Abstract
The transcription factors Sox4 and Sox11 are important regulators of diverse developmental processes including heart, lung, pancreas, spleen, and B-cell development. Here we have studied the role of the related Sox12 as the third protein of the SoxC group both in vivo and in vitro. Despite widespread Sox12 expression during embryonic development, Sox12-deficient mice developed surprisingly normally, so that they were born alive, showed no gross phenotypic abnormalities, and were fertile in both sexes. Comparison with the related Sox4 and Sox11 revealed extensive overlap in the embryonic expression pattern but more uniform expression levels for Sox12, without sites of particularly high expression. All three Sox proteins furthermore exhibited comparable DNA-binding characteristics and functioned as transcriptional activators. Sox12 was, however, a relatively weak transactivator in comparison to Sox11. We conclude that Sox4 and Sox11 function redundantly with Sox12 and can compensate its loss during mouse development. Because of differences in expression levels and transactivation rates, however, functional compensation is not reciprocal.
Transcription factors of the Sox family of high-mobility-group domain-containing proteins are important developmental regulators in all animals for which they have been studied so far (3, 31). Their role has been particularly well analyzed for vertebrates and, among vertebrates, for mammals. Here, 20 different Sox proteins exist whose functions are required for a wide range of processes from early blastocyst development to the development of various tissues and organ systems (22). These Sox proteins can furthermore be classified according to sequence similarity in several subgroups. Members of a common group usually are not only related in sequence but also expressed in a strongly overlapping manner and tend to exhibit at least partially redundant functions at sites of coexpression (32).
Group C consists of Sox4, Sox11, and Sox12, which are jointly referred to as SoxC proteins. Both Sox4 and Sox11 are widely expressed during embryogenesis, with particularly high levels in the developing nervous system (5, 7, 11, 20, 23, 29). Sox4 plays essential roles in heart outflow tract formation and early B-cell, late T-cell, endocrine pancreas, and osteoblast development (15, 16, 23, 34). As a result of the impaired development of the endocardial ridges into the semilunar valves and the outlet portion of the ventricular septum of the heart, Sox4-deficient mice die around 14 days postcoitum (dpc) from circulatory failure, with B-cell development being stalled at the pro-B-cell stage (23). Sox11, on the other hand, is required for body wall closure and spleen formation as well as proper development of the eye, the skeleton, the lung, and the heart (25). Sox11-deficient mice die at birth from congenital cyanosis caused by severe heart defects including common arterial trunk and ventricular septation defects. Other phenotypic defects at the time of death include dysgenesis of the anterior eye segment, various cranial and noncranial skeletal malformations, persistent herniation of the gut, asplenia, and hypoplasia of several other organs including the stomach, the pancreas, and the lung (25, 35). In the developing nervous system, Sox4 and Sox11 have largely redundant functions in the establishment of neuronal properties (1) and in glial maturation (8, 11, 17).
In contrast to what is the case for these two SoxC proteins, very little is known about Sox12. There is a single report on the human SOX12 gene (at the time still referred to as SOX22), which was mapped to human chromosome 20p13 (9). In that same report, SOX12 was found to be expressed in many developing human tissues and organs, including the developing peripheral nervous system and central nervous system (CNS). In this respect, SOX12 expression closely resembled the expression of its SoxC gene relatives.
To analyze its function, we here deleted the Sox12 gene in the mouse. From the phenotypes of Sox4- and Sox11-deficient mice and the widespread expression of Sox12, we expected embryonic to perinatal lethality but found Sox12-deficient mice to be surprisingly normal on a gross morphological level and accordingly viable and fertile. To put this unexpected phenotype in a better perspective, we performed an extended analysis of the embryonic expression sites and biochemical properties of Sox12. This analysis led us to conclude that Sox12 is functionally similar to but less prominent than Sox4 and Sox11 and possibly functions during mouse embryogenesis by modulating the activities of the related Sox4 and Sox11.
MATERIALS AND METHODS
Plasmids.
Genomic sequence from the Sox12 locus of C57BL/6J mice was obtained from the bacterial artificial chromosome clone RP23-396N8. A 9.1-kb XbaI/ScaI fragment spanning the Sox12 gene was modified such that a loxP site was introduced into the XhoI site of the 5′-untranslated region 268 bp upstream of the start codon. Additionally, a neomycin resistance cassette flanked by loxP and FLP recombination target (FRT) sites and followed by an internal ribosome entry site-enhanced green fluorescent protein (IRES-EGFP) cassette was inserted into the HindIII site of the 3′-untranslated region, and the modified Sox12 gene was introduced between the XbaI/SalI sites of pTV-0 (Fig. 1A).
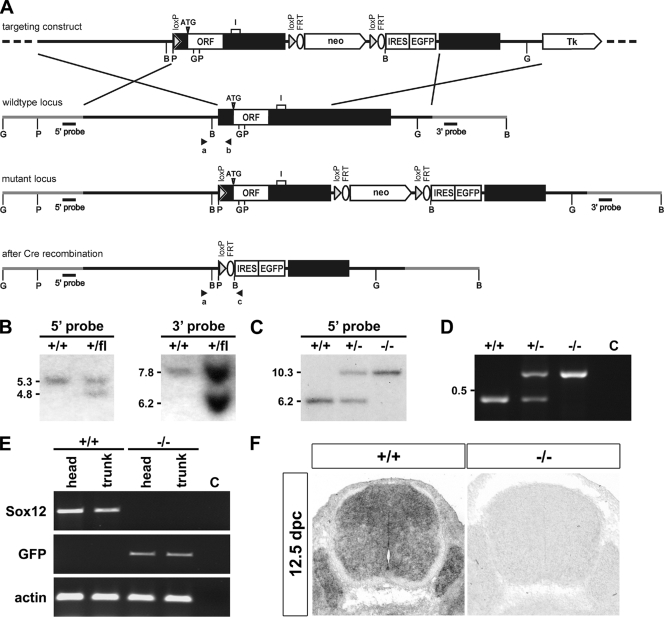
FIG. 1.
Targeted disruption of Sox12 in mice. (A) Schematic representation of the targeting construct (top), the Sox12 wild-type locus (upper middle), and the mutant locus before Cre recombination in ES cells (lower middle) and after Cre recombination in mice (bottom). The transcribed region of the Sox12 gene is shown as a box, and flanking regions are shown as bars. Sox12 coding sequences (open reading frames [ORF]), including the position of the start codon (ATG), an intron (I) predicted in the annotated mouse genome, and neomycin resistance cassette (neo), IRES-EGFP cassette, loxP, and FRT sites, are highlighted. Tk, thymidine kinase. Regions of homology, in which recombination between wild-type locus and targeting vector occurred, are marked by crossed lines. Restriction sites for BamHI (B), BglII (G), and PstI (P) are shown as well as the locations of 5′ and 3′ probes and of primers a, b, and c for genotyping (arrowheads). (B) Southern blot analysis of DNA from ES cells before (+/+) and after (+/flox [fl]) successful homologous recombination. Correct integration of the targeting construct was verified after digestion with PstI by use of the 5′ probe and after digestion with BamHI by use of the 3′ probe. The sizes of fragments corresponding to the wild type and the targeted allele are given in kb on the left of the panels. (C) Southern blot analysis of DNA from adult wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice after Cre-mediated deletion. The loss of all sequences between the first and third loxP sites, including the complete Sox12 open reading frame, was verified after digestion with BglII by use of the 5′ probe. The sizes of fragments corresponding to the wild type and the targeted allele are given in kb on the left of the panel. (D) Genotyping PCR on DNA from adult wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice. The lower band of 468 bp is indicative of the wild-type allele, the upper band of 580 bp of the Sox12 deletion allele. (E) Analysis of Sox12 expression in heads and trunks of 12.5-dpc-old wild-type (+/+) and Sox12-deficient (−/−) embryos by RT-PCR using primers specific for Sox12, GFP, and β-actin. (F) In situ hybridization of transverse sections from the trunks of wild-type (+/+) and Sox12-deficient (−/−) embryos at 12.5 dpc by use of an antisense riboprobe complementary to part of the 3′-untranslated region of Sox12.
