Abstract
The ability of the MBD4 glycosylase to excise a mismatched base from DNA has been assessed in vitro using DNA substrates with different extents of cytosine methylation, in the presence or absence of reconstituted nucleosomes. Despite the enhanced ability of MBD4 to bind to methylated cytosines, the efficiency of its glycosylase activity on T/G mismatches was slightly dependent on the extent of methylation of the DNA substrate. The reduction in activity caused by competitor DNA was likewise unaffected by the methylation status of the substrate or the competitor. Our results also show that MBD4 efficiently processed T/G mismatches within the nucleosome. Furthermore, the glycolytic activity of the enzyme was not affected by the positioning of the mismatch within the nucleosome. However, histone hyperacetylation facilitated the efficiency with which the bases were excised from the nucleosome templates, irrespective of the position of the mismatch relative to the pseudodyad axis of symmetry of the nucleosome.
Diverse organisms, prokaryotic and eukaryotic, methylate selected cytosines in their genomes (reviewed by Bestor [19]). In bacteria, methylation is usually associated with restriction-modification systems which protect the cell against attack from bacteriophages. Cytosine methylation is associated with genome defense in some eukaryotes, but it also plays vital roles in development and in the control of gene expression, to the extent that the homozygous knockout of cytosine methyltransferase genes in the mouse is an embryonic lethal mutation (40). Despite its importance, cytosine methylation imposes a genetic and epigenetic cost because 5-methylcytosines deaminate into thymine (reviewed in references 53 and 67). Unrepaired, the resulting T/G mismatches lead to CG-to-TA transition mutations. There is also evidence from work with Escherichia coli that 5-methylcytosines are more prone to mispairing during DNA replication than unmethylated bases and/or are less well repaired and that they are particularly susceptible to other forms of endogenous and exogenous damage which cause them to mispair (24, 54). A similar situation exists for eukaryotes (reviewed in reference 51).
Whatever the cause, mutations at methylated cytosines may influence phenotype in two ways: (i) by introducing nonsense or missense mutations which alter the functions of proteins and (ii) by demethylating promoter regions and thereby altering patterns of gene expression. As one line of defense against the genetic and epigenetic effects of mutations associated with methylated cytosines, cells have developed repair systems specific to T/G mismatches. Cytosine methyltransferases methylate both strands of palindromic recognition sequences (CpG or CNG sequences in eukaryotes and CCWGG sequences in E. coli); consistent with their role in preventing mutations at 5-methylcytosines, the DNA repair systems show a strong preference for T/G mismatches which arise in methylase recognition sequences (31, 32, 46).
E. coli initiates repair with a single-stranded endonuclease, Vsr (32), while eukaryotes use one or more glycosylases (reviewed in reference 67). Human cells have two T/G-specific glycosylases, TDG (thymine DNA glycosylase) (46) and MBD4 (methyl-binding domain protein 4) (18, 31). While TDG is strictly a glycosylase, MBD4 is a bipartite protein containing a C-terminal glycosylase domain and an N-terminal methyl-binding domain. The latter domain has recently been shown to be a transcriptional inhibitor, functioning equally well with and without the associated glycosylase domain (37). Surprisingly, the activity of the glycosylase is likewise unaffected by the presence of the methyl-binding domain (51). Despite the aforementioned preference of the enzyme for methylase recognition sites, there is some disagreement about whether the activity of the intact protein is affected by the presence or absence of a methyl group on the DNA strand opposite the mismatched T (52, 65, 66).
Structural considerations make it unlikely that the methyl-binding and the glycosylase domains can bind simultaneously to the same dinucleotide. Thus, it has been hypothesized previously that the methyl-binding domain serves to target MBD4 to highly methylated regions of the genome, making it readily available in the event of a T/G mismatch (52, 71). To test this hypothesis, we have assayed MBD4 glycosylase activity in vitro using T/G-containing substrates with multiple, methylatable cytosines. We have also determined the effect on enzyme activity of competition from excess, methylated, homoduplex DNA. Our results indicate that high local concentrations of methyl groups do not affect enzyme activity.
In eukaryotes, DNA repair systems operate on a chromatin template, not naked DNA (6). Therefore, we have performed MBD4-binding and glycosylase assays with T/G mismatches contained in nucleosomes assembled on both methylated and nonmethylated DNA. Results from initial studies of excision repair on a chromatin substrate indicated that the accessibility of enzymes to the DNA damage site is restricted by the presence of chromosomal proteins (70). They also suggested that UV-induced damage in the internucleosomal linker DNA regions is the first to be repaired (22). These regions may also be the first to be damaged; small alkylating agents exhibit a preference for linker DNA (59). In recent years, extensive work on the activities of base excision repair (BER) and nucleotide excision repair (NER) enzymes on nucleosomal substrates has been carried out (see references 35 and 55 for recent reviews). In general, the findings of these studies have supported the early notion that the histone octamer decreases the efficiency of the repair enzymes.
Interestingly, an analysis of BER on a nucleosome substrate using the human uracil glycosylases UNG2 (uracil [nuclear] DNA glycosylase 2) and SMUG1 (single-strand-selective monofunctional uracil DNA glycosylase 1) showed that both enzymes are able to remove uracil from nucleosomes independently of the position or rotational setting of the base (48). Uracil removal is inhibited by the presence of the histone octamer, but histone acetylation does not have any major effect. This finding is in contrast with observations regarding NER on site-specifically platinum-modified nucleosomes, in which the posttranslational modifications present in native histones enhance the repair reaction (68). However, as in BER, the rotational orientations of cyclobutane-thymine dimers have little effect on the rates of NER (60). Recently, uracil DNA glycosylase (UDG) was shown to catalyze BER on a nucleosome array template comprising 12 tandem repeats of a 208-bp segment (45); however, there have not yet been any studies of the effect of DNA methylation on nucleosomes in the glycosylase-mediated repair of base pair mismatches.
The loss of MBD4 is associated with the predisposition to some types of cancer (12). Nevertheless, no study has yet addressed the nucleosome constraints imposed on the MBD4 glycosylase, and the effect of global DNA methylation on the glycosylase activity of MBD4 remains undetermined. Thus, in this work we studied the roles of methylation and chromatin, individually and together.
MATERIALS AND METHODS
Protein expression and purification.
Human cDNA was prepared from HeLa cells. The MBD4 gene was amplified from the cDNA by PCR and cloned into pET28a (Novagen). Recombinant, N-terminally His-tagged protein was expressed in E. coli BL21(DE3) cells (Novagen) by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Preliminary attempts at protein expression resulted in a truncated protein, due to frameshifting at a run of A's within the MBD4 gene. This same frameshift mutation is common in human tumor cells (12). The problem was eliminated by converting two of the three AAA lysine codons into AAG by site-directed mutagenesis. The full-length protein was purified using Talon resin (BD Biosciences). Protein was eluted with 150 mM imidazole. Fractions were dialyzed against a solution of 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 15% glycerol, 60 mM NaCl, and 5 mM β-mercaptoethanol and further purified with a HiTrap SP HP column (Amersham Bioscience) using a 60 mM to 1 M NaCl gradient in a buffer consisting of 50 mM sodium phosphate (pH 8.3) and 1 mM EDTA. Fractions containing the pure protein were dialyzed against a solution of 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 15% glycerol, 60 mM NaCl, and 5 mM β-mercaptoethanol. A pTYB-MeCP2 vector containing cDNA for human MeCP2A (methyl-CpG-binding protein 2A; isoform e2) was a generous gift from Christopher Woodcock (47). Recombinant MeCP2 was expressed and purified as described previously (28).
DNA templates.
