Abstract
The Notch signal pathway plays multifaceted roles to promote or suppress tumorigenesis. The Notch1 receptor intracellular domain (N1IC), the activated form of the Notch1 receptor, activates the c-myc proto-oncogene. The complex of N1IC and transcription factor YY1 binds to the human c-myc promoter to enhance c-myc expression in a CBF1-independent manner. Here we demonstrated that N1IC interacted with the c-Myc-regulating proteins α-enolase and c-myc promoter binding protein 1 (MBP-1). Both α-enolase and MBP-1 suppressed the N1IC-enhanced activity of the c-myc promoter in a CBF1-independent manner. The YY1 response element in front of the P2 c-myc promoter was essential and sufficient for the modulation of c-myc by N1IC and α-enolase or MBP-1. Furthermore, N1IC, YY1, and α-enolase or MBP-1 but not CBF1 bound to the c-myc promoter through associating with the YY1 response element. Hemin-induced erythroid differentiation was suppressed by N1IC in K562 cells. This suppression was relieved by the expression of α-enolase and MBP-1. In addition, both α-enolase and MBP-1 suppressed the N1IC-enhanced colony-forming ability through c-myc. These results indicate that the activated Notch1 receptor and α-enolase or MBP-1 cooperate in controlling c-myc expression through binding the YY1 response element of the c-myc promoter to regulate tumorigenesis.
The Notch signal pathway is involved in the control of several cellular functions, including cell fate decision, proliferation, differentiation, apoptosis, and tumorigenesis (for reviews, see references 3, 5, 20, 26, 34, and 39). Notch genes encode evolutionarily conserved receptors that may be exhibited as an oncogene or tumor suppressor to modulate tumorigenesis (for reviews, see references 22, 29, and 34). Depending on the cellular context and cross talk with other signal pathways in various tumor types, the multifaceted roles of Notch signaling function via their effects on differentiation, cellular metabolism, cell cycle progression, angiogenesis, self-renewal, and immune response (29, 30, 34, 47). However, the mechanisms which underlie the Notch signal pathway controlling tumorigenesis are not yet fully understood.
Notch receptors are single-span transmembrane proteins with several functional domains. In the prevailing model of the Notch signal pathway, the Notch receptor is activated after ligand binding and is cleaved to release and translocate its intracellular domain into the nucleus. The Notch receptor intracellular domain modulates its target genes through both C promoter binding factor 1 (CBF1, also called RBP-Jκ)-dependent and -independent pathways (29).
Recently, several reports have documented that the Notch signal pathway enhances the expression of c-myc (19, 24, 28, 30, 35, 36, 44). In our previous study (24), the activated form of Notch1 receptor (the Notch1 receptor intracellular domain [N1IC]) was found to activate c-myc expression via its association with the transcription factor Ying Yang 1 (YY1). Furthermore, the N1IC-YY1-associated complex binds to the c-myc promoter and activates c-Myc expression in a CBF1-independent pathway.
We also had screened cellular factors associated with the activated Notch1 receptor to gain insight into the molecular mechanism of Notch signaling (45, 46). The α-enolase was identified as a candidate N1IC-associated protein. The α-enolase gene encodes both a glycolytic enzyme (17) and a transcriptional suppressor, the c-myc promoter binding protein 1 (MBP-1). MBP-1 is produced by an alternative translation initiation codon from the α-enolase gene but does not have the enzyme activity of enolase (11, 37). c-myc expression is suppressed by α-enolase and MBP-1 through binding to the major c-myc promoter, P2 (7, 11, 31, 32, 37). Both α-enolase and MBP-1 were shown to play an important role in tumorigenesis. They exert a tumor-suppressive effect on breast carcinoma (33), non-small-cell lung cancer (6, 14), prostate tumors (15, 16), and neuroblastoma (10), whereas α-enolase expression is upregulated in hepatitis C virus-related hepatocellular carcinoma (38).
It was demonstrated that the induction of c-myc expression contributes to tumorigenesis (for reviews, see references 1, 2, 9, and 13). A significant proportion (10% to 15%) of genes in both the human and Drosophila genomes are regulated by c-Myc (12, 23, 27). Therefore, it is important to dissect the regulatory mechanism of c-myc expression by Notch signaling, α-enolase, and MBP-1.
MATERIALS AND METHODS
Plasmids and plasmid construction.
The pcDNA-HA-N1IC expression construct contains cDNA encoding the human Notch1 receptor intracellular domain with an N-terminal hemagglutinin (HA) tag (46). The pCMV-YY1 expression construct contains cDNA encoding the full-length YY1 (25, 46). The cDNAs of α-enolase and MBP-1 were cloned by reverse transcription-PCR from RNA of HeLa cells and were confirmed by sequencing. The expression constructs pcDNA-HA-α-enolase and pcDNA-HA-MBP-1 contain the cDNAs encoding α-enolase and MBP-1 with N-terminal HA tags. The expression construct pSG5Flag-RBP-VP16 (a gift of E. Manet) contains cDNA encoding the constitutively active RBP-Jκ mutant. The pGST-α-enolase and pGST-MBP-1 plasmids direct the expression of glutathione S-transferase (GST) fusion proteins with α-enolase and MBP-1, respectively.