The mouse Sox12 open reading frame was also inserted between the HindIII and BamHI sites of pCMV5, yielding the mammalian expression plasmid pCMV/Sox12. Analogous pCMV5-based expression plasmids for Sox4, Sox11, Oct6, and Brn2 as well as the luciferase reporter plasmids 3×SX-luc and 3×FXO-luc were described previously (11, 12, 24). Additionally, we PCR amplified a 622-bp fragment spanning positions −566 to +54 of the Tubb3 promoter (1) from mouse genomic DNA and inserted it between the SacI and BglII sites of the luciferase reporter plasmid pGL2 (Promega). For in ovo electroporation experiments, cDNAs for Sox12, Sox11, and Sox4 were inserted behind the chicken β-actin promoter and upstream of an IRES-GFP cassette into pCAGGS-IRES-nls-GFP (gift of M. Cheung and J. Briscoe, NIMR, London, England).
Gene targeting and generation of mouse mutants.
The targeting vector was linearized with ClaI before electroporation into the F1 embryonic stem (ES) cell line V6.5 (C57BL/6J × 129Sv) (19), which was then selected with G418 (200 μg per ml) and ganciclovir (2 μM). Selected ES cell clones were screened by Southern blotting with a 320-bp 5′ probe which recognized a 5.3-kb fragment of the wild-type allele and a 4.8-kb fragment of the targeted allele in genomic DNA digested with PstI (Fig. 1A and B). Appropriate integration of the 3′ end of the targeting construct was verified using a 581-bp 3′ probe on ES cell DNA digested with BamHI. This probe hybridized to a 6.2-kb fragment of the targeted allele as opposed to a 7.8-kb fragment of the wild-type allele (Fig. 1A and B). Targeted ES cells were injected into C57BL/6J blastocysts to generate chimeras, and chimeras were crossed with C57BL6/J mice carrying the EIIa-Cre transgene (13) to achieve germ line transmission and simultaneous Cre-mediated deletion of the Sox12 open reading frame and the neomycin resistance cassette from the targeted allele. Both were verified by Southern blotting using the 320-bp 5′ probe, which recognized a 6.2-kb fragment of the wild-type allele and a 10.3-kb fragment of the Cre-deleted allele in genomic DNA digested with BglII (Fig. 1C). Homozygous mutant mice were generated by heterozygote intercrosses. Genotyping was routinely performed by PCR analysis using a common upper primer located 685 to 667 bp upstream of the start codon (5′-GGA GAA CAG ATG GGC AGC G-3′) and two lower primers located 236 to 218 bp upstream of the start codon in Sox12 (5′-GGC CAG TAG AGC TCC TCC G-3′) and 53 to 71 bp downstream of the start of the IRES sequence (5′-CGC ACA CCG GCC TTA TTC C-3′). DNA was obtained from tail tips or, in the case of embryos, from yolk sacs. PCR was performed with 20-μl reaction mixtures containing standard buffer, 10% dimethyl sulfoxide, and 0.25 μM of each primer. The cycling conditions consisted of an initial 2-min denaturing step at 94°C followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 62°C, and 30 seconds at 72°C. A 468-bp fragment was indicative of the wild-type allele and a 580-bp fragment of the targeted allele (Fig. 1D). All analyses described in this work were carried out with littermates of the F2 to F4 generations.
Chicken in ovo electroporation.
Fertilized chick eggs were obtained from Lohmann (Cuxhaven, Germany) and incubated in a humidified incubator at 37.8°C. Embryos were staged according to the work of Hamburger and Hamilton (6). pCAGGS-IRES-nls-GFP-based expression plasmids for Sox4, Sox11, and Sox12 were injected at a concentration of 2 μg/μl into the lumina of Hamburger and Hamilton stage 10 or 11 neural tubes. Then, electrodes were placed at either side of the neural tube and electroporation was carried out using a BTX ECM830 electroporator delivering five 50-millisecond pulses of 30 V. Transfected embryos were allowed to develop for 24 h or 48 h before dissection and analysis.
Histological staining procedures, in situ hybridization, and immunohistochemistry.
Mouse embryos were isolated between 9.5 dpc and 18.5 dpc from staged pregnancies. Chicken embryos were retrieved 1 or 2 days after in ovo electroporation. After 1-h to overnight fixation in 4% paraformaldehyde and embedding in tissue freezing medium (Leica, Bensheim, Germany), embryos were transversely or sagittally cut into 10- to 14-μm sections on a cryotome (Leica, Bensheim, Germany). In situ hybridizations were performed on 14-μm sections by use of digoxigenin-labeled antisense probes corresponding to positions 2015 to 3142 of Sox4 (accession no. XM_344594) and positions 543 to 2035 of Sox11 (accession no. NM_053349); two different antisense probes were used for Sox12, one corresponding to positions 559 to 1293 and one to positions 1932 to 2620 (accession no. NM_011438). For immunohistochemistry, the following primary antibodies were used on 10-μm sections: anti-Tubb3 mouse monoclonal (1:5,000 dilution; Covance), anti-Sox4 guinea pig antiserum (1:1,000 dilution; generated against a peptide spanning amino acids 240 to 333 of rat Sox4), anti-Sox12 rabbit antiserum (1:1,000 dilution; generated against a peptide spanning amino acids 5 to 31 of mouse Sox12 and affinity purified), and anti-Sox11 guinea pig antiserum (1:1,000 dilution; generated against a peptide spanning amino acids 204 to 285 of rat Sox11). Detection of immunoreactivity was with secondary antibodies conjugated to Cy3 immunofluorescent dyes (Dianova). Immunoreactivity and GFP autofluorescence were analyzed and documented using a DMIRB inverted microscope (Leica) equipped with a cooled Spot charge-coupled-device camera (Diagnostic Instruments, Sterling Heights, MI) or an MZFLIII stereomicroscope (Leica) equipped with an Axiocam (Zeiss).
RNA preparation and RT-PCR analyses.
RNA was prepared from whole wild-type and Sox12-deficient embryos at 12.5 dpc as well as from several organs of wild-type and Sox12-deficient embryos at 14.5 dpc, 16.5 dpc, and 18.5 dpc by use of Trizol reagent (Invitrogen). Two micrograms of total RNA from each sample was reverse transcribed in a total volume of 25 μl into cDNA by use of oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (New England Biolabs). From each cDNA solution, 0.2 μl was amplified with primer pairs specific for each of the three SoxC genes and β-actin. The following primers were used: for Sox4, 5′-AGC TTC TTG GCT TCC TAC CT-3′ and 5′-TTG GTA GCC GGA GTA TCT TC-3′, corresponding to positions 168 and 428 of accession no. NM_009238, yielding a product of 261 bp; for Sox11, 5′-ACC CAC AGA AAC CAT TAC GG-3′ and 5′-CTT CCT CAC CAT GGA AAA CG-3′, corresponding to positions 3652 and 4014 of accession no. NM_009234, yielding a product of 363 bp; for Sox12, 5′-CAG TCT GGG GAC AGA GTT CC-3′ and 5′-GAC CAG TGG AGC AGA CTT GG-3′, corresponding to positions 2224 and 2624 of accession no. NM_011438, yielding a product of 401 bp; for β-actin, 5′-CCT GGG CATG GAG TCC TG-3′ and 5′-GGA GCA ATG ATC TTG ATC TTC-3′, yielding a product of 202 bp; and for GFP, 5′-CGG TCA CGA ACT CCA GCA GG-3′ and 5′-GCG ACG TAA ACG GCC ACA AG-3′, yielding a product of 618 bp. PCRs were usually performed in a quantitative manner on a Roche LightCycler according to the manufacturer's instructions by use of the LightCycler-FastStart DNA master SYBR green kit with an annealing temperature of 60°C, and amounts of all PCR products were normalized to those for β-actin. For the nonquantitative standard reverse transcription-PCR (RT-PCR) shown in Fig. 1E, 5′-CGA GGT TAC CGA GAT GAT CG-3′ and 5′-GAG GGA TGG TTC AAG CTG AG-3′ (corresponding to positions 1224 and 1813 of NM_011438) were used as Sox12-specific primers, yielding a product of 590 bp.