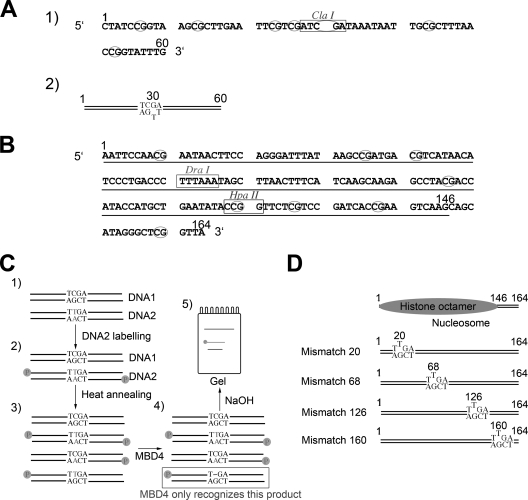
Oligonucleotides used in template construction are listed in Table 1. Protein fractions were tested for activity using a 30-bp synthetic substrate containing a G/T mismatch in a CpG sequence context (primers KS1 and KS3). A homoduplex with a T-A base pair (primers KS2 and KS3) was used as the control. To assay the effect of methylation, we used a 60-bp substrate (Fig. 1A) with six CpG sites, three on either side of the T/G mismatch (primers KS4 and KS5). The T-containing strands (primers KS3 and KS5) were tagged with an infrared label (LI-COR Biosciences) to allow detection. Pairs of oligonucleotides were mixed together in a 1:1 ratio, heated to 90°C, and cooled slowly to room temperature; annealing efficiency was determined by native polyacrylamide gel electrophoresis (PAGE). The methylation of CpG sites was carried out using the SssI methylase; the extent of methylation was determined using the methylation-sensitive restriction enzyme ClaI. Methylated and unmethylated, linearized plasmid DNA was used for competition experiments.
TABLE 1.
Oligonucleotides used in constructing templates
| Oligonucleotide | Sequencea |
|---|---|
| KS1 | ATGAAAGTTAATCGATTTAAAGGGTCAGGG |
| KS2 | ATGAAAGTTAATCAATTTAAAGGGTCAGGG |
| KS3 | CCCTGACCCTTTAAATTGATTAACTTTCAT |
| KS4 | CTATCCGGTAAGCGCTTGAATTCGTCGATCGATAAATAATTGCGCTAACCGGTATTTG |
| KS5 | CAAATACCGGTTAAAGCGCAATTATTTATCGATTGACGAATTCAAGCGCTTACCGGATAG |
| CH20F | GAATTCCAACGAATAACTTCGAGGGATTTAT |
| MIS20F | GAATTCCAACGAATAACTTTGAGGGATTTAT |
| CH68-F | ACCCTTTAAATCGATTAACTTTCATCA |
| CH68-R | TGATGAAAGTTAATCGATTTAAAGGGT |
| MIS68-F | ACCCTTTAAATTGATTAACTTTCATCA |
| MIS68-R | TGATGAAAGTTAATCAATTTAAAGGGT |
| 5SF | GAATTCCAACGAATAACTT |
| 5SR | CCCGAGGAATTCGGTATT |
Underlined letters correspond to mismatches.
FIG. 1.
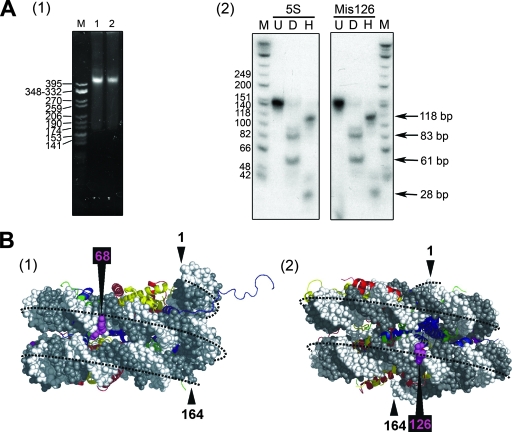
Preparation of T/G mismatch DNA templates used for the analysis of MBD4 glycosylase activity. (A) Sequence of a 60-bp synthetic DNA duplex used for this analysis. CpG dinucleotides are circled. The ClaI site used to assess the methylation of CpGs is shown. (B) Nucleotide sequence of the 164-bp fragment of the 5S rRNA gene of the sea urchin L. variegatus used for the nucleosome reconstitution work. The 1 and 164 positions are identified, and the main position occupied by the histone octamer in nucleosomes reconstituted from this DNA template is underlined. (C) Steps: 1, two versions (DNA1 and DNA2) of the 164-bp fragment depicted in panel B were constructed with the sequence altered in one to introduce a mismatch; 2, unlabeled (DNA1) and labeled (DNA2) DNA constructs were mixed in stoichiometric amounts; 3, DNA fragments in the mixture were melted and reannealed and were used in nucleosome reconstitution and glycosylase assays; 4 and 5, the glycosylase activity of MBD4 was assessed by NaOH cleavage of the resulting unpaired nucleotide. (D) Schematic representation of the constructs generated by the introduction of mismatches into the 164-bp sequence of the 5S rRNA gene fragment. The position occupied by the histone octamer in these constructs is also shown (Fig. 6).
The DNA templates used for the nucleosome reconstitution experiments were derived by PCR from P5S208-12, which consists of 12 tandemly repeated copies of a 196-bp fragment of the 5S rRNA gene from the sea urchin Lytechinus variegatus (Fig. 1B), connected by a 12-bp linker (58). The sea urchin 5S rRNA gene contains a strong histone octamer positioning signal in reconstituted nucleosomes (25). The plasmid was kindly provided to us by the late R. T. Simpson. Fragments of 196 bp (for the naked-DNA glycosylase assay) and 164 bp (for both the naked-DNA glycosylase assay and nucleosome reconstitution) were used (Fig. 1B). T/G mismatches were introduced, individually, at several positions along the 164-bp fragment by PCR according to the strategy schematically depicted in Fig. 1C and D. Briefly, two double-stranded DNA molecules, one having a CG base pair at the site where the mismatch was to be introduced (DNA1) (Fig. 1C) and a second one having a TA base pair introduced by PCR at the same position (DNA2) (Fig. 1C), were prepared. Upon the radioactive labeling of DNA2, the two DNA molecules were mixed and subjected to melting and reannealing to produce a DNA mixture containing a mismatched double-stranded DNA molecule for use in further experiments (nucleosome reconstitution and MBD4 glycosylase assays).
DNA fragments containing the T/G mismatch were prepared using the following primer combinations (Table 1): for CH20, CH20F and 5SR; for MIS20, MIS20F and 5SR; for CH68, set 1 (5SF and CH68-R) and set 2 (CH68-F and 5SR); and for MIS68, set 1 (5SF and MIS68-R) and set 2 (MIS68-F and 5SR). CH68 PCR products 1 and 2 and MIS68 PCR products 1 and 2 were mixed and incorporated in subsequent PCRs using 5SF and 5SR primers. The PCR products thus obtained were cloned into a TOPO TA cloning kit (Invitrogen).
The DNA templates were radiolabeled with [γ-32P]ATP by using T4 polynucleotide kinase (8). Mismatched DNA templates were then obtained from mixtures of labeled and unlabeled DNA templates that were incubated at 95°C for 15 min and allowed to reanneal by gradual cooling to generate the mismatch (Fig. 1C and corresponding legend). These T/G mismatch-containing DNA constructs were used for glycosylase assays and nucleosome reconstitution. Versions of the mismatch-containing DNA templates methylated at the CpG sites were obtained by treatment with SssI methylase (New England Biolabs), and the completeness of the methylation was assessed by digestion with the HpaII (methylation-sensitive) restriction enzyme (New England Biolabs).
Nucleosome reconstitution.
Histone octamers were prepared from chicken erythrocytes, and acetylated histone octamers were obtained from chicken MSB cells transformed with Marek's disease virus (3) and grown in the presence of sodium butyrate (62, 69). Chicken MSB cells were a kind gift from Vaughn Jackson.