The promoter DNA fragments were constructed in front of the luciferase gene in the pGL2-basic vector for all reporter plasmids containing various lengths of the human c-myc promoter. The c-myc promoter DNA was derived from pLB1530 (24), a kind gift from L. M. Boxer. Reporter plasmid pc-Myc-Pro-Luc contains the c-myc promoter (nucleotides [nt] −2328 to +961 in relation to the P2 promoter). The reporter plasmids KNM-Luc, SNM-Luc, NNM-Luc, KXM-Luc, KPM-Luc, and PXM-Luc contain c-myc promoter DNA fragments from the KpnI to NaeI, SmaI to NaeI, NotI to NaeI, KpnI to XhoI, KpnI to PvuII, and PvuII to XhoI sites, respectively. The P0-Luc and P1-Luc reporter plasmids, with P0 and P1 c-myc promoters, contain c-myc promoter DNA fragments from the KpnI to SmaI and SmaI to XhoI sites, respectively. The reporter plasmid KXdPSM-Luc contains the c-myc promoter from the KpnI to XhoI sites in which the DNA fragment from the PvuII to SmaI sites was deleted. The reporter plasmid KXM-YY1 (mt)-Luc contains the c-myc promoter from the KpnI to XhoI sites in which the putative YY1 response element located at nt −244 to −240 in relation to the P2 promoter was mutated from CCATA to TTATA. The reporter plasmid pYY1-RE-Luc containing the wild-type YY1 response elements was described previously (25). All constructs were verified by sequencing.
To knock down the endogenous Notch1 receptor and α-enolase or MBP-1, the following target sequences were constructed in the small interfering RNA (siRNA) vector pLKO.1: Notch1 receptor, 5′-GCCGAACCAATACAACCCTCT-3′; α-enolase or MBP-1, 5′-CCGGCGTTCAATGTCATCAAT-3′ (no. 22) and 5′-CGTGAACGAGAAGTCCTGCAA-3′ (no. 24). An siRNA vector against luciferase (pLKO.1-shLuc) was used as a negative control for knockdown validation. The pPGK-GFP expression construct contains cDNA encoding the green fluorescent protein (GFP) in the pLKO.1 vector as a control. To knock down the endogenous c-Myc, the target sequence 5′-GGTCAGAGTCTGGATCACC-3′ was constructed in the siRNA vector pSilencer 3.1-H1 neo.
Cell culture and transfection.
Human erythroleukemia K562 cells, acute T-cell leukemia Jurkat cells, acute T-cell lymphoblastic lymphoma SUP-T1 cells, and cervical carcinoma HeLa cells were cultured in RPMI 1640 and Dulbecco modified Eagle medium with 10% fetal bovine serum. Established K562 cells expressing the HA-N1IC fusion protein (K562/HA-N1IC) and their control cells (K562/pcDNA3) were described previously (46). Established c-Myc-knockdown HeLa cells (HeLaMyci4 and -5) and their control cells (HeLa control) were described previously (8).
K562 and HeLa cells were transiently transfected with the SuperFect transfection reagent (Qiagen) and the calcium phosphate coprecipitation method, respectively (45, 46). Jurkat and SUP-T1 cells were transiently transfected by electroporation (46). For luciferase reporter gene assay, K562 cells (5 × 105) were seeded onto six-well plates and transiently transfected for 2 days. Luciferase activities were measured using the dual-luciferase reporter assay system (Promega), and then Renilla luciferase activity was used to normalize for transfection efficiency.
For Western blot analysis, HeLa cells (2 × 106) were seeded onto 10-cm dishes and transiently transfected for 2 days. In chromatin immunoprecipitation (ChIP) experiments, K562/HA-N1IC cells (1 × 106) were transfected with 10 μg of the indicated reporter plasmids, and cells were harvested 24 hours after transfection. In erythroid differentiation experiments, K562 cells (5 × 105) were transfected and then induced to differentiate with hemin at 2 days after transfection. For colony-forming assay, K562 cells (5 × 105) and HeLa cells (3 × 105) were transfected with the indicated combination of plasmids with relevant controls for 2 days.
DAPT {N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester} (Sigma-Aldrich) at the indicated concentrations in dimethyl sulfoxide or an equal volume of dimethyl sulfoxide was added for treatment.
Western blot analysis.
Nuclear extracts and whole-cell lysates were prepared as previously described (46). Laemmli's sample buffer was added to the cell lysates or immunoprecipitated pellets, heated at 95°C for 5 min, and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blotting was performed with anti-HA, anti-α-enolase, anti-Notch1 C terminus (C-ter), anti-YY1, anti-c-Myc (Santa Cruz), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Biogenesis) antibodies.
Coimmunoprecipitation.
As described previously (46), 5 μl of anti-α-enolase or anti-Notch1 C-ter antibodies and 50 μl of a 50% (vol/vol) slurry of protein A-Sepharose were added into 450 μl of NETN buffer and then rotated at 4°C for more than 1 hour to prepare the slurry of antibody-conjugated protein A-Sepharose. Immunoprecipitation was performed by rotating mixtures of cell lysates and mouse immunoglobulin G (IgG)-bound protein A-Sepharose (control) or antibody (anti-α-enolase or anti-Notch1 C-ter)-conjugated protein A-Sepharose at 4°C for at least one hour.