Transfections and luciferase assays.
HEK 293 cells and Neuro2a cells were kept on 100-mm dishes in Dulbecco's modified Eagle medium containing 10% fetal calf serum in the presence of penicillin and streptomycin. For luciferase assays, cells were seeded on 35-mm dishes and transfected with polyethyleneimine for HEK 293 cells or Superfect (Qiagen, Hilden, Germany) for Neuro2a cells by use of 375 ng of the luciferase reporter plasmid and 15 to 750 ng of the effector plasmids per dish (11). The total amount of plasmid was kept constant using empty pCMV vector where needed. Cells were harvested 48 h after transfection, and extracts were assayed for luciferase activity (12). For the production of recombinant proteins, HEK 293 cells were transfected in 100-mm dishes with 10 μg of a pCMV-based expression plasmid.
Protein extracts, electrophoretic mobility shift assay, and Western blotting.
Whole-cell extracts were prepared and used for electrophoretic mobility shift assays using the 32P-labeled FXO and SX oligonucleotides as probes and poly(dGC) as a nonspecific competitor under standard conditions. Double-stranded oligonucleotides containing the S23 site of the Tubb3 promoter in the wild type or an altered S23M version with a mutated Sox binding site (1) served as additional probes. Where indicated, anti-Sox12 antiserum or rabbit antisera generated against amino acids 27 to 202 of rat Brn2 (24) or full-length mouse Oct6 (18) were also added.
Using the mentioned antibodies against SoxC and class III POU proteins and a monoclonal directed against acetylated α-tubulin (1:2,000 dilution; Sigma), Western blotting was performed on whole-cell extracts as described previously (11).
RESULTS
Targeted mutagenesis of Sox12.
The mouse Sox12 gene is localized within the H1 region on mouse chromosome 2, which is syntenic to human chromosome 20p13, the region to which human SOX12 had previously been mapped (9). To delete the Sox12 gene, we generated a targeting construct that contained a loxP site in the 5′-untranslated region (Fig. 1A). Additionally, a neomycin resistance cassette with flanking loxP and FRT sites was inserted into the 3′-untranslated region, followed by an IRES-EGFP cassette. After successful homologous recombination of this targeting construct (Fig. 1B), genetically modified ES cells were transferred into blastocysts. Resulting chimeras were intercrossed with EIIa-Cre transgenic mice (13) to achieve deletion of Sox12 coding sequences and the neomycin resistance cassette during germ line transmission (Fig. 1A and C). Successful deletion was verified by Southern blotting and genomic PCR (Fig. 1C and D). Those Sox12 sequences that were located between the loxP sites in the targeted allele were furthermore not detected by RT-PCR in cDNA prepared from 12.5-dpc-old Sox12−/− embryos, whereas they were clearly present in cDNA from wild-type littermates (Fig. 1E). RT-PCR primers were furthermore placed in such a way that they encompassed a region annotated as a short intron of 264 bp in the mouse genome (www.ensembl.org; transcript identification ENSMUST00000063332) (Fig. 1A). The size of the PCR product obtained with cDNA from wild-type embryos indicates, however, that the putative intronic sequences are part of the transcript. This confirms that Sox12 is also a monoexonic gene, as previously proposed for the other SoxC genes (31). Instead of Sox12 sequences, we detected GFP coding sequences in the cDNA of 12.5-dpc-old Sox12−/− embryos (Fig. 1E). However, GFP transcript levels in Sox12−/− embryos were lower than Sox12 transcript levels in the wild type. This may explain why we failed to detect significant GFP autofluorescence for Sox12−/− mice (data not shown). In situ hybridizations on sections of 12.5-dpc-old embryos with an antisense riboprobe directed against the 3′-untranslated region additionally confirmed the lack of Sox12 expression in the newly generated Sox12−/− mutant (Fig. 1F).
Absence of overt phenotypic alterations in Sox12-deficient mice.
When Sox12+/− mice were intercrossed on a mixed genetic background, all three expected genotypes were obtained at the time of birth and later at the time of weaning. Genotyping of weaned mice furthermore revealed that Sox12−/− mice were present at Mendelian ratios (Fig. 2A), arguing that Sox12 is dispensable for survival during embryogenesis and the first postnatal weeks. This contrasts dramatically with the results obtained for other SoxC genes, as neither Sox4-deficient nor Sox11-deficient mice were viable after birth (23, 25).
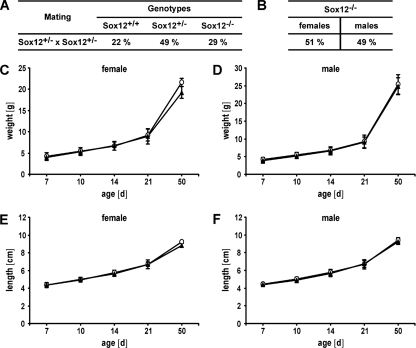
FIG. 2.
Genotype distribution and growth parameters of Sox12-deficient mice after birth. (A) Genotype distribution in Sox12+/− intercrosses as determined at the time of weaning. More than 15 litters were counted. Distribution is shown as the percentage of total littermates. (B) Percent gender distribution of Sox12−/− progeny from Sox12+/− intercrosses. (C and D) Body weights of wild-type (open circles) and homozygous Sox12-deficient (filled triangles) female (C) and male (D) littermates (≥5 for each genotype and sex) were determined during the first 50 days after birth. (E and F) Body lengths from nose to anus were measured for age-matched wild-type (open circles) and homozygous Sox12-deficient (filled triangles) female (E) and male (F) mice during the first 50 days after birth (≥5 for each genotype and sex). Data are means ± standard deviations. No statistically significant differences were detected by use of Student's t test.
Approximately half of the Sox12−/− mice were males and approximately half were females, arguing that gender distribution was not skewed in the absence of Sox12 (Fig. 2B). Crossing of Sox12−/− males with Sox12−/− females furthermore produced litters of normal size (data not shown). We thus have to conclude that Sox12−/− mice are not only viable but also fertile, at least on the mixed genetic background typical for newly generated knockout mice.
We also assessed the growth parameters of Sox12−/− mice during the first 2 months of postnatal development. Already at the earliest time of analysis, male and female Sox12−/− mice were indistinguishable from their wild-type littermates in terms of their weight (Fig. 2C and D). Male and female Sox12−/− mice also gained weight normally during the following weeks. Additionally, we failed to detect any significant differences in body length at birth or during the following weeks of postnatal development in either gender (Fig. 2E and F). Sox12−/− mice and their wild-type littermates were furthermore impossible to tell apart by their outer appearance in the cage. Inspection of the main inner organs did not reveal any obvious alterations in location, shape, or size (data not shown). We thus conclude that neither mouse development nor essential postnatal body functions require Sox12.
Sox12 expression during embryonic development.