Nucleosome reconstitution was carried out as described in reference 42. Briefly, [γ32P]ATP-end-labeled (8) versions of the DNA templates described above, mixed with an approximately 10-fold excess of random-sequence 146-bp DNA fragments in a solution of 2 M NaCl, 20 mM Tris-HCl (pH 7.5), and 0.1 mM EDTA buffer, were added to histone octamers in the same buffer (histone/DNA ratio, 1.0/1.13 [wt/wt]). The random-sequence 146-bp DNA fragments were obtained from chicken erythrocyte nucleosome core particles (8). Nucleosome reconstitution was then allowed to proceed by salt gradient dialysis reconstitution (61) as described elsewhere (10).
Gel electrophoresis.
Acetic acid-urea-Triton X-100 electrophoresis was carried out as described elsewhere (1). However, in this instance the urea concentration was 5.6 M and the Triton X-100 concentration was 4.4 mM. Sodium dodecyl sulfate (SDS)-PAGE was performed according to the method of Laemmli (38).
Native PAGE (acrylamide/bisacrylamide ratio, 29:1 [wt/wt]) for DNA and nucleosome analyses (using different acrylamide concentrations as indicated in the figure legends and a mixture of 20 mM sodium acetate, 1 mM EDTA, 20 mM Tris-HCl [pH 7.2]) was carried out as described elsewhere (39, 72). Denaturing 8% PAGE (acrylamide/bisacrylamide ratio, 19:1 [wt/wt]) was carried out in the presence of 8.3 M urea in a buffer containing 90 mM Tris, 90 mM boric acid, and 2 mM EDTA.
Electrophoretic mobility shift assays.
Recombinant proteins (MBD4 and MeCP2) were incubated in mixtures of 10 mM Tris-HCl (pH 7.5), 150 μg of bovine serum albumin (BSA), 0.1% NP-40, 3 mM dithiothreitol, 5% glycerol, and 150 mM NaCl for 10 min at room temperature. Increasing amounts of protein were then mixed with nucleosomes prepared as described above, and the mixtures were incubated for 20 min at room temperature. The nucleosome-protein complexes were analyzed by native PAGE.
Nucleosome positioning.
Nucleosome positioning was determined as originally described (25). Nucleosomes reconstituted onto the 196- and 164-bp (mismatch) DNA templates were trimmed down to 146-bp nucleosome core particles by digestion with micrococcal nuclease (6 U/mg of DNA; Worthington Biochemical Corp.) for 20 min at 37°C in a buffer containing 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), and 1 mM CaCl2. The reaction was stopped by the addition of 10 mM EDTA and 1% SDS, and the mixture was further incubated at 37°C for 10 min (to completely dissociate the histones from the DNA). After the addition of sucrose to a concentration of 10%, the solution was loaded onto a native 4% polyacrylamide gel and the 146-bp DNA band was excised and extracted from the gel overnight as described previously (42). The DNA thus obtained was digested with HpaII and DraI restriction enzymes (New England Biolabs), and the digested products were analyzed by native 6% PAGE.
Glycosylase assay.
Purified MBD4 protein was assayed for glycosylase activity against labeled DNA or nucleosomes. Two types of experiments were performed: the addition of a fixed concentration (50 nM) of MBD4 for various incubation times and the addition of various MBD4 concentrations for a fixed period of 2.5 h (see the figures and legends for details). All incubations were at 37°C with a reaction buffer consisting of 10 mM HEPES-KOH (pH 7.5), 0.5 mM dithiothreitol, 0.5 mM EDTA, and 0.5 mg of BSA/ml. β-Mercaptoethanol was then added to a final concentration of 10 mM, and the reaction mixture was further incubated for 10 min. The cleavage of the DNA backbone at the abasic site resulting from glycosylase activity was carried out by treatment with 0.1 N NaOH at 90°C for 15 min, and the cleavage products were analyzed by denaturing 8% PAGE in a buffer of 90 mM Tris, 90 mM boric acid, and 2 mM EDTA. In the case of the nucleosome assays, native PAGE was carried out at the end of the incubation period to ensure that the nucleosome structure remained intact during the process.
RESULTS
MBD4 is a highly folded methyl-CpG-binding domain protein with a distinctive chromatin-binding pattern.
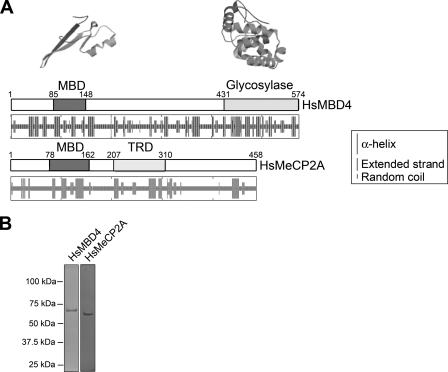
Human MBD4, also known as methyl-CpG-binding endonuclease 1 (MED1), is a 574-amino-acid protein (see Fig. 2) that belongs to a family of methyl-CpG-binding domain proteins (56) that includes MeCP2, a 486-amino-acid protein. Mutations in MBD4 have been linked previously to cancer (12), while those in MeCP2 have been linked to Rett syndrome (4, 9). Figure 2A shows a structural comparison of MBD4 and MeCP2. Both proteins are characterized by the presence of a well-defined common methyl-CpG-binding N-terminal domain combined with a second domain: a functional transcription repression domain in MeCP2 and a glycosylase domain at the C terminus in MBD4. In MeCP2, the protein regions flanking the two functional domains are predicted to have low levels of secondary structure, in agreement with recent experimental reports showing that the protein is intrinsically disordered (2). In contrast, the prediction analysis shown in Fig. 2A suggests a much more organized structure for MBD4.
FIG. 2.
MBD4 and MeCP2, two methylated-DNA-binding proteins with different structures. (A) The box drawings are a schematic representation of the functional domain organization of human MeCP2 and MBD4. The secondary structures predicted from the amino acid sequences by using the hierarchical neural network (23) are shown underneath. The tertiary-structure organizations of the methyl-CpG-binding and glycosylase domains of human MBD4 as predicted from the crystallographic data on the methyl-CpG-binding domain (MBD) of human MBD1 (50) and the mouse glycosylase domain of MBD4 (71) are shown above the corresponding schematic linear representations. Tertiary-structure predictions were carried out using the SWISS-MODEL server (57). TRD, transcription repression domain; Hs, Homo sapeins. (B) SDS-PAGE analysis of the recombinant MBD4 and MeCP2 forms used in this work. The numbers on the left-hand side of the gel correspond to the molecular masses of the proteins as indicated by a marker.
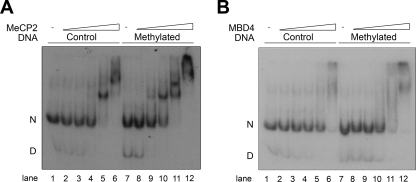
Thus, although MBD4 and MeCP2 share a methyl-CpG-binding domain, not only their functional domains but also their overall structural organization patterns are different. Hence, we considered that the ways in which they bind to DNA and chromatin might be different. Figure 3 shows that this is indeed the case. Although both proteins exhibit a significant binding preference for nucleosomes containing methylated DNA (Fig. 3, compare lanes 3 to 6 with lanes 9 to 12), MeCP2 exhibits specific binding behavior which contrasts with the nonspecific behavior of MBD4. As pointed out above, the different tertiary-structure conformations of these monomeric proteins are likely the reason for such differences.
FIG. 3.