Immunofluorescence staining and confocal microscopy.
For immunofluorescence staining, cells (1 × 104) were washed and then resuspended in 300 μl of phosphate-buffered saline. They were spun onto silane-coated slides with a cytofuge (Thermo Cytospin 4; Shandon) at 400 rpm for 5 min. After air drying for 10 min, cells on slides were fixed with acetone-methanol (1:1) at −20°C for 5 min. The cells were then incubated with primary rabbit anti-α-enolase antibody or goat anti-Notch1 C-ter antibody (Santa Cruz) and subsequently with secondary Alexa Fluor 488-conjugated donkey anti-goat IgG or Alexa Fluor 568-conjugated donkey anti-rabbit IgG (Molecular Probes). For nuclear staining, cells were further incubated with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma-Aldrich). After staining, the cells were mounted with antibleaching reagent (Dako), and the localizations of the Notch1 receptor intracellular domain and α-enolase or MBP-1 were examined by confocal laser scanning microscopy (Leica TCS SP2).
GST fusion protein pull-down assay.
GST-MBP-1 and GST-α-enolase fusion proteins expressed from the pGST-MBP-1 and pGST-α-enolase expression constructs were induced and purified as described previously (46). Whole-cell extracts of K562/HA-N1IC, K562/pcDNA3, and SUP-T1 cells were prepared in NETN buffer. A 50% (vol/vol) slurry of glutathione-agarose resin prebound with 0.5 μg of GST or GST fusion proteins was incubated with 500 μg of whole-cell extracts for the pull-down assay at 4°C for 2 h as described previously (46).
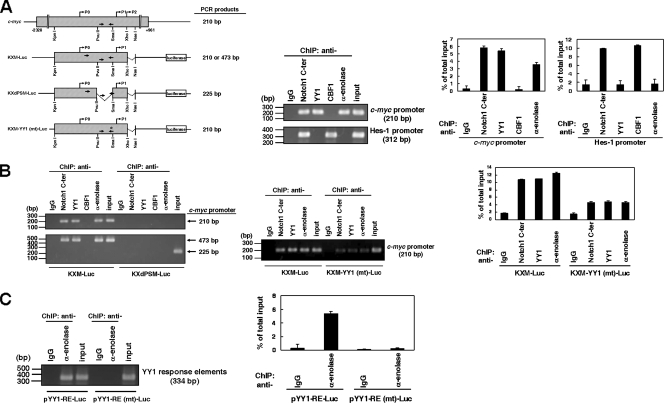
ChIP assay.
For the ChIP assay to amplify the DNA fragments in chromosomal or plasmid DNAs, nuclear or whole-cell extracts were prepared as described before (43, 46). The ChIP assay of K562/HA-N1IC cells with or without transfection was performed using protein A-Sepharose-bound antibodies, including anti-α-enolase, anti-Notch1 C-ter, anti-YY1, and anti-CBF1 (Chemicon) antibodies. The specific primers 5′-GAGGAGCAGCAGAGAAAGG-3′ and 5′-TCCCCCACGCCCTCTGC-3′ for PCR amplification were used to amplify a 210-bp DNA fragment (nt −409 to −200 in relation to the P2 promoter) of the c-myc promoter including chromosomal DNA and reporter plasmids KXM-Luc and KXM-YY1 (mt)-Luc. The specific primers 5′-CAAGACCAAAGCGGAAAGAA-3′ and 5′-GGATCCTGTGTGATCCCTAGGC-3′ for PCR amplification were used to amplify a 312-bp DNA fragment of the Hes-1 promoter in chromosomal DNA. The specific primers 5′-CCTCTATCATTCCTCCC-3′ and 5′-TCCCCCACGCCCTCTGC-3′ for PCR amplification were used to amplify 473- and 225-bp DNA fragments (nt −672 to −200 in relation to the P2 promoter) of the c-myc promoter in reporter plasmids KXM-Luc and KXdPSM-Luc, respectively. The GLprimer1 and GLprimer2 primers for PCR amplification were used to amplify a 334-bp DNA fragment containing the thymidine kinase promoter of herpes simplex virus type 1 and YY1 response elements in the reporter plasmids pYY1-RE-Luc and pYY1-RE (mt)-Luc. The percentages of immunoprecipitated promoter fragments were quantified by real-time PCR using Sybr green as described elsewhere (24) and normalized to total input DNA.
Erythroid differentiation and benzidine staining.
The transfected K562 cells were treated with 40 μM hemin (Sigma-Aldrich) for 2 days to induce erythroid differentiation. The cells (1 × 105) were then washed twice with ice-cold phosphate-buffered saline and resuspended in 27 μl of ice-cold phosphate-buffered saline. The benzidine stock solution contains 0.2% (wt/vol) benzidine dihydrochloride (Sigma-Aldrich) in 0.5 M glacial acetic acid. After addition of 3 μl of benzidine solution containing hydrogen peroxide (0.3% final concentration), the cells were further incubated for 10 min at room temperature. The dark-blue-stained cells were quantitated as benzidine-positive cells under light microscopy. At least 100 cells were counted in triplicate for each experiment.
