 2A). Furthermore, at E8.5, we found PGCs along their migratory path in the hindgut endoderm (Fig. 2A and B). Notably, we observed no detectable effects on the clustering of PGCs or in their migratory behavior. Similarly, there were no significant differences in the number of PGCs in wt and mutant embryos from E7.5 to E8.5, as judged by counting Stella/PGC7-positive cells using Z series of confocal sections (Mann-Whitney U test) (Fig. 2C). Furthermore, we found appropriate expression of germ line marker genes TNAP, Oct4, and stella/PGC7 (data not shown).
FIG. 2.
PGC specification, migration, and gonadal colonization are unperturbed in IfitmDel mutants. (A) Projection image of whole-mount immunohistochemistry on IfitmDel−/− and wt littermates at E7.5 (posterior view) and E8.5 (lateral view). PGCs are stained with anti-Stella/PGC7 antibody (green). The left panel shows a lateral view of an embryo of the appropriate age; arrowheads indicate localization of PGCs. al, allantois. (B) Cryosections of E8.5 IfitmDel−/− and wt embryos show migrating PGCs within the hindgut (hg) endoderm stained with anti-Stella/PGC7 antibody (green). The right panel shows a higher-magnification confocal image. BF, bright field; IHC, immunohistochemistry. (C) Counts of Stella/PGC7-positive PGCs from IfitmDel−/−, IfitmDel+/−, and wt littermates at E7.5 and E8.5. MB, mid-bud; LB/eHF, late bud/early head fold; HF, head fold; lHF: late head fold; s, somite. (D) Immunohistochemistry on cryosections of E12.5 genital ridges from IfitmDel−/− and wt littermates using germ cell-specific Oct4 antibody and an antibody against Ifitm3. Note that Ifitm3 protein is absent in IfitmDel−/− genital ridges. Extranuclear Oct4 staining outside the outlined genital ridge (gr) is due to unspecific secondary antibody binding. (E) Germ cell density counts on E12.5 genital ridge cryosections from IfitmDel−/− and wt littermates show no significant difference (Mann-Whitney U test). (F) Immunohistochemistry on cryosections of E12.5 genital ridges from IfitmDel−/− and wt littermates for chromatin modifications K9H3me2 (red), K27H3me3 (red), and DNA methylation status (metCyt [red]). Germ cells are stained with Oct4 or Stella/PGC7 antibody (green). In panels A, B, and F, DNA is stained with DAPI or TOTO3 (blue). Bars, 100 μm (A, left panels), 200 μm (A, right panels), 300 μm (D), and 10 μm (F).
Homozygous IfitmDel germ cells were also found to colonize the genital ridges and initiate apparently normal development (Fig. 2D). We found no significant differences in the density of germ cells in wt and mutant genital ridges at E12.5 (Mann-Whitney U test) (Fig. 2E). Using antibody staining against Ifitm3 on IfitmDel genital ridges, we confirmed the absence of Ifitm3 protein in the mutants (Fig. 2D; see Fig. S4B in the supplemental material).
Apart from a characteristic gene expression profile, PGCs demonstrate a characteristic “signature” of chromatin modifications (3, 29; P. Hajkova and K. Ancelin, personal communication). We analyzed gonadal germ cells for methylation on histone 3 lysine 9 and histone 3 lysine 27 (29), and found these to be indistinguishable between homozygous and heterozygous IfitmDel and wt germ cells (Fig. 2F). Furthermore, we noted the absence of methylcytosine staining in mutant germ cells at E12.5 as seen in the germ line of wt embryos at this stage (Fig. 2F) (14).
Adult homozygous IfitmDel animals are fertile.