MeCP2 and MBD4 preferentially bind to methylated nucleosomes. Nucleosomes were reconstituted using [γ32P]ATP-end-labeled unmethylated (control) and methylated versions of a 5S rRNA gene fragment (see Materials and Methods) and were titrated with increasing amounts of either MeCP2 or MBD4. The electrophoretic shifts resulting from the complexes formed between these proteins and the nucleosomes were analyzed by electrophoresis in electrophoretic mobility shift assays. Approximately 30 to 40 nM concentrations of nucleosomes incubated for 20 min at room temperature with increasing amounts (indicated by triangles) of MeCP2 (A) or MBD4 (B) were analyzed by native 4% PAGE. Lanes: 2 and 8, 7.5 nM protein; 3 and 9, 15 nM protein; 4 and 10, 30 nM protein; 5 and 11, 60 nM protein; and 6 and 12, 120 nM protein. D, free DNA; N, nucleosomes; −, no protein.
DNA binding to methylated and unmethylated substrates.
Previous studies on the effect of methylation on MBD4 activity have used DNA substrates that are differentially methylated solely at the site of the T/G mismatch (the methylated cytosine being 5′ of the G). In effect, the substrates were either unmethylated or hemimethylated. We tested the effects of the full methylation of the DNA at CpG sequences both 5′ and 3′ of the mismatch. Since CpG sequences are most often part of CpG islands, whose methylation states vary during development or in accordance with differential gene expression, our assay is more representative of the situation in the eukaryotic cell than previous assays. The duplex DNA substrates were methylated with SssI after the annealing of the single-stranded DNA molecules into duplex form, so the cytosine 5′ of the G in the T/G mismatch was unmethylated.
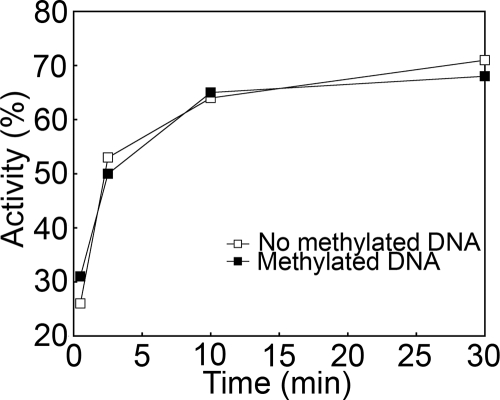
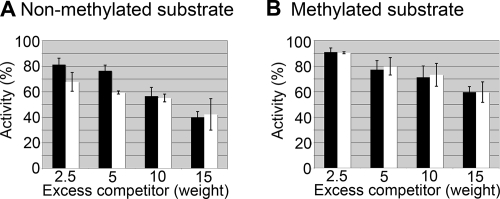
Figure 4 shows the results of the time course assay of glycosylase activity using the unmethylated and methylated versions of a synthetic DNA template depicted in Fig. 1A. This synthetic DNA template was used initially to optimize the enzymatic activity of our recombinant MBD4. Under these conditions, most of the substrate was cleaved within the first 10 min. Afterwards, similar experiments were carried out using a longer DNA template (Fig. 1B) which is amenable to nucleosome reconstitution. The results obtained with this template were very similar (results not shown). In addition, both templates showed that glycosylase activity was unaffected by the methylation of the surrounding CpG sites. We then tested to see whether incubation with competitor DNA, lacking mismatches, would affect glycosylase activity and whether the competition was affected by the methylation state of the heteroduplex substrate and/or the competitor DNA. Plasmid DNA, containing 402 CpG sites per molecule, was used as the competitor. Figure 5A shows that the level of glycosylase activity on a nonmethylated mismatch DNA substrate in the presence of a methylated competitor was lower than that in the presence of a nonmethylated competitor. Such a difference became negligible with large excess amounts of the competitor (above a fivefold excess by weight). However, when a methylated mismatch DNA substrate was used, the inhibitory effect of the DNA competitor was much smaller (Fig. 5B). In addition, the methylation state of the competitor DNA had no effect.
FIG. 4.
The methylation of CpG sites in synthetic duplexes does not affect the glycosylase activity of MBD4. Urea-denaturing PAGE analysis of the glycosylase activity was carried out with methylated and unmethylated versions of the 60-bp DNA fragment. An approximately 20 nM concentration of the 60-bp heteroduplex DNA was incubated with 50 nM MBD4 for various lengths of time. Reactions were run on a denaturing gel, and DNA species were detected by infrared scanning. Results are expressed as the percentage of cleavage.
FIG. 5.
The methylation of substrate and/or competitor DNA has a minimal effect on MBD4 glycosylase activity. An approximately 50 nM concentration of a nonmethylated (A) or methylated (B) 164-bp 5S rRNA gene fragment was incubated with 25 nM MBD4 for 7.5 min in the presence of various amounts of competitor DNA. Reactions were run on a denaturing gel, and DNA species were detected and measured using a phosphorimager. Results are expressed as the percentage of cleavage, normalized to the degree of cleavage of the substrate without competitor DNA. Amounts of excess (n-fold) of competitor CpG sites relative to substrate CpG sites are indicated. Black bars, nonmethylated competitor; white bars, methylated competitor.
The presence of a T/G mismatch does not alter nucleosome positioning.
We next asked what role chromatin plays in modulating the glycosylase activity of MBD4 and whether the methylation of the DNA template or histone acetylation plays a major role. To this end, we used nucleosomes reconstituted onto a 164-bp DNA fragment of the 5S rRNA gene from the sea urchin L. varieagatus (58). The 5S rRNA gene template has been used extensively as a model for previous structural and functional studies of chromatin (5, 29, 30, 33, 34, 58, 64) due to its sequence-intrinsic potential to assemble the histone octamer into a preferential major position (25) (Fig. 1D). In addition, this DNA fragment contains several CpG sites (Fig. 1B), making it amenable to the study of the effects of global CpG methylation on MBD4 activity. As shown in Fig. 6B, the T/G mismatches at positions 68 and 126 are located in the major and minor grooves, respectively, on the side of the double helix that is exposed to the buffer and, hence, should be easily accessible for MBD4 binding.
FIG. 6.
(A) DNA mismatches do not alter the position of the histone octamer. (1) Native 4% PAGE analysis of nucleosomes reconstituted onto a native 164-bp fragment of the 5S rRNA gene (lane 1) and the same DNA fragment containing a T/G mismatch at position 126 (lane 2) (Fig. 2). M is a CfoI-cut pBR322 marker. (2) The nucleosomes shown in lanes 1 and 2 of panel 1 were digested with micrococcal nuclease, and the 146-bp resistant fragment extracted from the nucleosome core particles generated in this way was analyzed by native 6% PAGE. Lanes: U, 146-bp DNA; D and H, the same fragment as that in lane U upon digestion with DraI and HpaII, respectively. M indicates a φX174 HinfI-digested marker. 5S, 5S rRNA gene fragment; Mis126, construct with mismatch at position 126. (B) The mismatched T/G sites at positions 68 and 126 (Fig. 1D) are exposed to the buffer in the positioned nucleosomes. Side views of the molecular structure of a 164-bp nucleosome with the core histone octamer positioned as depicted in Fig. 1D are shown. The DNA backbone is in white and gray, and the histones are represented as follows: H2A, yellow; H2B, red; H3, blue; and H4, green. The structure of the nucleosome was obtained by using the crystallographic coordinates of the nucleosome core particle from reference 41.
In the nucleosome reconstitution process, special care was taken to avoid the presence of free DNA in the final reconstituted product. Indeed, the reconstitution of nucleosomes by salt gradient dialysis (61) often leads to the presence of various amounts of unreconstituted free DNA, which must be removed by using a sucrose gradient. To avoid this complication, radioactively labeled mismatched template DNA was mixed with an excess of random-sequence 146-bp DNA (42) (see Materials and Methods). This step made it possible to use only relatively small amounts of mismatched DNA and resulted in the added benefit of providing competitive random-sequence DNA to prevent the nonspecific binding of MBD4.
It was also important to check that the introduction of a T/G mismatch did not affect the nucleosome reconstitution ability and positioning properties of the 5S rRNA gene template. Figure 6 shows the results obtained with the 164-bp fragment containing a mismatch at position 126. As can be seen clearly in Fig. 6A, panel 2, the mismatch did not affect the positioning of the histone octamer. The same results were obtained with all of the mismatch DNA templates depicted in Fig. 1D.