Colony-forming assay.
To assay for anchorage-independent growth in soft agar, transfected K562, HeLa, Jurkat, or SUP-T1 cells were harvested after 2 days of transfection, and 2,000 K562 cells, 4,000 HeLa cells, 2,000 Jurkat cells, or 2,000 SUP-T1 cells were mixed in 1 ml of 0.2% agar with complete medium and seeded onto a six-well plate containing 1.5 ml of 0.6% agar in complete medium. These cells were further incubated for 7 or 14 days, and 200 μl of medium was added every 3 days to prevent desiccation. After staining with 0.005% crystal violet in phosphate-buffered saline for 1 h, colonies larger than 0.1 mm in diameter were counted under the microscope from 10 random fields.
Real-time PCR analysis.
As described previously (24), total RNA was extracted using the Trizol reagent (Invitrogen), and real-time PCR analysis was performed. The 84-bp Hes-1 cDNA was amplified with the primers 5′-AGCGGGCGCAGATGAC-3′ and 5′-CGTTCATGCACTCGCTGAA-3′. The 71-bp Hey-1 cDNA was amplified with the primers 5′-GAAAAAGCCGAGATC-3′ and 5′-TAACCTTTCCCTCCT-3′. The 51-bp Hey-2 cDNA was amplified with the primers 5′-AGATGCTTCAGGCAACAGGG-3′ and 5′-CAAGAGCGTGTGCGTCAAAG-3′. The 478-bp c-Myc cDNA was amplified with the primers 5′-TACCCTCTCAACGACAGCAG-3′ and 5′-TCTTGACATTCTCCTCGGTG-3′. The 176-bp cDNA of the internal control GAPDH was amplified with the primers 5′-AAATCCCATCACCATCTTCC-3′ and 5′-TCACACCCATGACGAACA-3′. The relative quantification of mRNA expression level was normalized to that of GAPDH and corrected to a calibrator using the RelQuant software (Roche). All data are mean values and standard deviations from three or four independent experiments.
RESULTS
N1IC associates with α-enolase and MBP-1 in the nuclei of cells.
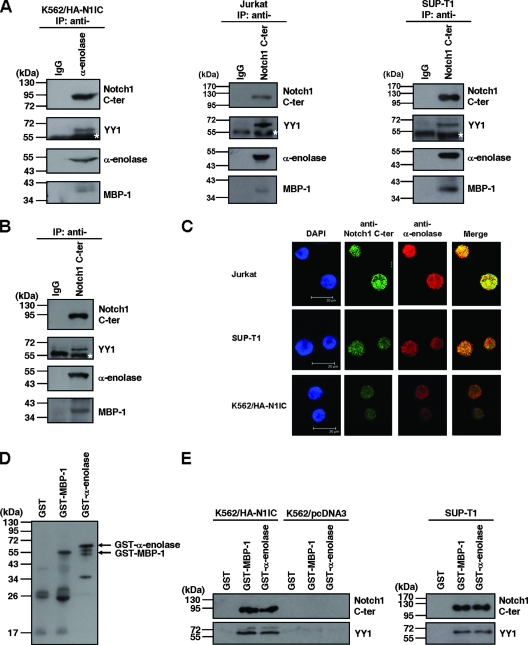
To dissect the molecular mechanism of Notch signaling in tumorigenesis, we had screened cellular factors associated with the activated Notch1 receptor (45, 46). α-Enolase was a candidate N1IC-associated protein. To confirm the association between N1IC and α-enolase in cells, coimmunoprecipitation was performed with N1IC-expressing K562 cells (K562/HA-N1IC). The α-enolase was immunoprecipitated by antibody against α-enolase but not IgG (Fig. 1A, left). After stripping and reprobing of this immunoprecipitated blot, N1IC and YY1 were also detected by anti-Notch1 C-ter and anti-YY1 antibodies, respectively. Furthermore, MBP-1 was also immunoprecipitated and detected by anti-α-enolase antibody.
FIG. 1.
N1IC associates with α-enolase and MBP-1. (A) Whole-cell extracts of N1IC-expressing K562/HA-N1IC cells (left), Jurkat cells (middle), and SUP-T1 cells (right) were prepared for coimmunoprecipitation using anti-IgG, anti-α-enolase, and anti-Notch1 C-ter antibodies. The precipitated proteins were resolved by SDS-PAGE and analyzed by Western blot analysis using anti-Notch1 C-ter antibody. The immunoblot was stripped and then reprobed with anti-YY1 and anti-α-enolase antibodies, sequentially. Note the anti-α-enolase antibody can recognize both α-enolase and MBP-1 (18). (B) Nuclear extracts of K562/HA-N1IC cells were prepared for coimmunoprecipitation using anti-IgG and anti-Notch1 C-ter antibodies. The precipitated proteins were analyzed by Western blot analysis using anti-Notch1 C-ter, anti-YY1, and anti-α-enolase antibodies. (C) The localizations of the Notch1 receptor intracellular domain and α-enolase or MBP-1 were assessed by immunofluorescence staining and confocal microscopy. Slides were incubated with goat anti-Notch1 C-ter or rabbit anti-α-enolase antibodies and subsequently with Alexa Fluor 488-conjugated donkey anti-goat IgG or Alexa Fluor 568-conjugated donkey anti-rabbit IgG. Cell nuclei were also stained by DAPI. (D) Purified GST, GST-MBP-1, and GST-α-enolase fusion proteins were analyzed by Western blot analysis using anti-α-enolase antibodies. (E) Whole-cell extracts of K562/HA-N1IC, K562/pcDNA3, and SUP-T1 cells were used for pull-down assay with purified GST, GST-MBP-1, and GST-α-enolase fusion proteins. The pull-down pellets were resolved by SDS-PAGE and analyzed by Western blotting using anti-Notch1 C-ter (upper panel) or anti-YY1 (lower panel) antibodies. The immunoblots shown here are representative of three or four independent experiments. IP, immunoprecipitated proteins. The white stars indicate the heavy chain of antibody used for immunoprecipitation.