To verify that IfitmDel mutants are fertile, we set up appropriate test matings with F1 wt mating partners (Fig. 3A). The average litter size produced by homozygous IfitmDel animals was not significantly different from litter sizes of heterozygous IfitmDel or wt control animals (Kruskal-Wallis test). Intercrosses between homozygous IfitmDel males and females also produced healthy litters of a normal size (data not shown). Consistent with these results, the testes and ovaries from homozygous IfitmDel mutants appeared normal (Fig. 3B and C). Homozygous IfitmDel ovaries contained follicles at various stages of development (primary, secondary, and Graafian follicles) and a number of corpi lutei. Mutant testes showed seminiferous tubules comprising cells of all stages of spermatogenic maturation. A cross section through the ducti efferentes epididymi showed large quantities of maturing spermatozoae (Fig. 3C). Taken together, these results confirm that loss of the Ifitm gene cluster does not affect late germ cell development or gametogenesis.
FIG. 3.
IfitmDel−/− animals are fertile. (A) Matings of IfitmDel mutant males and females to wt F1 mating partners produced normal-sized litters compared to control matings. S.D., standard deviation; S.E.M., standard error of the mean. (B) Ovaries and testes of adult IfitmDel−/− and wt control mice. (C) Paraffin sections of gonads from adult IfitmDel−/− and wt control mice stained with hematoxylin-eosin. cl, corpus luteum; I, primary follicle; II, secondary follicle; III, Graafian follicle; st, seminiferous tubule; de, ducti efferentes epididymi. Bars, 1 mm (B, upper panels; C, upper larger panels), 3 mm (B, bottom panels), 200 μm (C, bottom panels), and 100 μm (C, upper small panels).
In summary, homozygous IfitmDel embryos show normal specification of PGCs, which subsequently proliferate and migrate into the genital ridges as seen in wt embryos. Mutant gonadal germ cells undergo genome-wide DNA demethylation and demonstrate a normal epigenetic status. The mutation furthermore has no detectable effect on gametogenesis or fertility. Therefore, the five Ifitm genes appear to be dispensable for development of mouse germ cells.
Targeted mutation of Ifitm3 does not affect the germ line.
Since two previous studies (19, 33) suggested a role for mouse Ifitm genes in development, our results using an engineered deletion of the entire Ifitm gene cluster are unexpected. We reasoned that this disparity may occur if individual Ifitm family genes have opposing effects, which may be neutralized by deletion of all the family members. This hypothesis has some credence based on the findings of Tanaka et al., which indicated that single, but not combined, changes in the expression levels of individual members of the Ifitm family could affect germ cells (33). We decided, therefore, to target only Ifitm3, which has the most prominent expression in early PGCs and has been suggested to influence PGC migration (33).
We generated an Ifitm3 null allele through homologous recombination in ES cells by targeted insertion of EGFP 30 nucleotides downstream from and in frame with the Ifitm3 start codon (Fig. 4A). Correct targeting was confirmed by Southern blot analysis (Fig. 4B), and the mutant ES cells were then used to transmit the allele through the germ line onto the C57BL/6Jc/c background. Note that the puromycin selection cassette was excised from the allele using a germ cell-specific Cre deleter to generate the Ifitm3EGFP mutant allele. We found that intercrosses between heterozygous Ifitm3EGFP animals gave rise to homozygous Ifitm3EGFP mutants, which were indistinguishable from their heterozygous or wt littermates (35% Ifitm3EGFP/Ifitm3EGFP, 44% Ifitm3EGFP/wt, 21% wt; n = 63). Genotyping was done by genomic PCR (Fig. 4C). Similarly, homozygous Ifitm3EGFP embryos showed outwardly normal development (Fig. 4D and data not shown).
FIG. 4.