MBD4 glycosylase activity in the presence of a nucleosome.
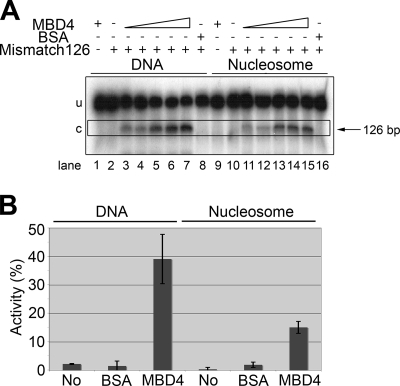
As shown in Fig. 7, the presence of a nucleosome decreased the efficiency of MBD4 but did not abolish its activity, which was significantly lower than that observed in the presence of the corresponding DNA template (see also Table 2). It is important that in the representation shown in Fig. 7B, the relative enzymatic activity corresponds to the absolute amounts of labeled DNA. Hence, the reduced MBD4 activity observed for the nucleosome corresponds to about 40% of the activity for the corresponding naked DNA, as only approximately 33% of the labeled DNA template was a substrate for the enzyme (Fig. 1C). Identical results were obtained with all the other mismatched DNA constructs depicted in Fig. 1D except for the construct with the mismatch at position 160, which showed no glycosylase activity regardless of its methylation state (Table 2). This result was attributed to the short distance of the mismatch from the end of the DNA fragment, which may have impaired MBD4 binding. Indeed, as can be seen in Table 2, we also observed no activity with the naked DNA template carrying a mismatch at this position. Similarly, a mismatch at position 20 yielded a lower level of activity, both on DNA and on nucleosomes, than mismatches at positions 68 and 126. This finding suggests again that in general the activity of MBD4 decreases with the proximity of the mismatch site to the end of the DNA template, regardless of the naked or chromatinized nature of the DNA. This property is most likely simply a reflection of the artifactual nature of the discrete sizes of the templates used in these in vitro experiments.
FIG. 7.
MBD4 glycosylase activity in the presence of a nucleosome. (A) Urea-denaturing PAGE analysis of the glycosylase activities on a 164-bp DNA construct with a mismatch at position 126 in its DNA and reconstituted nucleosome versions. Approximately 10 nM DNA was incubated with MBD4 at 10 nM (lanes 3 and 11), 20 nM (lanes 4 and 12), 40 nM (lanes 5 and 13), 80 nM (lanes 6 and 14), and 160 nM (lanes 1, 7, 9, and 15) or with 160 nM BSA (lanes 8 and 16) (see Materials and Methods for further details). Because the nucleosome reconstitution procedure was carried out in the presence of a 10-fold excess of a 146-bp random-sequence DNA fragment (see Materials and Methods), the assay of glycosylase activity on naked DNA was also carried out in the presence of a 10-fold excess of the same 146-bp random-sequence cold DNA. Triangles indicate increasing concentrations. u, uncut DNA substrate; c, cut DNA product; −, absent; +, present. (B) Bar plot representation of the glycosylase activity indicated in panel A for 160 nM MBD4. The activity is expressed in terms of the amount of the NaOH-cut 126-bp fragment as a percentage of the amount of the total starting mismatched DNA template (cut plus uncut). “No” indicates a 164-bp DNA template with no mismatch.
TABLE 2.
MBD4 glycosylase activities on different nucleosome substrates
| Mismatch position | Activitya on:
|
||
|---|---|---|---|
| DNA | Normal nucleosomes | Acetylated nucleosomes | |
| 20 | ++ | + | + |
| 68 | ++++ | ++ | +++ |
| 126 | ++++ | ++ | +++ |
| 160 | − | − | − |
++++, >25%; +++, 15 to 25%; ++, 5 to 15%; +, <5%; −, 0%.
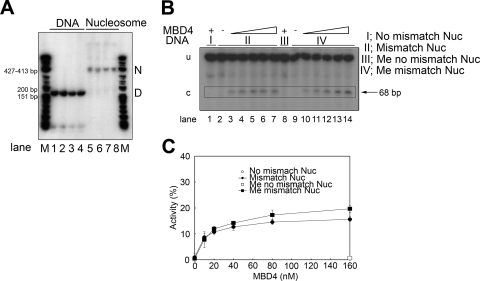
Figure 8 compares the glycosylase activities of MBD4 on methylated and nonmethylated nucleosomes reconstituted in the presence of excess nonmethylated 146-bp DNA (see Materials and Methods). The results indicate that the mismatch repair efficiency was subtly enhanced by the methylated state of the nucleosomal DNA template (Fig. 8). This finding is in contrast to observations regarding naked DNA templates in the absence of competitor DNA (Fig. 4) but similar to observations regarding the MBD4 activity on naked DNA templates when competitor DNA was used (Fig. 5, compare black bars).
FIG. 8.
MBD4 glycosylase activity on a nucleosome is unaffected by DNA methylation. (A) Native 4% PAGE analysis of the 164-bp DNA construct depicted in Fig. 1C and the corresponding nucleosomes reconstituted using this DNA template. Lanes: 1 and 5, homoduplex DNA; 2 and 6, DNA containing a T/G mismatch at position 68; 3 and 7, methylated homoduplex DNA; 4 and 8, DNA containing a T/G mismatch at position 68. M is a φX174 HinfI-digested marker. D and N indicate the electrophoretic mobilities of DNA and nucleosomes, respectively. (B) Same as panel A but with nucleosomes reconstituted onto the corresponding methylated and unmethylated versions of the 164-bp DNA with a T/G mismatch at position 68. Approximately 10 nM concentrations of nucleosome DNA equivalents were incubated with MBD4 at 10 nM (lanes 3 and 10), 20 nM (lanes 4 and 11), 40 nM (lanes 5 and 12), 80 nM (lanes 6 and 13), and 160 nM (lanes 1, 7, 8, and 14) (see Materials and Methods for further details). Sections I to IV correspond to the indicated versions of the 164-bp DNA template. Triangles represent increasing concentrations. Nuc, nucleosome; me, methylated; u, uncut DNA; c, cut DNA; −, absent; +, present. (C) Analysis of the MBD4 cleavage activity, with various amounts of protein, carried out on nucleosomes reconstituted on methylated and unmethylated versions of the 164-bp DNA. The activity is expressed in terms of the amount of the NaOH-cut 68-bp fragment as a percentage of the amount of the total mismatched DNA template. The data presented are the means (±1 standard deviation) of results obtained from three experiments.
Histone acetylation facilitates the glycosylase activity of MBD4.
Because of the potential regulatory role for histone acetylation in base mismatch repair observed in vivo (63), it was important to analyze histone acetylation in MBD4 mismatch repair by using nucleosomes containing acetylated histones.
The acetylation of the core histones has been shown previously to open the nucleosome conformation in a way that involves the dynamic release of the flanking DNA at the entry and exit sites into the nucleosome core particle and alters the conformation of the DNA (14, 27, 49). Therefore, we decided to check whether acetylation had any effect on the MBD4 glycosylase activity on a nucleosome template.