Alternatively, whole-cell extracts of N1IC-expressing gastric SC-M1 cells and HEK293 cells were also used to confirm the associations among these proteins. N1IC, YY1, α-enolase, and MBP-1 all were coimmunoprecipitated by anti-Notch1 C-ter antibody (data not shown). Furthermore, associations among these endogenous proteins were also observed in Jurkat and SUP-T1 cells by coimmunoprecipitaion using anti-Notch1 C-ter antibody (Fig. 1A, middle and right).
α-Enloase is localized in cytoplasm and nuclei (41), but MBP-1 is localized predominantly in nuclei (11). To check whether N1IC interacts with α-enolase and MBP-1 in nuclei, coimmunoprecipitation was also performed with nuclear extracts of K562/HA-N1IC cells using anti-Notch1 C-ter antibody (Fig. 1B). Besides YY1, the α-enolase and MBP-1 were coimmunoprecipitated with N1IC. Additionally, immunofluorescence staining of N1IC and α-enolase or MBP-1 was also performed and then analyzed by confocal microscopy. As described in a previous study (18), the polyclonal H-300 antibody against α-enolase antibody was used to detect α-enolase and MBP-1. N1IC and α-enolase or MBP-1 were colocalized in nuclei of Jurkat, SUP-T1, and K562/HA-N1IC cells (Fig. 1C). These results showed that N1IC is associated with YY1, α-enolase, and MBP-1 in the nuclei of cells.
To investigate whether the interaction between N1IC and α-enolase or MBP-1 is direct or indirect, both the GST-MBP-1 and GST-α-enolase fusion proteins were expressed and purified for GST pull-down assay. The partially purified GST-MBP-1 and GST-α-enolase fusion proteins were analyzed by Western blot analysis using anti-α-enolase antibody (Fig. 1D). Both GST-MBP-1 and GST-α-enolase fusion proteins were associated with N1IC and YY1 of K562/HA-N1IC cells (Fig. 1E, left). However, GST-MBP-1 and GST-α-enolase fusion proteins were not associated with endogenous YY1 of K562/pcDNA3 control cells. Additionally, the associations of GST-MBP-1 and GST-α-enolase fusion proteins with endogenous N1IC and YY1 were also confirmed in SUP-T1 cells (Fig. 1E, right). These results suggest that YY1 does not directly interact with α-enolase and MBP-1 in the absence of N1IC. N1IC is required for the associations among N1IC, YY1, and α-enolase or MBP-1.
The YY1 response element is important for transactivation of c-myc by N1IC.
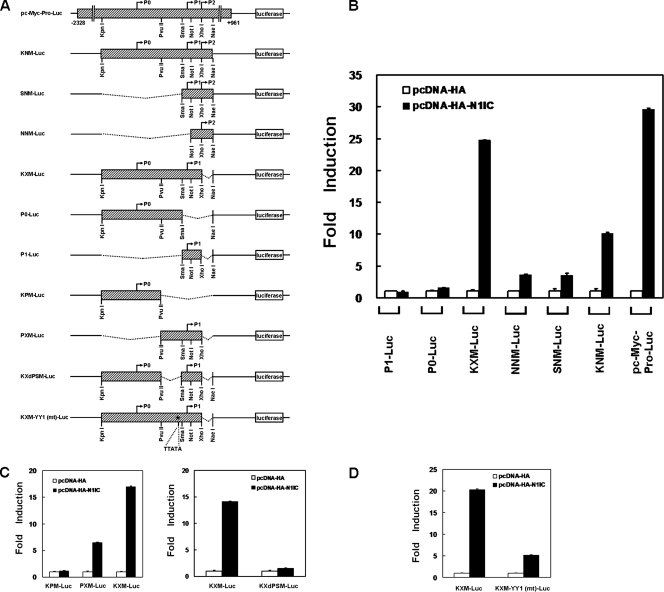
To assess the role of the cooperation between N1IC and α-enolase or MBP-1 in the control of the c-myc gene, we first searched the critical regions of the c-myc promoter for c-myc expression modulated by N1IC. Reporter plasmids containing various lengths of the human c-myc promoter were constructed for luciferase reporter gene assay (Fig. 2A).
FIG. 2.