To understand why the phenotypic consequences of Sox12 deletion are much less severe than those of Sox4 or Sox11 deletion, we studied the expression patterns of all three SoxC genes during embryogenesis. By quantitative RT-PCR, Sox12 was detectable in cDNA from many developing organs at 14.5 dpc, 16.5 dpc, and 18.5 dpc (Fig. 3A). Sox12 levels were generally highest in the CNS, the kidney, and the heart. Variations in Sox12 expression levels were, however, rather modest among organs. This indicates that Sox12 is widely expressed in the developing mouse embryo and at fairly uniform levels. In terms of widespread expression, Sox12 closely resembled Sox4 (Fig. 3C). Even the expression pattern revealed some similarities between the two Sox genes, as organs with higher levels of Sox12 usually also contained higher levels of Sox4. The differences in Sox4 expression levels among organs were, however, more pronounced than those seen for Sox12 (Fig. 3, compare panels C and A). Sox11, on the other hand, exhibited a preferential expression in the CNS and a low expression in most other organs at 14.5 dpc and 16.5 dpc (Fig. 3B). At 18.5 dpc, even the CNS expression had declined significantly. This agrees well with previous reports of a dramatic overall downregulation of Sox11 expression in the embryo from 14.5 dpc onwards (7, 11).
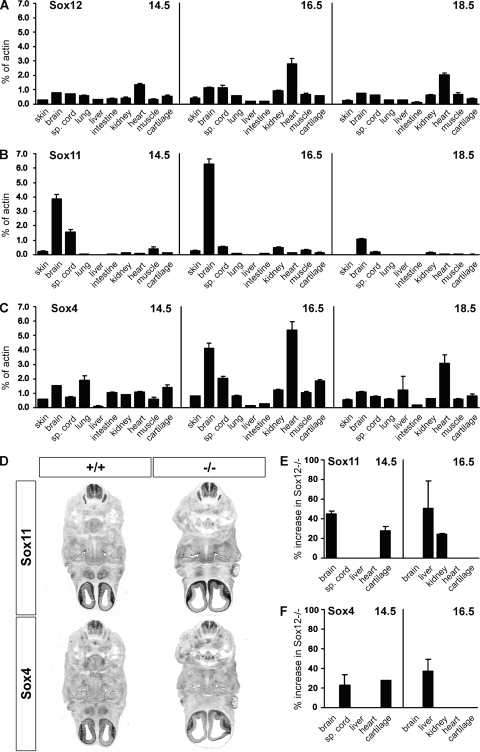
FIG. 3.
RT-PCR analysis of embryonic SoxC gene expression in wild-type and Sox12-deficient embryos. (A to C) Expression of Sox12 (A), Sox11 (B), and Sox4 (C) was determined by quantitative RT-PCR for several tissues of wild-type embryos at 14.5 dpc, 16.5 dpc, and 18.5 dpc, as indicated. sp., spinal. The amounts of PCR products were normalized to the β-actin level. (D) In situ hybridization of transverse sections from wild-type (+/+) and Sox12-deficient (−/−) embryos at 12.5 dpc by use of antisense riboprobes specific for Sox4 and Sox11. Hybridization signals were colorimetrically visualized as bluish precipitates. (E and F) Expression of Sox11 (E) and Sox4 (F) in select tissues of Sox12-deficient embryos at 14.5 dpc and 16.5 dpc was analyzed by quantitative RT-PCR with normalization to the expression level of the β-actin gene and comparison to the corresponding expression in the wild type. Increased expression levels in the Sox12-deficient mutant are presented as percentages.
Expression of Sox4 and Sox11 was also studied for Sox12−/− embryos by in situ hybridization. At 12.5 dpc, no significant differences from the wild type were apparent in levels or patterns of Sox4 and Sox11 expression (Fig. 3D). By quantitative PCR, however, we were able to detect modest, mostly transient upregulation of Sox4, Sox11, or both in select organs of Sox12−/− embryos at 14.5 dpc and 16.5 dpc (Fig. 3E and F). Some organs with increased SoxC transcript levels in Sox12−/− embryos displayed already high transcript levels for Sox4 or Sox11 in the wild type, whereas others did not. Compensatory upregulation of Sox4 and/or Sox11 expression could thus contribute to the lack of a phenotype for Sox12−/− embryos.
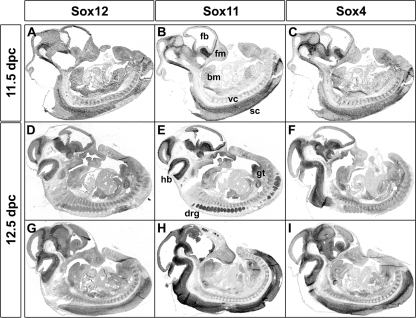
As the resolution of quantitative RT-PCR is limited, we next analyzed embryonic Sox12 expression by in situ hybridization and again included the related Sox4 and Sox11 genes in our study. At 11.5 dpc and 12.5 dpc, all three SoxC genes exhibited widespread expression in the developing embryo (Fig. 4). Expression sites for all three SoxC genes included the CNS along its entire length, the craniofacial mesenchyme, the branchial arch mesenchyme, the skeletogenic mesenchyme of the vertebral column, the dorsal root ganglia, the intestine, and the mesenchyme of the genital tubercle. For both Sox4 and Sox11, the in situ hybridization signal was highest in the CNS. Sox11 furthermore exhibited strong staining in the dorsal root ganglia. Compared to these clear variations in signal intensities, Sox12 signals exhibited a fairly uniform intensity.
FIG. 4.
Determination of early embryonic Sox12 expression by in situ hybridization. Adjacent sagittal sections of wild-type embryos at 11.5 dpc (A to C) and 12.5 dpc (D to I) were hybridized with antisense riboprobes specific for Sox12 (A, D, and G), Sox11 (B, E, and H), and Sox4 (C, F, and I). Sections in panels A to C and G to I are midsagittal, while those in panels D to F are more lateral. Hybridization signals were colorimetrically visualized as bluish precipitates. Abbreviations: bm, branchial arch mesenchyme; fb, forebrain; fm, facial mesenchyme; hb, hindbrain; drg, dorsal root ganglia; gt, genital tubercle; sc, spinal cord; vc, vertebral column mesenchyme.
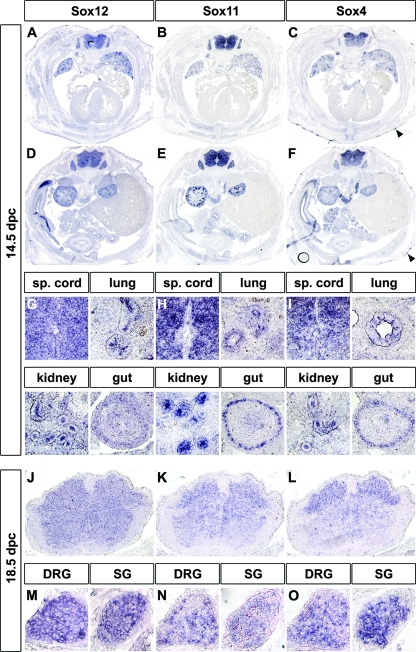
At 14.5 dpc, SoxC gene expression was less widespread than at earlier times. Nevertheless, side-by-side comparison of the expression patterns still revealed a striking overall similarity (Fig. 5A to I). Transverse sections of the trunk revealed the spinal cord, the dorsal root and sympathetic chain ganglia, the lung, the intestine, the kidney, and parts of the forming skeleton as major expression sites for all three SoxC genes (Fig. 5A to F). In the lung, expression was seen both in the mesenchyme and in the epithelium, with expression in the bronchial duct epithelia being stronger for all three SoxC genes than that in the surrounding lung (Fig. 5G to I). In the kidney, expression was detected in the ureteric tips as well as in the condensing metanephrogenic mesenchyme (Fig. 5G to I). Despite this overall similarity, there were also clear differences in the respective expression patterns. A strong expression in the thymus, in the endocardial cushions of the atrioventricular canal of the heart, or in the forming hair follicles was, for instance, characteristic of Sox4, as was a dorsal-to-ventral gradient in spinal cord expression levels (Fig. 5C and F and data not shown) that was still recognizable at 18.5 dpc (Fig. 5J to L). Spinal cord expression of Sox4 and Sox11 was furthermore lower in the ventricular zone than outside, whereas Sox12 expression was fairly uniform (Fig. 5G to I). Sox12 expression also lacked the pronounced expression in the gut epithelium that is observed both for Sox4 and Sox11. Other differences became visible only at later times of development. Whereas all three SoxC genes were expressed at 14.5 dpc in dorsal root ganglia and sympathetic ganglia at high amounts, Sox11 became dramatically reduced by 18.5 dpc (Fig. 5M to O).