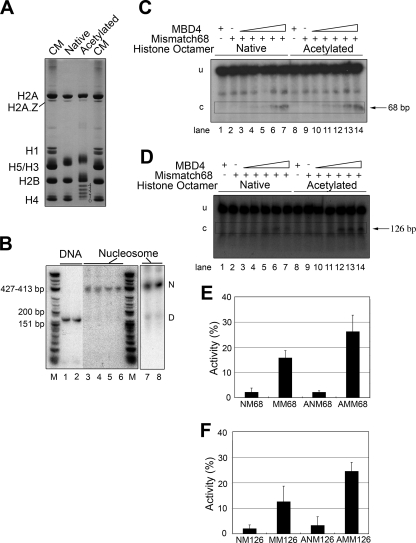
To this end, we prepared highly globally hyperacetylated histone octamers consisting of an average of 10 to 12 acetyl groups per octamer (11) (Fig. 9A). Nucleosomes were then reconstituted using a 164-bp DNA fragment containing a mismatch at either position 68 or 126 (Fig. 9B, lanes 1 and 2). Lanes 3 to 8 of Fig. 9B show that all the reconstituted nucleosomes, regardless of their histone composition or the positioning of the mismatch on the DNA template, had the same electrophoretic appearance, with essentially no free DNA present. Figure 9C and D show the glycosylase activities of MBD4 on acetylated and nonacetylated nucleosome templates having mismatches at bp 68 and 126 of their DNA, respectively. Figure 9E and F provide a representation of the analysis of the data presented in Fig. 9C and D. These results clearly show that histone acetylation significantly facilitates the glycosylase activity of MBD4. This effect was observed for both mismatch positions.
FIG. 9.
Histone acetylation facilitates the glycosylase activity of MBD4 on nucleosome substrates. (A) Acetic acid-urea-Triton X-100-PAGE analysis of core histones used for the reconstitution of acetylated nucleosomes. CM, chicken erythrocyte histone marker; native, histones from histone octamers obtained from chicken erthrocytes; acetylated, histones from histone octamers purified from chicken MSB cells (see Materials and Methods). In this type of gel, the proteins run with electrophoretic mobilities that depend on their charge/mass ratios. Accordingly, the neutralization of the charge upon the acetylation of lysine residues of histones results in decreased mobility that allows differentiation among forms with different extents of acetylation. This effect can be clearly visualized for histone H4, for which the numbers 0, 1, 2, 3, and 4 shown on the right-hand side of the gel correspond to non-, mono-, di-, tri-, and tetra-acetylated forms, respectively. (B) Native 4% PAGE analysis of the 164-bp DNA construct depicted in Fig. 1C and the corresponding nucleosomes reconstituted using this DNA template. Lanes: 1, 3, and 5, DNA with no mismatches; 2, 4, and 6, DNA containing a T/G mismatch at position 68; 3 and 4, nucleosomes reconstituted with native core histones; 5 and 6, nucleosomes reconstituted with acetylated core histones; 7, nucleosome reconstituted onto 164-bp DNA containing a T/G mismatch at position 126 and native core histones; 8, same as lane 7 except that acetylated histones were used for the nucleosome reconstitution. M is a φX174 HinfI-digested marker. D and N indicate the electrophoretic mobilities of DNA and nucleosomes, respectively. (C and D) Urea-denaturing PAGE analysis of the glycosylase activities on nucleosomes reconstituted with native and acetylated histones by using a 164-bp DNA construct with a T/G mismatch at either position 68 (C) or position 126 (D). Approximately 10 nM DNA was incubated with MBD4 at 10 nM (lanes 3 and 10), 20 nM (lanes 4 and 11), 40 nM (lanes 5 and 12), 80 nM (lanes 6 and 13), and 160 nM (lanes 1, 7, 8, and 14) (see Materials and Methods for further details). Triangles indicate increasing protein concentrations. −, absent; +, present. (E and F) Bar plot representations of the MBD4 glycosylase activity detected in the analyses presented in panels C and D, respectively. NM, no mismatch; MM, mismatch; ANM, acetylated no-mismatch-containing nucleosomes; AMM, acetylated mismatch-containing nucleosomes. Numbers after abbreviations correspond to the positions of the mismatches.
DISCUSSION
In contrast to MeCP2, which exhibits a discrete distribution of tertiary folded structures throughout an otherwise disordered sequence (2), MBD4 contains two well-defined N- and C-terminal folded domains linked by what is predicted to be a protein domain with abundant secondary structures (Fig. 2). The methyl-binding domain and highly folded catalytic domain (71) constitute approximately 60% of the overall sequence. Our preliminary analytical ultracentrifuge data support a folded organization of MBD4. Therefore, it is not surprising that these proteins, despite their common methyl-binding abilities, bind to DNA in different ways (Fig. 3).
However, the role of the methyl-CpG-binding domain, if any, in the catalytic activity of MBD4 has not been fully determined. The results presented in this paper conclusively show that DNA methylation enhances the binding of MBD4 to naked DNA and chromatinized DNA similarly to what has been previously reported for MeCP2 (47) and in agreement with the methylated-DNA-binding nature of these proteins. In the presence of competitor DNA (a situation closer to the in vivo setting), the enhanced binding caused by the full methylation of CpG regions surrounding a T/G mismatch appeared to cause a small but noticeable enhancement of MBD4 glycosylase activity (Fig. 5A and 8C), in agreement with results reported previously for hemimethylation immediately adjacent to the mismatch (52). Also, the enzymatic activity of the glycosylase domain of MBD4 itself has been shown, in some previous studies, to prefer T/G mismatches within the context of a CpG site when the site is hemimethylated (65). In the study by Turner and coworkers, the single-turnover rate of the enzyme was found to be 17% higher than that in the nonmethylated context (65). It is quite possible that given the folded structure of MBD4 (Fig. 2A), the methyl-CpG-binding domain and the catalytic site are in close proximity, acting in a synergistic way. Our competition data support the notion that the methylation of CpG sites acts to target the protein to regions of heavy cytosine methylation.
Although a decrease in the efficiency with which MBD4 was able to repair the T/G mismatch when the DNA template was assembled into nucleosomes was observed, the presence of a histone octamer did not prevent the repair activity. The extent of the reduction of glycosylase activity that we observed (Fig. 7B) was similar to that previously reported for other human glycosylases, such as UNG2 and SMUG1 (48), or for the UDG-apyrimidinic/apurinic endonuclease (APE) repair system (15, 16, 45). However, it was significantly less than that recently reported for 8-oxoguanine DNA glycosylase 1 (OGG1), which requires the involvement of the SWI/SNF chromatin-remodeling complex (44). This result is not surprising, as UNG2, SMUG1, UDG-APE, and MBD4 belong to the same structural family, which is different from that of OGG1 (73).
In agreement with the data in reference 48, the DNA repair activity on nucleosome templates was unaffected by the location of the T/G mismatch along the nucleosomal DNA superhelix (Fig. 7A and 8B). Since the two main translational positions used here for the T/G mismatch were facing away from the histones (Fig. 6B), we were unable to determine whether such rotational orientation had a major role in the MBD4 glycosylase activity. While the rotational orientation of the mismatched base pair had a noticeable effect with UDG-APE (16), it did not appear to have any major effect in the cases of UNG2 and SMUG1 (48). Therefore, our data suggest that the removal or displacement of the histone octamer is not required for efficient mismatch repair by MBD4 on a chromatin substrate.
The subtle but reproducible role of acetylation in enhancing the ability of MBD4 to exert its glycosylase activity (Fig. 9) is in contrast with the apparent lack of a role played by the histone tails in the UDG-APE system (15). However, our results are consistent with the notion that histone acetylation relaxes the nucleosome structure (7, 21). This idea suggests the possibility that repair occurs faster in the highly acetylated regions of the genome, which often are associated with transcriptional activity (21).