The YY1 response element is important for transactivation of c-myc by N1IC. (A) Schematic representation of luciferase reporter plasmids containing various lengths of the human c-myc promoter. The reporter plasmid pc-Myc-Pro-Luc contains the full-length c-myc promoter. The P0, P1, and P2 promoters transcribing the c-myc gene and restriction enzymes cleavage sites are shown. The hatched boxes represent the sequence of the c-myc promoter. The star indicates the position of the putative YY1 response element which was mutated in the KXM-YY1 (mt)-Luc reporter plasmid. (B) Reporter plasmids containing various lengths of the c-myc promoter were cotransfected with the N1IC expression construct (pcDNA-HA-N1IC) or its control vector (pcDNA-HA) into K562 cells. (C) Reporter plasmids (left, KPM-Luc, PXM-Luc, and KXM-Luc; right, KXdPSM-Luc and KXM-Luc) containing various lengths of the c-myc promoter were cotransfected with the pcDNA-HA-N1IC expression construct or pcDNA-HA control vector into K562 cells. (D) Both KXM-Luc and KXM-YY1 (mt)-Luc reporter plasmids containing wild-type or mutated YY1 response elements in the c-myc promoter were cotransfected with the pcDNA-HA-N1IC expression construct or pcDNA-HA control vector into K562 cells. Two days after transfection, luciferase activity was determined from whole-cell extracts, and the basal promoter activity of reporter construct was set to unity. Values are means and standard deviations from at least three independent experiments.
The activity of the reporter gene was enhanced after the cotransfection with N1IC-expressing plasmid (pcDNA-HA-N1IC) and pc-Myc-Pro-Luc or KNM-Luc reporter plasmids containing the P0, P1, and P2 c-myc promoters in K562 cells (Fig. 2B). The expression of N1IC also elevated reporter gene activity after transfection with the KXM-Luc reporter plasmid in which the P2 c-myc promoter was deleted. Thus, the P2 c-myc promoter is dispensable for the transactivation of c-myc by N1IC. Additionally, reporter gene activity was slightly promoted after the cotransfection of the N1IC-expressing construct and SNM-Luc reporter plasmid containing both the P1 and P2 c-myc promoters or with the NNM-Luc reporter plasmid containing the P2 c-myc promoter. However, the expression of N1IC did not significantly enhance reporter gene activity after the transfection with P0-Luc or P1-Luc reporter plasmids, containing P0 or P1 c-myc promoters, respectively.
To further map the critical regions of the c-myc promoter regulated by N1IC, the DNA fragment of this promoter in the KXM-Luc reporter plasmid was divided into two parts to construct the KPM-Luc (5′ half) and PXM-Luc (3′ half) reporter plasmids. Reporter gene activity was enhanced after the cotransfection with the N1IC construct and reporter plasmid PXM-Luc, but not KPM-Luc (Fig. 2C, left). Owing to the inability of N1IC to promote reporter gene activity of the P1-Luc reporter plasmid (Fig. 2B), it is possible that N1IC may activate c-myc through the DNA sequence between the PvuII and SmaI sites in the c-myc promoter of the PXM-Luc reporter plasmid. To evaluate this possibility, the DNA fragment of the c-myc promoter between the PvuII and SmaI sites in the KXM-Luc reporter plasmid was omitted and the KXdPSM-Luc reporter plasmid was constructed. The data showed that the expression of N1IC did not elevate reporter gene activity after transfection with the KXdPSM-Luc reporter plasmid (Fig. 2C, right). Therefore, the DNA sequence between the PvuII and SmaI sites in the c-myc promoter is necessary for its activation by N1IC.
As described previously (24), we also found that the activation of the c-myc promoter by N1IC depends on the formation of the N1IC-YY1-associated complex. There should be a YY1 response element in the DNA sequence between the PvuII and SmaI sites of the c-myc promoter. To check whether the putative YY1 response element in the KXM-Luc reporter plasmid is involved in activation of the c-myc promoter by N1IC, the reporter plasmid KXM-YY1 (mt)-Luc containing the mutated YY1 response element was constructed for reporter gene assay. The results showed that the enhancement of reporter gene activity by N1IC was decreased from 20-fold to 5-fold after the YY1 response element was mutated (Fig. 2D).
Both α-enolase and MBP-1 suppress the N1IC-enhanced activity of the c-myc promoter.
The N1IC-YY1-associated complex binds to the YY1 response element (24). As shown in Fig. 1, N1IC interacted with α-enolase and MBP-1 in the nuclei. In addition to binding the P2 promoter, it is possible that α-enolase and MBP-1 could regulate c-myc expression through associating with N1IC to bind the YY1 response element. To address this issue, a reporter gene assay was performed.