FIG. 5.
Determination of late-embryonic Sox12 expression in the trunk by in situ hybridization. Adjacent transverse sections of wild-type embryos were hybridized with antisense riboprobes specific for Sox12 (A, D, G, J, and M), Sox11 (B, E, H, K, and N), and Sox4 (C, F, I, L, and O) at 14.5 dpc (A to I) and 18.5 dpc (J to O). Sections were taken from the trunk at the level of the heart (A to C) or the kidney (D to F). Hybridization signals were colorimetrically visualized as bluish precipitates. The arrowheads in panels C and F point to hair follicles that strongly express Sox4. Magnifications from the overviews (A to F) show spinal (sp.) cord, lung, kidney, and gut at 14.5 dpc (G to I) as well as spinal cord (J to L), dorsal root ganglia (DRG), and sympathetic chain ganglia (SG) at 18.5 dpc (M to O).
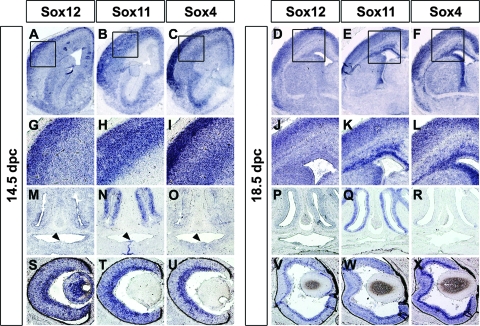
Analysis of the head and brain revealed the same picture of overall similarity in expression patterns among the three SoxC genes but also differences in many details. Thus, all three SoxC genes were strongly expressed in the developing cerebral cortex, thalamus, hippocampus, and cerebellar cortex (Fig. 6A to F and data not shown). In the developing cerebral cortex, Sox12 and Sox4 were enriched in the dorsal cortical plate at 14.5 dpc, whereas Sox11 was not (Fig. 6G to I). At 18.5 dpc, both Sox11 and Sox4 exhibited strong expression in the subventricular zone (Fig. 6K and L). Sox12, by contrast, did not show this subventricular zone enrichment (Fig. 6J). All three SoxC genes were furthermore expressed in many cell types of the developing eye, but lens expression at 14.5 dpc was particularly strong for Sox12 (Fig. 6S to X). The olfactory epithelium, on the other hand, showed predominant expression of Sox11 at both 14.5 dpc and 18.5 dpc (Fig. 6 M to R). Sox11 was also the only SoxC gene significantly expressed at 14.5 dpc at the sites where the two palatal shelves fuse (Fig. 6N).
FIG. 6.
Determination of late-embryonic Sox12 expression in the head by in situ hybridization. Adjacent coronal sections of wild-type embryos were hybridized with antisense riboprobes specific for Sox12 (A, D, G, J, M, P, S, and V), Sox11 (B, E, H, K, N, Q, T, and W), and Sox4 (C, F, I, L, O, R, U, and X) at 14.5 dpc (A, B, C, G, H, I, M, N, O, S, T, and U) and 18.5 dpc (D, E, F, J, K, L, P, Q, R, V, W, and X). Hybridization signals were colorimetrically visualized as bluish precipitates. Shown is a telencephalic hemisphere at low resolution (A to F) as well as high magnifications of the developing cortex (G to L), the olfactory epithelium (M to R), and the eye (S to X). The fusing palatal shelves at 14.5 dpc are marked by arrowheads (M to O).
DNA-binding properties of Sox12.
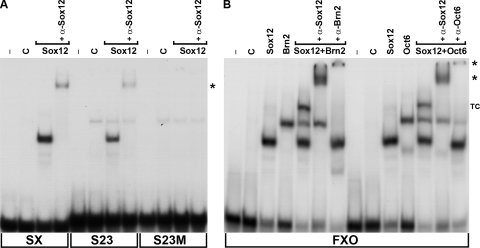
Having established that all three SoxC genes show highly similar and overlapping expression patterns during mouse embryogenesis, we next studied how the DNA-binding activity of Sox12 would compare with that of its close relatives Sox4 and Sox11. For this purpose, electrophoretic mobility shift analyses were performed. Very few target genes have so far been identified for SoxC proteins. As a consequence, the number of known bona fide response elements that could be used for DNA binding studies is relatively low. We and others have previously shown that Sox4 and Sox11 bind to a Sox consensus binding site from the CD3ɛ enhancer in the context of the SX oligonucleotide and mediate reporter gene activation through this site (11, 12, 30). Therefore, we used this site in electrophoretic mobility shift assays in addition to site S23 from the Tubb3 gene as a bona fide response element from a neural SoxC target gene (1). Extracts from HEK 293 cells transfected with an expression plasmid for Sox12 yielded a protein-DNA complex with the SX as well as the S23 oligonucleotide (Fig. 7A). That this complex indeed contained Sox12 was evident from the fact that it was not obtained in extracts from untransfected HEK 293 cells. Antibodies directed against Sox12 supershifted this complex, providing additional proof of the presence of Sox12 in this complex. In contrast, no complex was obtained when the Sox consensus recognition motif was mutated in the S23 oligonucleotide (S23M in Fig. 7A). As previous binding studies with Sox4 and Sox11 led to similar results (1, 11), all three SoxC proteins appear to recognize the same sites.
FIG. 7.
DNA-binding activity of Sox12. (A) The radiolabeled SX site and oligonucleotides containing the S23 site from the Tubb3 promoter in the wild type (S23) or the mutant version (S23M) were incubated in electrophoretic mobility shift assays with extracts from mock-transfected HEK 293 cells (C) or HEK 293 cells ectopically expressing Sox12 as indicated. Supershifted complexes obtained after incubation with a Sox12-specific antibody are marked by an asterisk. −, no extract added. (B) A radiolabeled FXO oligonucleotide with adjacent binding sites for Sox and POU proteins was incubated with extracts from mock-transfected HEK 293 cells or from HEK 293 cells ectopically expressing Sox12, Brn2, or Oct6 as indicated. In the presence of Sox12 and a POU protein, additional complexes were obtained. The presence of both proteins in this ternary complex (TC) was verified by addition of antibodies directed against Sox12 (α-Sox12), Brn2 (α-Brn2), and Oct6 (α-Oct6). Supershifted complexes are marked by asterisks.
Sox12 also bound to the FXO oligonucleotide (Fig. 7B), another site to which the binding of Sox4 and Sox11 has been shown previously (11). This FXO oligonucleotide contained not only the Sox recognition element from the FGF4 enhancer (36) but also an adjacent binding site for POU proteins. It has been previously reported that Sox4 and Sox11 each formed ternary complexes with class III POU proteins such as Oct6 and Brn2 on this oligonucleotide and that this joint binding on target gene enhancers might be crucial for the synergistic activation of gene expression by SoxC and POU proteins in the developing neural tube (11, 27, 33).
Therefore, we also checked Sox12 binding to the FXO oligonucleotide in the presence of either Oct6 or Brn2 (Fig. 7B). When incubated alone with the oligonucleotide, both Sox12 and the POU protein formed complexes of characteristic mobility. In the presence of both Sox12 and Oct6 or Brn2, a complex of lower mobility additionally appeared. This novel complex was supershifted with antibodies directed against Sox12 or against the respective POU protein, confirming its identity as a ternary complex containing DNA as well as Sox and POU protein (Fig. 7B). We conclude that Sox12 is as capable of ternary complex formation with class III POU proteins as are Sox11 and Sox4.
Transactivation capacities of Sox12.