Disruption of the Ifitm3 gene locus through targeted insertion of EGFP does not affect development and germ cell lineage establishment. (A) Schematic of the targeting strategy used to generate the Ifitm3EGFP allele. (B) Southern blot analysis of targeted ES cell clones prior to Cre-mediated excision. (C) PCR-based genotyping of Ifitm3EGFP mutant mice on tail extracts. (D) Ifitm3EGFP/Ifitm3EGFP mutants and wt littermates at E12.5. (E) Projection image of whole-mount immunohistochemistry on Ifitm3EGFP/Ifitm3EGFP and Ifitm3EGFP/wt littermates at E8.5 (lateral view). PGCs are stained with anti Stella/PGC7 antibody (green). The left panel shows lateral view of an embryo of the appropriate age; arrowheads indicate localization of PGCs. al, allantois. (F) Cryosections of E8.5 Ifitm3EGFP/Ifitm3EGFP and wt embryos show migrating PGCs within the hindgut (hg) endoderm stained for alkaline phosphatase (AP). (G) Immunohistochemistry on cryosections of Ifitm3EGFP mutant and wt genital ridges at E12.5 for germ cell marker Oct4. Extranuclear Oct4 staining outside the outlined genital ridge (gr) is due to unspecific secondary antibody binding. ms, mesonephros. (G) Immunohistochemistry on genital ridges of E12.5 Ifitm3EGFP/wt and Ifitm3EGFP/Ifitm3EGFP mice, for GFP (green), Ifitm3 (red), Oct4 (green), and Stella/PGC7 (red), respectively. In panels E and H DNA is stained with DAPI (blue). Bars, 2.25 mm (D), 200 μm (E), 100 μm (F), 300 μm (G), and 10 μm (H).
We next examined PGC specification in homozygous Ifitm3EGFP mutants but again found no detectable effect, since Stella/PGC7-positive PGCs were found in homozygous Ifitm3EGFP embryos at E7.5 (data not shown). Furthermore, the PGCs from Ifitm3EGFP homozygous mutants initiated and completed migration within the hindgut endoderm to populate the genital ridges at E12.5 (Fig. 4E and F). These gonadal germ cells showed expression of Oct4 and Stella/PGC7, similar to the case for germ cells found in heterozygous Ifitm3EGFP or wt embryos (Fig. 4G and H). As expected, no homozygous Ifitm3EGFP germ cells showed wt Ifitm3 protein, but they were positive for GFP (Fig. 4H). Adult homozygous Ifitm3EGFP mutants were fertile and gave rise to healthy litters. The average litter sizes from matings to wt C57BL/6 mice were 8.87 for Ifitm3EGFP/Ifitm3EGFP mutants (15 litters/7 animals tested; standard deviation, 3.2; standard error of the mean, 0.83) and 7.83 for wt controls (6 litters/4 animals tested; standard deviation, 3.06; standard error of the mean, 1.25). In summary, there were no detectable effects on embryogenesis or germ cell development in Ifitm3 null animals.
DISCUSSION
Previous studies showed that the closely related Ifitm gene family members exhibit wide-ranging expression, including prominent and overlapping expression in developing germ cells and neighboring somatic tissues (16, 26, 32, 33). We here show that deletion of the entire Ifitm gene cluster, thereby compensating for any possible redundancy of function, does not, however, result in a detectable effect during development to adulthood. Pursuing this finding, we also generated a second mutation of Iftm3 alone, first due to the genes' particularly marked expression in the proximal epiblast at the time these cells acquire germ cell competence (16, 26). During this process, Ifitm3 in particular might play a critical role by promoting homotypic adhesion among PGC precursors (26). Second, and more importantly, we reasoned that deletion of a single gene in the cluster could disrupt possible interactions between cells mediated by different Ifitm family members, which might not be seen when the whole gene cluster is deleted. However, as already apparent for the cluster deletion, we observed no effect in Ifitm3EGFP mutants on germ cell development or indeed elsewhere, as normal and fertile homozygous adults were obtained.
Specifically concerning germ cell development, our observations have important implications for the proposed model of “homing and repulsion” mediated by Ifitm genes (33). This model suggests that different Ifitms regulate interactions between germ cells and neighboring somatic cells during the migration of early PGCs from mesoderm into endoderm (33). Tanaka and colleagues propose that Ifitm3 activity acts as an attractive guidance cue for PGC localization and counteracts the repulsive activity of Ifitm1 on PGCs. It is based on the suggested downregulation of Ifitm1 expression in migrating PGCs at E8.5 (33). We, however, do not observe such a downregulation of Ifitm1 in PGCs using single-cell cDNA amplification by PCR (see Fig. S1B in the supplemental material). In addition, loss of function of Ifitm3, either as part of the family cluster or individually, did not perturb the migration or further development of germ cells or indeed embryos to adulthood.