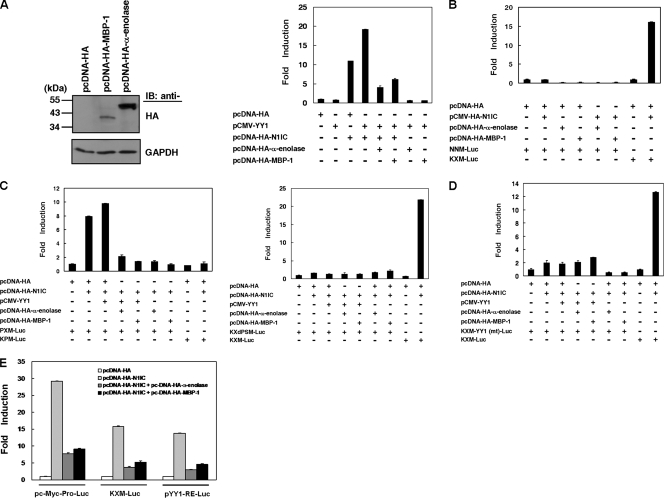
Both α-enolase- and MBP-1-expressing plasmids (pcDNA-HA-α-enolase and pcDNA-HA-MBP-1) were constructed and transfected into HeLa cells. The exogenous expression of these two proteins were detected by Western blot analysis using anti-HA antibody (Fig. 3A, left), and then the KXM-Luc reporter plasmid containing the c-myc promoter was cotransfected with YY1, N1IC, and α-enolase or MBP-1 expression constructs into K562 cells. In agreement with our previous report (24), the expression of YY1 further elevated the N1IC-enhanced activity of the reporter gene containing the c-myc promoter. This elevation of reporter gene activity was suppressed by the expression of α-enolase or MBP-1 (Fig. 3A, right). Both α-enolase and MBP-1 also inhibited the N1IC-enhanced activity of the reporter gene containing the c-myc promoter after transfection with the pc-Myc-Pro-Luc reporter plasmid containing the full-length c-myc promoter (Fig. 3E). However, the reporter gene activity was not significantly enhanced after cotransfection of the N1IC-expressing construct and the NNM-Luc reporter plasmid containing the P2 c-myc promoter (Fig. 3B).
FIG. 3.
Both α-enolase and MBP-1 suppress the N1IC-enhanced activity of the c-myc promoter through the YY1 response element. (A) The MBP-1- and α-enolase-expressing constructs (pcDNA-HA-MBP-1 and pcDNA-HA-α-enolase) were transfected into HeLa cells. Two days after transfection, MBP-1, α-enolase, and GAPDH were detected by Western blot analysis using anti-HA and anti-GAPDH antibodies. IB, proteins detected by immunoblotting. The KXM-Luc reporter plasmid was cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. (B) NNM-Luc and KXM-Luc reporter plasmids were cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. (C) The PXM-Luc or KPM-Luc (left) and KXdPSM-Luc or KXM-Luc (right) reporter plasmids were cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. (D) The KXM-YY1 (mt)-Luc and KXM-Luc reporter plasmids were cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. (E) The pc-Myc-Pro-Luc, KXM-Luc, and pYY1-RE-Luc reporter plasmids were cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. Luciferase reporter gene activity was determined as described in the legend to Fig. 2.
The YY1 response element is essential and sufficient for modulation of the c-myc promoter by N1IC, YY1, α-enolase, and MBP-1.
Based on the results described above, we further searched the critical regions of the c-myc promoter modulated by N1IC, YY1, and α-enolase or MBP-1. The PXM-Luc, KPM-Luc, KXdPSM-Luc, and KXM-Luc reporter plasmids were cotransfected with the indicated expression constructs into K562 cells for reporter gene assay (Fig. 3C). The expression of N1IC and YY1 enhanced reporter gene activity after transfection of the PXM-Luc reporter plasmid but not the KPM-Luc and KXdPSM-Luc reporter plasmids. This enhancement was suppressed by the expression of both α-enolase and MBP-1.
To investigate the role of the YY1 response element in the control of the c-myc promoter by N1IC, YY1, and α-enolase or MBP-1, the KXM-YY1 (mt)-Luc reporter plasmid and expression constructs of the indicated proteins were cotransfected for reporter gene assay (Fig. 3D). The results showed that the mutation of the YY1 response element dramatically abolished the effect of N1IC, YY1, and α-enolase or MBP-1 on the control of the c-myc promoter.
A reporter gene assay was also performed to check whether the expression of N1IC and α-enolase or MBP-1 regulates the activity of a reporter gene with YY1 response elements. The pYY1-RE-Luc reporter plasmid containing YY1 response elements was cotransfected with N1IC- and α-enolase- or MBP-1-expressing constructs into K562 cells. With YY1 response elements, the expression of α-enolase or MBP-1 suppressed N1IC-enhanced activity of the reporter gene (Fig. 3E).
N1IC cooperates with α-enolase and MBP-1 in regulating c-myc promoter activity in a CBF1-independent manner.