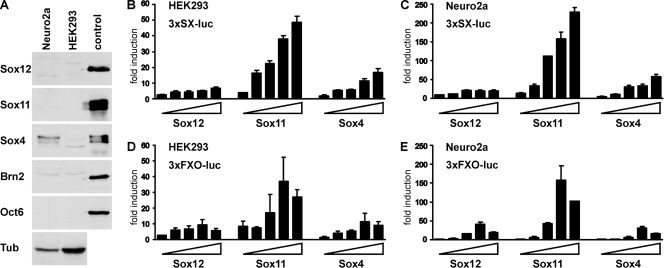
Whether these similar DNA-binding properties would also translate into comparable transactivation capacities was analyzed in luciferase reporter gene assays with transiently transfected HEK 293 cells and Neuro2a cells. The luciferase reporter gene was under the control of artificial promoters that consisted of a minimal promoter combined with multimerized SX or FXO binding sites (12). Of the analyzed SoxC proteins, only Sox4 was endogenously present in significant amounts in Neuro2a cells, and all were absent from HEK 293 cells (Fig. 8A).
FIG. 8.
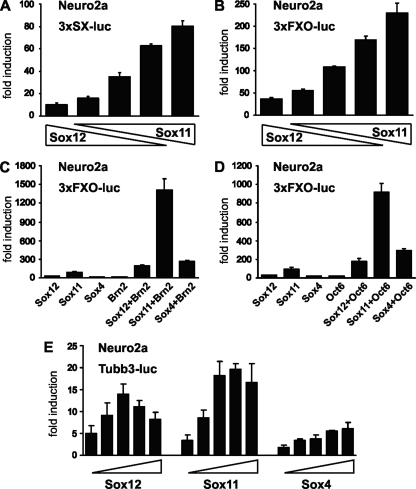
Transactivation capacity of Sox12. (A) Endogenous amounts of SoxC and class III POU proteins in Neuro2a and HEK 293 cells were determined by Western blotting. Extracts from HEK 293 cells transfected with corresponding plasmids served as the positive control. Extract quality was checked with an antibody directed against acetylated α-tubulin (Tub). (B to E) Transient transfections were performed in HEK 293 cells (B and D) and Neuro2a cells (C and E) with the 3×SX-luc (B and C) and 3×FXO-luc (D and E) reporter plasmids (all 375 ng) in the presence of increasing amounts of Sox12, Sox11, and Sox4 (15 ng, 50 ng, 125 ng, 375 ng, and 750 ng). All luciferase activities were determined in two independent experiments, each performed in duplicate, and results are presented as induction levels ± standard errors of the mean.
When the 3×SX-luc reporter was transfected with increasing amounts of Sox4, we obtained a dose-dependent reporter gene activation, with induction rates increasing from 2-fold to 17-fold for HEK 293 cells and from 5-fold to 58-fold for Neuro2a cells (Fig. 8B and C). Induction rates were even higher for Sox11, increasing from 4-fold to 49-fold for HEK 293 cells and from 14-fold to 229-fold for Neuro2a cells. This confirmed previous findings that Sox11 is usually a stronger transcriptional activator than Sox4 (11). When Sox12 was cotransfected, the 3×SX-luc reporter was activated between 2-fold and 7-fold in HEK 293 cells and between 8-fold and 20-fold in Neuro2a cells (Fig. 8B and C). In case of the 3×SX-luc reporter, Sox12 was thus the weakest transactivator of all SoxC proteins in the two tested cell lines.
When the 3×FXO-luc reporter was used instead, kinetics and absolute values of transactivation differed (Fig. 8D and E). With this reporter, activation rates first increased but then dropped again at the highest concentration of the Sox expression plasmid. These kinetics were similarly observed for all three SoxC proteins. Again, clear differences in reporter gene activation were obtained with the three different Sox proteins. In HEK 293 cells, Sox12 activated the 3×FXO-luc reporter up to 9-fold compared to a 12-fold induction for Sox4 and a 37-fold induction for Sox11 (Fig. 8D). In Neuro2a cells, maximal activation rates were 41-fold for Sox12, 158-fold for Sox11, and 30-fold for Sox4 (Fig. 8E). Sox12 and Sox4 were thus again weaker activators of this reporter than Sox11, but the abilities to activate were comparable between Sox4 and Sox12, with Sox4 yielding slightly higher rates than Sox12 for HEK 293 cells and vice versa for Neuro2a cells.
We also investigated how the presence of Sox12 would impact the ability of Sox11 to activate expression of the 3×SX-luc and 3×FXO-luc reporters (Fig. 9A and B). For this purpose, reporter plasmids were transfected with constant total amounts of expression plasmids but different ratios of Sox11 versus Sox12. In both Neuro2a and HEK 293 cells, reporter gene activity reflected the ratio in which expression plasmids were transfected. Induction rates steadily increased with a higher contribution of Sox11 and reached their maximum when only Sox11 was present (Fig. 9A and B and data not shown). From these results, it can be concluded that Sox12 does not actively repress the activity of other SoxC proteins. Considering the differences in transactivation capacities among SoxC proteins, however, their relative ratio could be a major factor in determining the exact transcription rate of target genes.
FIG. 9.
Influence of Sox12 on the transactivation capacities of other SoxC and POU proteins. Transient transfections were performed in Neuro2a cells with 3×SX-luc (A), 3×FXO-luc (B to D), or a reporter plasmid in which the luciferase gene is under control of the Tubb3 promoter (E). Fixed amounts of reporter plasmid (375 ng) were cotransfected with constant total amounts of expression plasmids (375 ng) but various contributions of Sox12 and Sox11 (ratios of 10:0, 8:2, 5:5, 2:8, and 0:10) for panels A and B as indicated below the bars, whereas they were cotransfected with various combinations of expression plasmids for Sox12, Sox11, Sox4, Brn2, and Oct6 or empty vector (400 ng each) for panels C and D. The Tubb3 luciferase reporter plasmid shown in panel E was transfected in the presence of increasing amounts of Sox12, Sox11, and Sox4 (15 ng, 50 ng, 125 ng, 375 ng, and 750 ng). All luciferase activities were determined in three independent experiments, each performed in duplicate. Luciferase activities are given as induction levels ± standard errors of the mean.
We also compared the ability of Sox12 to synergistically activate reporter gene expression with that of the other SoxC proteins. For these studies, the 3×FXO-luc reporter was cotransfected with different combinations of SoxC and class III POU proteins in Neuro2a cells (Fig. 9C and D). None of the class III POU proteins was endogenously expressed in these cells (Fig. 8A). As previously published (11), the 3×FXO-luc reporter was activated much more strongly by combinations of POU protein and Sox11 than by either protein alone. Combinations of Sox11 and Brn2, for instance, led to a 1,400-fold activation, whereas transfection with Sox11 or Brn2 alone activated reporter gene expression only 89-fold or 18-fold, respectively (Fig. 9C). For combinations with Oct6, a 915-fold induction rate was determined (Fig. 9D). This means that activation rates were 13 times higher for Sox11 in combination with Brn2 and 8.5 times higher in combination with Oct6 than the respective additive rates obtained with either transcription factor alone.
Importantly, synergistic effects were also observed when POU proteins were cotransfected with Sox4 or Sox12 (Fig. 9C and D). In combination with Sox4, synergistic activation rates were 7.7 to 8.6 times higher than the sum of activation rates for each transcription factor alone. In case of Sox12, synergistic activation rates were 3.9 to 4.3 times higher than the sum. Similar results were also obtained when HEK 293 instead of Neuro2a cells were used (data not shown). We therefore conclude that all SoxC proteins, including Sox12, have the capacity to function synergistically with class III POU proteins, although Sox12 may be less efficient than Sox4 and Sox11.