Furthermore, we show that loss of Ifitm1 in the context of deletion of all family members has no detectable phenotypic effects. This contrasts with reports which suggested that silencing of Ifitm1 by RNAi leads to defects in somite epithelialization, paraxial mesoderm organization, and PGC guidance (19, 33). This discrepancy likely is a result of the different technical approaches used. While our data are based on loss of function through targeted mutation, previous studies employed “knockdown” by RNAi in ES cell-derived embryos as well as overexpression approaches using cultured embryos (19, 33). In view of our findings, the conclusions drawn from RNAi-mediated silencing require further verification involving evidence that the Ifitm1 knockdown was specific to Ifitm1 to rule out off target effects or ultimately a genetic loss of function of Ifitm1 alone.
There are other key differences in the methods of investigation used in the two studies. Tanaka and colleagues (33) based their conclusions of Ifitm1 essentiality for PGC migration on observations in Ifitm1 RNAi-silenced embryos derived from transgenic ES cell/tetraploid embryo aggregations. Depending on the strain and passage of the ES cells used, embryos generated by this approach often show a developmental delay, and the majority of them do not survive to term (21, 35; B. Payer and S. Barton, personal communication). Since the initial PGC migration from mesoderm into endoderm is a dynamic process that occurs over a short developmental time window, even a slight delay in embryonic development may affect this migration process. However, our studies on mutant embryos developing in vivo do not show any long-term consequences of a possible delay in PGC migration, even assuming that this may occur in some cases. Overall, it is difficult to extrapolate from short-term phenotypic effects caused by either overexpression or knockdown of particular Ifitm genes, restricted mainly to 24 h to 30 h of embryo culture or 1 day of in vivo development, to their long-term consequences. Notably, the proposed role of Ifitm3 in particular was largely based on overexpression of the gene in ectopic tissues. In conclusion, we suggest that the previous findings leading to the “homing and repulsion” model (33) should be reconsidered in view of our strong evidence that as an entity, the clustered Ifitm genes as well as Ifitm3 alone are dispensable for all aspects of PGC development and gametogenesis.
Although we did not detect any effects of the loss of the conserved Ifitm genes on germ cells or development, we did observe an upregulation of Ifitm3 in embryonic fibroblasts in response to treatment with type I interferons (see Fig. S1A in the supplemental material) (16, 32). While interferons have not been reported to play a major role during embryogenesis, they are the key cytokines involved in the regulation of the immune system (31). Both type I and II interferons have the defining ability of conferring an antiviral state on cells, inhibiting viral cell entry, transcription, initiation of translation, and maturation, assembly, and release of viral particles (31). Mouse cells expressing human IFITM1 were shown to be less permissive for vesicular stomatitis virus, demonstrating that Ifitm family members can possess intrinsic antiviral activity (2). Furthermore, mouse Ifitms are present in a variety of immune cells, where they associate among other with the tetraspanin protein CD81, known as a functional component of signaling complexes involved in antigen-specific B-cell activation (30). Also, activation of mouse Ifitm3 has been reported to occur in pancreatic cells during acute-phase caerulin-induced pancreatitis and following systemic lipopolysaccharide treatment and in intestinal cells upon Salmonella infection (25). These results indicate that upregulation of Ifitm genes may be a standard response of tissues during systemic or local immune system stimulation. Therefore, while B- and T-cell counts in unchallenged homozygous IfitmDel mutants appear normal, the lack of Ifitm genes could potentially affect the ability of IfitmDel mutant mice to cope with different pathogenic challenges. Future work is needed to establish whether the Ifitm gene family, while dispensable for development, may be necessary for a functional immune response.
In summary, our data demonstrate that although they are widely expressed at embryonic and adult stages, mutations in the Ifitm gene family do not result in an overt detectable effect on embryonic development, viability, or fertility, contrary to previous reports. It is possible that the loss of Ifitm gene function during development may be compensated for by redundancy through probably sequence-unrelated genes.