The structure of MBD4, consisting of an N-terminal methyl-binding domain and a C-terminal glycosylase domain, provides an interesting conundrum, because the two domains can act independently of each other. The methyl-binding domain binds regions of heavy methylation, most likely CpG islands associated with promoter regions of transcriptionally silenced genes. This domain has recently been shown to act as a transcriptional repressor, with and without the associated glycosylase domain (37). Likewise, the glycosylase domain can process T/G mismatches in TpG/CpG sequence contexts independent of the methyl-binding domain and with at best a minimal influence from hemimethylation. Our study has shown that the methylation of CpGs surrounding the mismatch has a slight measurable effect on the glycosylase activity of the intact protein and, furthermore, that competitor DNA reduces glycosylase activity depending on the methylation status of the T/G-containing heteroduplex DNA or the competitor DNA. However, it should be noted that the density of CpGs in the DNA fragments used in our experiments may have been insufficient to have an effect and that the surrounding sequence contexts may have been suboptimal. Both factors have been shown previously to affect the binding of other members of the methyl-binding domain family of proteins (13, 43). Alternatively, it is possible that the binding affinity of MBD4 for the T/G mismatch was so much stronger than its affinity for methylated CpGs that the glycosylase activity was relatively unaffected by nearby methylated sequences.
Despite the apparent independence of the two domains in all of the experiments done to date, common sense suggests that their structural juxtaposition must have a functional significance. Most studies, including ours, have been based on the hypothesis that local methylation could enhance glycosylase activity by increasing DNA binding. This enhancement would serve to target DNA repair to regions of the DNA where mismatches caused by the deamination of 5-methylcytosine would be most likely to occur. However, experimental evidence to date does not support the hypothesis. Instead, Kondo and coworkers (37) have turned the argument around, hypothesizing that the glycosylase enhances transcriptional repression by MBD4 by repairing T/G mismatches to CG, thus maintaining methylation density. Based on our finding that the glycosylase activity of MBD4 is enhanced by the acetylation of histones, we propose the following modification to this hypothesis.
The hyperacetylation of histones is associated with transcriptionally active regions of the genome, independent of DNA methylation (20). If the DNA was methylated, T/G mismatches caused by the deamination of 5-methylcytosine would be expected to weaken the DNA structure, making localized melting of the two strands more frequent and thereby increasing the opportunities for transcriptional initiation. It is known that deamination occurs more frequently in single-stranded DNA than in double-stranded DNA (26) and that C-to-T mutations are enhanced by transcription (17). Thus, the initial deamination event could precipitate a cycle of increased transcription followed by further deamination, continuing until the promoter region was sufficiently demethylated to result in constitutive gene expression. One way to prevent this cascade would be to ensure that transcription was prevented while the repair of the initial T/G mismatch was carried out. MBD4 has the necessary properties for such a process, being both a repair enzyme and a transcriptional repressor. As shown in this study, its activity would be enhanced by histone acetylation associated with transcriptional activity. MBD4 could also recruit methylases to the repaired CpG and the histone modification enzymes (e.g., histone deacetylases) to the promoter region; both methylases and histone modification enzymes are associated with the transcriptional silencing of genes (reviewed in reference 36). These hypotheses remain to be tested.
Acknowledgments
We are very grateful to José Maria Eirín-López for his help in preparing Fig. 6B. We are also very grateful to Deanna Dryhurst and to Anita A. Thambirajah for carefully reading the manuscript and for their valuable comments.
This work was supported by the Canadian Institutes of Health Research grants MOP 68884 (to C.G.C.) and MOP 57718 (to J.A.) and by a Michael Smith Health Research Foundation postdoctoral fellowship to T.I.
Footnotes
Published ahead of print on 2 June 2008.
REFERENCES
- 1.Abbott, D. W., V. S. Ivanova, X. Wang, W. M. Bonner, and J. Ausió. 2001. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 27641945-41949. [DOI] [PubMed] [Google Scholar]
- 2.Adams, V. H., S. J. McBryant, P. A. Wade, C. L. Woodcock, and J. C. Hansen. 2007. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 28215057-15064. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17105-116. [PubMed] [Google Scholar]
- 4.Amir, R. E., I. B. Van den Veyver, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23185-188. [DOI] [PubMed] [Google Scholar]
- 5.An, W., S. H. Leuba, K. van Holde, and J. Zlatanova. 1998. Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc. Natl. Acad. Sci. USA 953396-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ataian, Y., and J. E. Krebs. 2006. Five repair pathways in one context: chromatin modification during DNA repair. Biochem. Cell Biol. 84490-504. [DOI] [PubMed] [Google Scholar]
- 7.Ausió, J., and D. W. Abbott. 2004. The role of histone variability in chromatin stability and folding, vol. 39. Elsevier, Amsterdam, The Netherlands.
- 8.Ausió, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206451-463. [DOI] [PubMed] [Google Scholar]
- 9.Ausió, J., D. B. Levin, G. V. De Amorim, S. Bakker, and P. M. Macleod. 2003. Syndromes of disordered chromatin remodeling. Clin. Genet. 6483-95. [DOI] [PubMed] [Google Scholar]
- 10.Ausió, J., and S. C. Moore. 1998. Reconstitution of chromatin complexes from high-performance liquid chromatography-purified histones. Methods (San Diego) 15333-342. [DOI] [PubMed] [Google Scholar]
- 11.Ausió, J., and K. E. van Holde. 1986. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry 251421-1428. [DOI] [PubMed] [Google Scholar]
- 12.Bader, S., M. Walker, and D. Harrison. 2000. Most microsatellite unstable sporadic colorectal carcinomas carry MBD4 mutations. Br. J. Cancer 831646-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballestar, E., and A. P. Wolffe. 2001. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 2681-6. [DOI] [PubMed] [Google Scholar]
- 14.Bauer, W. R., J. J. Hayes, J. H. White, and A. P. Wolffe. 1994. Nucleosome structural changes due to acetylation. J. Mol. Biol. 236685-690. [DOI] [PubMed] [Google Scholar]
- 15.Beard, B. C., J. J. Stevenson, S. H. Wilson, and M. J. Smerdon. 2005. Base excision repair in nucleosomes lacking histone tails. DNA Repair (Amsterdam) 4203-209. [DOI] [PubMed] [Google Scholar]
- 16.Beard, B. C., S. H. Wilson, and M. J. Smerdon. 2003. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl. Acad. Sci. USA 1007465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beletskii, A., and A. S. Bhagwat. 1998. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem. 379549-551. [PubMed] [Google Scholar]
- 18.Bellacosa, A., L. Cicchillitti, F. Schepis, A. Riccio, A. T. Yeung, Y. Matsumoto, E. A. Golemis, M. Genuardi, and G. Neri. 1999. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl. Acad. Sci. USA 963969-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bestor, T. H. 1990. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos. Trans. R. Soc. Lond. B 326179-187. [DOI] [PubMed] [Google Scholar]
- 20.Brinkman, A. B., S. W. Pennings, G. G. Braliou, L. E. Rietveld, and H. G. Stunnenberg. 2007. DNA methylation immediately adjacent to active histone marking does not silence transcription. Nucleic Acids Res. 35801-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calestagne-Morelli, A., and J. Ausio. 2006. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem. Cell Biol. 84518-527. [DOI] [PubMed] [Google Scholar]
- 22.Cleaver, J. E. 1977. Nucleosome structure controls rates of excision repair in DNA of human cells. Nature 270451-453. [DOI] [PubMed] [Google Scholar]
- 23.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25147-150. [DOI] [PubMed] [Google Scholar]