Finally, we compared the abilities of all three SoxC proteins to activate the neural Tubb3 target gene promoter. In contrast to the artificial promoters, the Tubb3 promoter was activated only in Neuro2a and not in HEK 293 cells (Fig. 9E and data not shown). In agreement with studies on artificial promoters, Sox11 was again the strongest activator and induced Tubb3 promoter activity up to 20-fold. Sox12, on the other hand, caused up to 14-fold inductions, whereas Sox4-dependent promoter activation reached a plateau at 6-fold inductions.
Functional comparison of SoxC proteins during chicken spinal cord development.
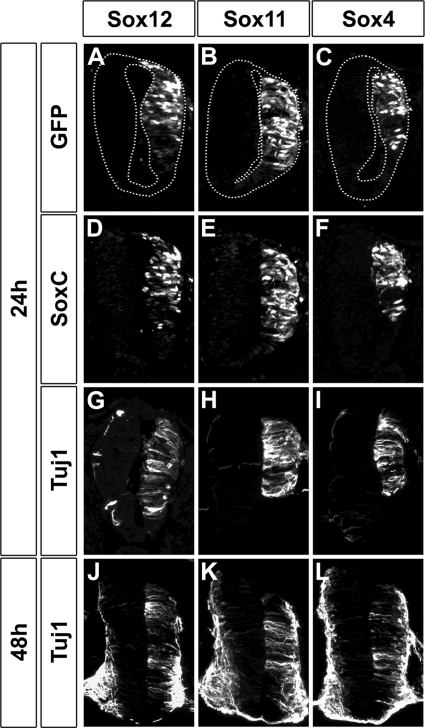
Whether these findings for tissue culture would also translate to the in vivo situation was analyzed by experiments involving electroporation into the chicken neural tube. It was shown previously that overexpression of Sox11 and Sox4 led to precocious and increased expression of Tubb3 (1). When we repeated these experiments with Sox11 and Sox4 (Fig. 10E and F), precocious Tubb3 expression was readily visible 24 h after electroporation (Fig. 10H and I) on the electroporated side, which was marked by the GFP additionally introduced during the electroporation event (Fig. 10B and C). Even 48 h after electroporation, Tubb3 expression was still increased on the electroporated side, although endogenous Tubb3 expression had started (Fig. 10K and L). Furthermore, Tubb3 expression was higher in neural tubes electroporated with Sox11 than in Sox4-electroporated neural tubes, arguing that Sox11 may also be a stronger transactivator than Sox4 in vivo. When Sox12 was electroporated (Fig. 10D), we also observed increased Tubb3 expression on the electroporated side (Fig. 10A) 24 h and 48 h after the electroporation (Fig. 10G and J). Furthermore, the Tubb3 amounts induced by Sox12 electroporation were comparable to those achieved by Sox4 electroporations but consistently lower than those seen for electroporations with Sox11. These results highlight the fact that all three SoxC proteins have transactivation capacities but may nevertheless differ in the efficiencies with which they activate specific targets.
FIG. 10.
Sox12 activity during early chicken spinal cord development. Immunohistochemistry against Sox12 (D), Sox11 (E), Sox4 (F), and Tubb3 (G to L) was performed on transverse sections of chicken embryos electroporated with expression plasmids for Sox12 (A, D, G, and J), Sox11 (B, E, H, and K), or Sox4 (C, F, I, and L) 24 h (A to I) or 48 h (J to L) after the electroporation event. The electroporated side is oriented to the right and marked by GFP autofluorescence (A to C).
DISCUSSION
Similar to its close relatives Sox4 and Sox11, Sox12 is widely expressed during embryonic development in mammals, as shown previously for humans (9) and confirmed here for the mouse. Considering the significant and multiple developmental defects in Sox4-deficient and Sox11-deficient mice (23, 25), the absence of an overt defect in embryonic and early postnatal development in Sox12-deficient mice was unexpected. Instead of the embryonic lethality of Sox4-deficient mice at 14 dpc or the perinatal lethality of Sox11-deficient mice, Sox12-deficient mice were born alive and had a grossly normal appearance. Postnatal growth was similarly unaffected and Sox12-deficient mice were fertile in both sexes, arguing that Sox12 is largely dispensable for embryonic and early postnatal development as well as for postnatal homeostasis. It has to be noted, however, that our Sox12-deficient mice were still on a mixed genetic background. Thus, the possibility that phenotypes will eventually become apparent on a pure genetic background after extensive backcrossing cannot be excluded.
Comparison of the Sox12 expression pattern with those of Sox4 and Sox11 pointed to a strongly overlapping pattern for all three SoxC genes. Although closer inspection revealed differences, it is thus plausible that one reason for the lack of a developmental phenotype for Sox12-deficient mice is functional compensation by coexpressed Sox4 and/or Sox11. In agreement with such an assumption, compensatory increases of Sox4 and Sox11 expression were observed for some organs of Sox12-deficient embryos. In other organs, however, Sox4 or Sox11 expression remained unchanged, arguing that the exact requirements for functional compensation are organ specific.
If functional compensation is invoked, however, it must be nonreciprocal, with Sox4 and Sox11 being able to compensate for the loss of Sox12 but not vice versa. After all, there are severe developmental defects in the Sox4- and the Sox11-deficient animals (23, 25) despite the continued presence of Sox12.
This situation is somewhat reminiscent of previous findings for the SoxE proteins Sox8, Sox9, and Sox10. These three proteins also exhibit significant coexpression during embryogenesis. Loss of Sox9 or Sox10 furthermore leads to severe developmental defects in several cell types, including chondrocytes and several neural crest derivatives, and results in midembryonic-to-perinatal lethality (2, 4), whereas Sox8-deficient mice are born normally and exhibit comparably mild defects during postnatal development (26). In the case of SoxE proteins, this nonreciprocal ability for compensation appears to be caused by differences in expression level as well as by functional differences between the three proteins (10, 14).
In case of SoxC proteins, differences in embryonic expression may also be part of the reason for the nonreciprocal functional compensation. For one, quantitative RT-PCR and in situ hybridizations indicate that Sox12 expression levels are probably a bit lower than expression levels for Sox4 and Sox11. More importantly, Sox12 expression is fairly uniform throughout the embryo. Taking into account that organs with aberrant development in the Sox4- or Sox11-deficient mice are among the prominent expression sites for the respective Sox protein, the absence of preferential high-level expression sites may help to explain the lack of a phenotype for Sox12-deficient mice. We would furthermore predict that Sox12 upregulation in Sox4- and Sox11-deficient mice does not exist or is not sufficient for compensation.
A nonreciprocal functional redundancy among SoxC proteins may additionally be caused by differences in functional properties. Our studies argue that these differences are subtle. SoxC proteins bound similar target sequences in vitro and used these sites for the activation of target gene promoters both in tissue culture and in the chicken neural tube, either alone or in combination with class III POU proteins. In agreement with their similar DNA-binding and transactivation properties, all three SoxC proteins share extensive sequence conservation not only within their DNA-binding HMG domain but also within an approximately 35-amino-acid stretch at the carboxy terminus (7, 9, 11). This conserved carboxy-terminal region likely represents the transactivation domain, which in the case of Sox11 has been previously mapped to the carboxy-terminal third (11).
Importantly, we found no evidence for a function of Sox12 as a transcriptional repressor. Its relation to Sox4 and Sox11 is therefore very different from the one of the SoxB1 proteins Sox1, Sox2, and Sox3 to the SoxB2 proteins Sox14 and Sox21 (32). These two groups of related Sox proteins have been shown to have antagonistic relationships with SoxB1 proteins activating and SoxB2 proteins repressing the same target genes (21, 28).
Nevertheless, differences in the transactivation capacities exist. At least in comparison to Sox11, Sox12 was a very weak transactivator, and Sox12 also exhibited a relatively modest synergistic effect with class III POU proteins compared to both Sox11 and Sox4. Whether these differences in transactivation capacities and synergies are caused by subtle differences in the HMG domains or conserved carboxy-terminal transactivation domains or rather by the nonconserved regions in the respective proteins remains to be analyzed in future experiments. The fact that Sox12 contributes less to the overall activation of SoxC target genes than coexpressed Sox11 may, however, be functionally relevant, as its loss may have much less impact than the loss of Sox11, even if expression levels were comparable. Through coexpression, Sox12 furthermore modulates the expression rates of genes that are under the control of Sox11 and Sox4. The function of Sox12 is thus likely more subtle than the functions of its relatives, and its impact on embryonic development may be revealed only with compound mouse mutants from which other SoxC genes have been additionally deleted.