Supplementary Material
Acknowledgments
We thank W. Skarnes and L. van der Weyden for helpful discussions and G. Hynes and J. Riseborough for animal care. We are grateful to T. Nakano, T. Jenuwein, and A. Niveleau for reagents; to F. Tang for single-cell cDNAs; and to B. Payer for helpful comments on the manuscript.
U.C.L. is supported by a Wellcome Trust Ph.D. studentship (grant 065601). D.J.A. is supported by Cancer Research UK and The Wellcome Trust. M.A.S. is funded by the BBSRC, The Wellcome Trust, and the EU Epigenome Programme. Open access to the article is funded by the Wellcome Trust.
Footnotes
Published ahead of print on 27 May 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, D. J., P. J. Biggs, T. Cox, R. Davies, L. van der Weyden, J. Jonkers, J. Smith, B. Plumb, R. Taylor, I. Nishijima, Y. Yu, J. Rogers, and A. Bradley. 2004. Mutagenic insertion and chromosome engineering resource (MICER). Nat. Genet. 36867-871. [DOI] [PubMed] [Google Scholar]
- 2.Alber, D., and P. Staeheli. 1996. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J. Interferon Cytokine Res. 16375-380. [DOI] [PubMed] [Google Scholar]
- 3.Ancelin, K., U. C. Lange, P. Hajkova, R. Schneider, A. J. Bannister, T. Kouzarides, and M. A. Surani. 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8623-630. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, R., T. K. Copeland, H. Scholer, J. Heasman, and C. Wylie. 2000. The onset of germ cell migration in the mouse embryo. Mech. Dev. 9161-68. [DOI] [PubMed] [Google Scholar]
- 5.Andreu, P., S. Colnot, C. Godard, P. Laurent-Puig, D. Lamarque, A. Kahn, C. Perret, and B. Romagnolo. 2006. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 661949-1955. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury, L. E., V. S. Goldmacher, and T. F. Tedder. 1993. The CD19 signal transduction complex of B lymphocytes. Deletion of the CD19 cytoplasmic domain alters signal transduction but not complex formation with TAPA-1 and Leu 13. J. Immunol. 1512915-2927. [PubMed] [Google Scholar]
- 7.Bradbury, L. E., G. S. Kansas, S. Levy, R. L. Evans, and T. F. Tedder. 1992. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 1492841-2850. [PubMed] [Google Scholar]
- 8.Clark, J. M., and E. M. Eddy. 1975. Fine structural observations on the origin and associations of primordial germ cells of the mouse. Dev. Biol. 47136-155. [DOI] [PubMed] [Google Scholar]
- 9.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2769-779. [DOI] [PubMed] [Google Scholar]
- 10.Evans, S. S., R. P. Collea, J. A. Leasure, and D. B. Lee. 1993. IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J. Immunol. 150736-747. [PubMed] [Google Scholar]
- 11.Evans, S. S., D. B. Lee, T. Han, T. B. Tomasi, and R. L. Evans. 1990. Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood 762583-2593. [PubMed] [Google Scholar]
- 12.Friedman, R. L., S. P. Manly, M. McMahon, I. M. Kerr, and G. R. Stark. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38745-755. [DOI] [PubMed] [Google Scholar]
- 13.Ginsburg, M., M. H. Snow, and A. McLaren. 1990. Primordial germ cells in the mouse embryo during gastrulation. Development 110521-528. [DOI] [PubMed] [Google Scholar]
- 14.Hajkova, P., S. Erhardt, N. Lane, T. Haaf, O. El-Maarri, W. Reik, J. Walter, and M. A. Surani. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 11715-23. [DOI] [PubMed] [Google Scholar]
- 15.Imai, T., and O. Yoshie. 1993. C33 antigen and M38 antigen recognized by monoclonal antibodies inhibitory to syncytium formation by human T cell leukemia virus type 1 are both members of the transmembrane 4 superfamily and associate with each other and with CD4 or CD8 in T cells. J. Immunol. 1516470-6481. [PubMed] [Google Scholar]