- 24.Doiron, K. M., J. Lavigne-Nicolas, and C. G. Cupples. 1999. Effect of interaction between 5-azacytidine and DNA (cytosine-5) methyltransferase on C-to-G and C-to-T mutations in Escherichia coli. Mutat. Res. 42937-44. [DOI] [PubMed] [Google Scholar]
- 25.Dong, F., J. C. Hansen, and K. E. van Holde. 1990. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc. Natl. Acad. Sci. USA 875724-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frederico, L. A., T. A. Kunkel, and B. R. Shaw. 1990. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 292532-2537. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Ramirez, M., C. Rocchini, and J. Ausió. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 27017923-17928. [DOI] [PubMed] [Google Scholar]
- 28.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 27832181-32188. [DOI] [PubMed] [Google Scholar]
- 29.Hansen, J. C., and A. P. Wolffe. 1992. Influence of chromatin folding on transcription initiation and elongation by RNA polymerase III. Biochemistry 317977-7988. [DOI] [PubMed] [Google Scholar]
- 30.Hayes, J. J., and A. P. Wolffe. 1993. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 906415-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrich, B., U. Hardeland, H. H. Ng, J. Jiricny, and A. Bird. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401301-304. [DOI] [PubMed] [Google Scholar]
- 32.Hennecke, F., H. Kolmar, K. Brundl, and H. J. Fritz. 1991. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature 353776-778. [DOI] [PubMed] [Google Scholar]
- 33.Howe, L., and J. Ausió. 1998. Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes. Mol. Cell. Biol. 181156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe, L., M. Iskandar, and J. Ausió. 1998. Folding of chromatin in the presence of heterogeneous histone H1 binding to nucleosomes. J. Biol. Chem. 27311625-11629. [DOI] [PubMed] [Google Scholar]
- 35.Jagannathan, I., H. A. Cole, and J. J. Hayes. 2006. Base excision repair in nucleosome substrates. Chromosome Res. 1427-37. [DOI] [PubMed] [Google Scholar]
- 36.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 3189-97. [DOI] [PubMed] [Google Scholar]
- 37.Kondo, E., Z. Gu, A. Horii, and S. Fukushige. 2005. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol. Cell. Biol. 254388-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 39.Li, A., A. H. Maffey, W. D. Abbott, N. Conde e Silva, A. Prunell, J. Siino, D. Churikov, A. O. Zalensky, and J. Ausio. 2005. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry 442529-2535. [DOI] [PubMed] [Google Scholar]
- 40.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69915-926. [DOI] [PubMed] [Google Scholar]
- 41.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 42.Maffey, A. H., T. Ishibashi, C. He, X. Wang, A. R. White, S. C. Hendy, C. C. Nelson, P. S. Rennie, and J. Ausio. 2007. Probasin promoter assembles into a strongly positioned nucleosome that permits androgen receptor binding. Mol. Cell. Endocrinol. 26810-19. [DOI] [PubMed] [Google Scholar]
- 43.Meehan, R. R., J. D. Lewis, S. McKay, E. L. Kleiner, and A. P. Bird. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58499-507. [DOI] [PubMed] [Google Scholar]
- 44.Menoni, H., D. Gasparutto, A. Hamiche, J. Cadet, S. Dimitrov, P. Bouvet, and D. Angelov. 2007. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A. Bbd nucleosomes. Mol. Cell. Biol. 275949-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakanishi, S., R. Prasad, S. H. Wilson, and M. Smerdon. 2007. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res. 354313-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neddermann, P., P. Gallinari, T. Lettieri, D. Schmid, O. Truong, J. J. Hsuan, K. Wiebauer, and J. Jiricny. 1996. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 27112767-12774. [DOI] [PubMed] [Google Scholar]
- 47.Nikitina, T., X. Shi, R. P. Ghosh, R. A. Horowitz-Scherer, J. C. Hansen, and C. L. Woodcock. 2007. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 27864-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsen, H., T. Lindahl, and A. Verreault. 2002. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 215943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norton, V. G., B. S. Imai, P. Yau, and E. M. Bradbury. 1989. Histone acetylation reduces nucleosome core particle linking number change. Cell 57449-457. [DOI] [PubMed] [Google Scholar]
- 50.Ohki, I., N. Shimotake, N. Fujita, J. Jee, T. Ikegami, M. Nakao, and M. Shirakawa. 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105487-497. [DOI] [PubMed] [Google Scholar]
- 51.Petronzelli, F., A. Riccio, G. D. Markham, S. H. Seeholzer, M. Genuardi, M. Karbowski, A. T. Yeung, Y. Matsumoto, and A. Bellacosa. 2000. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J. Cell. Physiol. 185473-480. [DOI] [PubMed] [Google Scholar]
- 52.Petronzelli, F., A. Riccio, G. D. Markham, S. H. Seeholzer, J. Stoerker, M. Genuardi, A. T. Yeung, Y. Matsumoto, and A. Bellacosa. 2000. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem. 27532422-32429. [DOI] [PubMed] [Google Scholar]
- 53.Pfeifer, G. P. 2006. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301259-281. [DOI] [PubMed] [Google Scholar]
- 54.Pitsikas, P., J. M. Patapas, and C. G. Cupples. 2004. Mechanism of 2-aminopurine-stimulated mutagenesis in Escherichia coli. Mutat. Res. 55025-32. [DOI] [PubMed] [Google Scholar]
- 55.Reed, S. H. 2005. Nucleotide excision repair in chromatin: the shape of things to come. DNA Repair (Amsterdam) 4909-918. [DOI] [PubMed] [Google Scholar]
- 56.Roloff, T. C., H. H. Ropers, and U. A. Nuber. 2003. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson, R. T., F. Thoma, and J. M. Brubaker. 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42799-808. [DOI] [PubMed] [Google Scholar]
- 59.Smerdon, M. J., and A. Conconi. 1999. Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol. 62227-255. [DOI] [PubMed] [Google Scholar]
- 60.Svedruzic, Z. M., C. Wang, J. V. Kosmoski, and M. J. Smerdon. 2005. Accommodation and repair of a UV photoproduct in DNA at different rotational settings on the nucleosome surface. J. Biol. Chem. 28040051-40057. [DOI] [PubMed] [Google Scholar]
- 61.Tatchell, K., and K. E. Van Holde. 1977. Reconstitution of chromatin core particles. Biochemistry 165295-5303. [DOI] [PubMed] [Google Scholar]
- 62.Thambirajah, A. A., D. Dryhurst, T. Ishibashi, A. Li, A. H. Maffey, and J. Ausio. 2006. H2A. Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 28120036-20044. [DOI] [PubMed] [Google Scholar]
- 63.Tini, M., A. Benecke, S. J. Um, J. Torchia, R. M. Evans, and P. Chambon. 2002. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9265-277. [DOI] [PubMed] [Google Scholar]
- 64.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 184629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner, D. P., S. Cortellino, J. E. Schupp, E. Caretti, T. Loh, T. J. Kinsella, and A. Bellacosa. 2006. The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res. 667686-7693. [DOI] [PubMed] [Google Scholar]
- 66.Valinluck, V., P. Liu, J. I. Kang, Jr., A. Burdzy, and L. C. Sowers. 2005. 5-Halogenated pyrimidine lesions within a CpG sequence context mimic 5-methylcytosine by enhancing the binding of the methyl-CpG-binding domain of methyl-CpG-binding protein 2 (MeCP2). Nucleic Acids Res. 333057-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walsh, C. P., and G. L. Xu. 2006. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 301283-315. [DOI] [PubMed] [Google Scholar]
- 68.Wang, D., R. Hara, G. Singh, A. Sancar, and S. J. Lippard. 2003. Nucleotide excision repair from site-specifically platinum-modified nucleosomes. Biochemistry 426747-6753. [DOI] [PubMed] [Google Scholar]
- 69.Wang, X., S. C. Moore, M. Laszckzak, and J. Ausió. 2000. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 27535013-35020. [DOI] [PubMed] [Google Scholar]
- 70.Wilkins, R. J., and R. W. Hart. 1974. Preferential DNA repair in human cells. Nature 24735-36. [DOI] [PubMed] [Google Scholar]
- 71.Wu, P., C. Qiu, A. Sohail, X. Zhang, A. S. Bhagwat, and X. Cheng. 2003. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem. 2785285-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yager, T. D., and K. E. van Holde. 1984. Dynamics and equilibria of nucleosomes at elevated ionic strength. J. Biol. Chem. 2594212-4222. [PubMed] [Google Scholar]
- 73.Yang, W. 2008. Structure and mechanism for DNA lesion recognition. Cell Res. 18184-197. [DOI] [PubMed] [Google Scholar]