Acknowledgments
We thank M. Cheung and J. Briscoe for the pCAGGS-IRES-nls-GFP plasmid.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to E.S. (So251/3-1).
Footnotes
Published ahead of print on 27 May 2008.
REFERENCES
- 1.Bergsland, M., M. Werme, M. Malewicz, T. Perlmann, and J. Muhr. 2006. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 203475-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., J. M. Deng, Z. Zhang, R. R. Behringer, and B. de Crombrugghe. 1999. Sox9 is required for cartilage formation. Nat. Genet. 2285-89. [DOI] [PubMed] [Google Scholar]
- 3.Bowles, J., G. Schepers, and P. Koopman. 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227239-255. [DOI] [PubMed] [Google Scholar]
- 4.Britsch, S., D. E. Goerich, D. Riethmacher, R. I. Peirano, M. Rossner, K. A. Nave, C. Birchmeier, and M. Wegner. 2001. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 1566-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, M., M. Abu-Elmagd, H. Clevers, and P. J. Scotting. 2000. Roles of Sox4 in central nervous system development. Mol. Brain Res. 79180-191. [DOI] [PubMed] [Google Scholar]
- 6.Hamburger, H., and H. L. Hamilton. 1953. A series of normal stages in the development of the chick embryo. J. Morphol. 8849-92. [PubMed] [Google Scholar]
- 7.Hargrave, M., E. Wright, J. Kun, J. Emery, L. Cooper, and P. Koopman. 1997. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 21079-86. [DOI] [PubMed] [Google Scholar]
- 8.Hoser, M., S. L. Baader, M. R. Bosl, A. Ihmer, M. Wegner, and E. Sock. 2007. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J. Neurosci. 275495-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jay, P., I. Sahly, C. Goze, S. Taviaux, F. Poulat, G. Couly, M. Abitbol, and P. Berta. 1997. Sox22 is a new member of the Sox gene family, mainly expressed in human nervous tissue. Hum. Mol. Gen. 61069-1077. [DOI] [PubMed] [Google Scholar]
- 10.Kellerer, S., S. Schreiner, C. C. Stolt, M. R. Bösl, and M. Wegner. 2006. Functional equivalency of transcription factors Sox8 and Sox10 is tissue-specific. Development 1332875-2886. [DOI] [PubMed] [Google Scholar]
- 11.Kuhlbrodt, K., B. Herbarth, E. Sock, J. Enderich, I. Hermans-Borgmeyer, and M. Wegner. 1998. Cooperative function of POU proteins and Sox proteins in glial cells. J. Biol. Chem. 27316050-16057. [DOI] [PubMed] [Google Scholar]
- 12.Kuhlbrodt, K., B. Herbarth, E. Sock, I. Hermans-Borgmeyer, and M. Wegner. 1998. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18237-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 935860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maka, M., C. C. Stolt, and M. Wegner. 2005. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 277155-169. [DOI] [PubMed] [Google Scholar]
- 15.Mavropoulos, A., N. Devos, F. Biemar, E. Zecchin, F. Argenton, H. Edlund, P. Motte, J. Martial, and B. Peers. 2005. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev. Biol. 285211-223. [DOI] [PubMed] [Google Scholar]
- 16.Nissen-Meyer, L. S., R. Jemtland, V. T. Gautvik, M. E. Pedersen, R. Paro, D. Fortunati, D. D. Pierroz, V. A. Stadelmann, S. Reppe, F. P. Reinholt, A. Del Fattore, N. Rucci, A. Teti, S. Ferrari, and K. M. Gautvik. 2007. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J. Cell Sci. 1202785-2795. [DOI] [PubMed] [Google Scholar]
- 17.Potzner, M., C. Griffel, E. Lütjen-Drecoll, M. R. Bösl, M. Wegner, and E. Sock. 2007. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol. Cell. Biol. 275316-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renner, K., H. Leger, and M. Wegner. 1994. The POU-domain protein Tst-1 and papovaviral T-antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc. Natl. Acad. Sci. USA 916433-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rideout, W. M., T. Wakayama, T. Wutz, K. Eggan, L. Jackson-Grusby, J. Dausman, R. Yanagimachi, and R. Jaenisch. 2000. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet. 24109-110. [DOI] [PubMed] [Google Scholar]
- 20.Rimini, R., M. Beltrame, F. Argenton, D. Szymczak, F. Cotelli, and M. E. Bianchi. 1999. Expression patterns of zebrafish sox11A, sox11B and sox21. Mech. Dev. 89167-171. [DOI] [PubMed] [Google Scholar]
- 21.Sandberg, M., M. Källstrom, and J. Muhr. 2005. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 8995-1001. [DOI] [PubMed] [Google Scholar]
- 22.Schepers, G. E., R. D. Taesdale, and P. Koopman. 2002. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor families. Dev. Cell 3167-170. [DOI] [PubMed] [Google Scholar]
- 23.Schilham, M. W., M. A. Oosterwegel, P. Moerer, J. Ya, P. A. J. Deboer, M. Vandewetering, S. Verbeek, W. H. Lamers, A. M. Kruisbeek, A. Cumano, and H. Clevers. 1996. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380711-714. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber, J., J. Enderich, E. Sock, C. Schmidt, C. Richter-Landsberg, and M. Wegner. 1997. Redundancy of class III POU proteins in the oligodendrocyte lineage. J. Biol. Chem. 27232286-32293. [DOI] [PubMed] [Google Scholar]
- 25.Sock, E., S. D. Rettig, J. Enderich, M. R. Bösl, E. R. Tamm, and M. Wegner. 2004. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 246635-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sock, E., K. Schmidt, I. Hermanns-Borgmeyer, M. R. Bösl, and M. Wegner. 2001. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol. Cell. Biol. 216951-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka, S., Y. Kamachi, A. Tanouchi, H. Hamada, N. Jing, and H. Kondoh. 2004. Interplay of SOX and POU factors in regulation of the nestin gene in neural primordial cells. Mol. Cell. Biol. 248834-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchikawa, M., Y. Kamachi, and H. Kondoh. 1999. Two distinct subgroups of group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 84103-120. [DOI] [PubMed] [Google Scholar]
- 29.Uwanogho, D., M. Rex, E. J. Cartwright, G. Pearl, C. Healy, P. J. Scotting, and P. T. Sharpe. 1995. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 4923-36. [DOI] [PubMed] [Google Scholar]
- 30.van de Wetering, M., M. Oosterwegel, K. van Norren, and H. Clevers. 1993. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 123847-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegner, M. 1999. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 271409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegner, M., and C. C. Stolt. 2005. From stem cells to neurons and glia: a soxist's view of neural development. Trends Neurosci. 28583-588. [DOI] [PubMed] [Google Scholar]
- 33.Wiebe, M. S., T. K. Nowling, and A. Rizzino. 2003. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 27817901-17911. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, M. E., K. Y. Yang, A. Kalousova, J. Lau, Y. Kosaka, F. C. Lynn, J. Wang, C. Mrejen, V. Episkopou, H. C. Clevers, and M. S. German. 2005. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes 543402-3409. [DOI] [PubMed] [Google Scholar]
- 35.Wurm, A., E. Sock, R. Fuchshofer, M. Wegner, and E. R. Tamm. 2008. Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Exp. Eye Res. -907.86895. [DOI] [PubMed] [Google Scholar]
- 36.Yuan, H. B., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 92635-2645. [DOI] [PubMed] [Google Scholar]