- 16.Lange, U. C., M. Saitou, P. S. Western, S. C. Barton, and M. A. Surani. 2003. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev. Biol. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson, K. A., and W. J. Hage. 1994. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 18268-91. [DOI] [PubMed] [Google Scholar]
- 18.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199417-423. [DOI] [PubMed] [Google Scholar]
- 19.Lickert, H., B. Cox, C. Wehrle, M. M. Taketo, R. Kemler, and J. Rossant. 2005. Dissecting Wnt/beta-catenin signaling during gastrulation using RNA interference in mouse embryos. Development 1322599-2609. [DOI] [PubMed] [Google Scholar]
- 20.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 908424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohinata, Y., B. Payer, D. O'Carroll, K. Ancelin, Y. Ono, M. Sano, S. C. Barton, T. Obukhanych, M. Nussenzweig, A. Tarakhovsky, M. Saitou, and M. A. Surani. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436207-213. [DOI] [PubMed] [Google Scholar]
- 23.Payer, B., M. Saitou, S. C. Barton, R. Thresher, J. P. Dixon, D. Zahn, W. H. Colledge, M. B. Carlton, T. Nakano, and M. A. Surani. 2003. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 132110-2117. [DOI] [PubMed] [Google Scholar]
- 24.Reid, L. E., A. H. Brasnett, C. S. Gilbert, A. C. Porter, D. R. Gewert, G. R. Stark, and I. M. Kerr. 1989. A single DNA response element can confer inducibility by both alpha- and gamma-interferons. Proc. Natl. Acad. Sci. USA 86840-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ropolo, A., R. Tomasini, D. Grasso, N. J. Dusetti, M. C. Cerquetti, J. L. Iovanna, and M. I. Vaccaro. 2004. Cloning of IP15, a pancreatitis-induced gene whose expression inhibits cell growth. Biochem. Biophys. Res. Commun. 3191001-1009. [DOI] [PubMed] [Google Scholar]
- 26.Saitou, M., S. C. Barton, and M. A. Surani. 2002. A molecular programme for the specification of germ cell fate in mice. Nature 418293-300. [DOI] [PubMed] [Google Scholar]
- 27.Sato, M., T. Kimura, K. Kurokawa, Y. Fujita, K. Abe, M. Masuhara, T. Yasunaga, A. Ryo, M. Yamamoto, and T. Nakano. 2002. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 11391-94. [DOI] [PubMed] [Google Scholar]
- 28.Sato, S., A. S. Miller, M. C. Howard, and T. F. Tedder. 1997. Regulation of B lymphocyte development and activation by the CD19/CD21/CD81/Leu 13 complex requires the cytoplasmic domain of CD19. J. Immunol. 1593278-3287. [PubMed] [Google Scholar]
- 29.Seki, Y., K. Hayashi, K. Itoh, M. Mizugaki, M. Saitou, and Y. Matsui. 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 278440-458. [DOI] [PubMed] [Google Scholar]
- 30.Smith, R. A., J. Young, J. J. Weis, and J. H. Weis. 2006. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 7113-121. [DOI] [PubMed] [Google Scholar]
- 31.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, S. S., and Y. Matsui. 2002. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech. Dev. 119(Suppl 1)S261-S267. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, S. S., Y. L. Yamaguchi, B. Tsoi, H. Lickert, and P. P. Tam. 2005. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell 9745-756. [DOI] [PubMed] [Google Scholar]
- 34.van der Weyden, L., D. J. Adams, L. W. Harris, D. Tannahill, M. J. Arends, and A. Bradley. 2005. Null and conditional semaphorin 3B alleles using a flexible puroDeltatk loxP/FRT vector. Genesis 41171-178. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Z. Q., F. Kiefer, P. Urbanek, and E. F. Wagner. 1997. Generation of completely embryonic stem cell-derived mutant mice using tetraploid blastocyst injection. Mech. Dev. 62137-145. [DOI] [PubMed] [Google Scholar]
- 36.Zheng, B., M. Sage, E. A. Sheppeard, V. Jurecic, and A. Bradley. 2000. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol. 20648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.