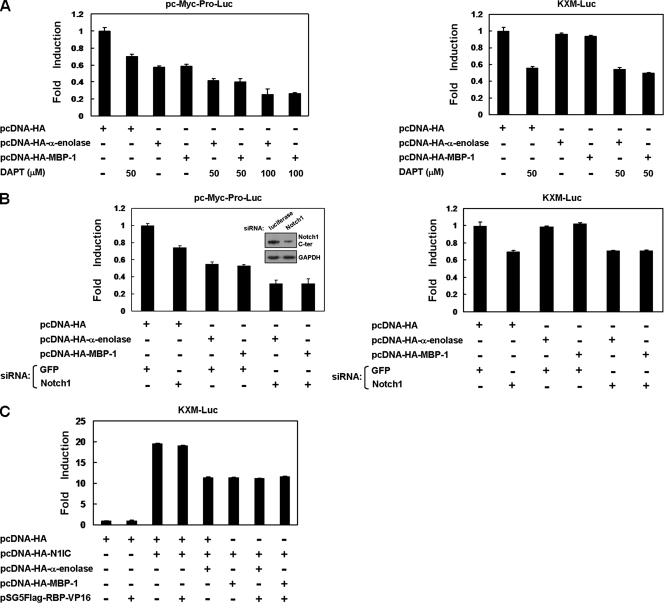
The cooperation of N1IC and α-enolase or MBP-1 in modulating c-myc promoter activity was also evaluated by suppression of endogenous Notch signaling for reporter gene assay. When K562 cells were transfected with the pc-Myc-Pro-Luc reporter plasmid containing the full-length c-myc promoter, reporter gene activity was inhibited by treatment with a Notch signaling inhibitor (DAPT), knockdown of the Notch1 receptor, and expression of α-enolase or MBP-1 (Fig. 4A, left, and B, left). The expression of α-enolase or MBP-1 further suppressed this reporter gene activity inhibited by treatment with DAPT and knockdown of the Notch1 receptor.
FIG. 4.
N1IC cooperates with α-enolase and MBP-1 to regulate the c-myc promoter in a CBF1-independent manner. (A) Reporter plasmids pc-Myc-Pro-Luc (left) and KXM-Luc (right) were cotransfected with MBP-1- and α-enolase-expressing constructs (pcDNA-HA-MBP-1 and pcDNA-HA-α-enolase) into K562 cells treated with 50 or 100 μM DAPT. (B) Reporter plasmids pc-Myc-Pro-Luc (left) and KXM-Luc (right) were cotransfected with expression constructs of the indicated proteins and siRNA vector against Notch1 receptor or its control vector (pPGK-GFP) into K562 cells. The upper inset shows results of Notch1 receptor knockdown after transfection with siRNA vector in N1IC-expressing K562/HA-N1IC cells by Western blot analysis. (C) The KXM-Luc reporter plasmid was cotransfected with plasmids expressing the indicated proteins or their control vector into K562 cells. Luciferase reporter gene activity was determined as described in the legend to Fig. 2.
As demonstrated in Fig. 2B, the P2 c-myc promoter is dispensable for the transactivation of c-myc by N1IC. After transfection with the KXM-Luc reporter plasmid in which the P2 c-myc promoter was deleted, reporter gene activity in K562 cells was decreased by treatment with DAPT and knockdown of the Notch1 receptor but not by the expression of α-enolase or MBP-1 (Fig. 4A, right, and B, right). The expression of α-enolase or MBP-1 also did not further affect this reporter gene activity suppressed by treatment with DAPT or knockdown of the Notch1 receptor.
It was demonstrated that N1IC enhances the c-myc promoter activity through a CBF1-independent pathway (24). To elucidate whether the cooperation of N1IC and α-enolase or MBP-1 in modulating c-myc promoter activity is CBF1 independent, the pSG5Flag-RBP-VP16 expression construct was transfected into cells to express a constitutively active RBP-Jκ-VP16 fusion protein. The KXM-Luc reporter plasmid was cotransfected with N1IC-, α-enolase- or MBP-1-, and RBP-Jκ-VP16 fusion protein-expressing constructs into K562 cells for reporter gene assay (Fig. 4C). Reporter gene activity regulated by N1IC and α-enolase or MBP-1 was not further affected by the exogenous RBP-Jκ-VP16 fusion protein. Therefore, the modulation of c-myc promoter activity by cooperation of N1IC and α-enolase or MBP-1 is CBF1 independent.
N1IC, YY1, and α-enolase or MBP-1, but not CBF1, bind to the c-myc promoter through the YY1 response element.
The aforementioned data showed that N1IC, YY1, and α-enolase or MBP-1 regulated the c-myc promoter through the YY1 response element. We surmised that these proteins might bind to the DNA of the c-myc promoter to modulate reporter gene activity in the context of living cells. To study this possibility, we examined the DNA binding abilities of these proteins on the c-myc promoter by ChIP assay using anti-IgG, anti-Notch1 C-ter, anti-YY1, anti-CBF1, and anti-α-enolase antibodies in K562/HA-N1IC cells. The immunoprecipitated DNA was used to amplify the 210-bp PCR products of the c-myc promoter and the 312-bp PCR products of the promoter of Hes-1, a target gene of the CBF1-dependent Notch signal pathway. The amplified 210-bp PCR products of the c-myc promoter were present in the samples immunoprecipitated with anti-Notch1 C-ter, anti-YY1, and anti-α-enolase antibodies but not in those immunoprecipitated with anti-IgG and anti-CBF1 antibodies (Fig. 5A). Furthermore, the amplified 312-bp PCR products of the Hes-1 promoter were present in samples immunoprecipitated with anti-Notch1 C-ter and anti-CBF1 antibodies but not in those immunoprecipitated with anti-YY1 and anti-α-enolase antibodies. The percentages of immunoprecipitated promoter fragments were also quantified by real-time PCR. These results suggest that N1IC, YY1, and α-enolase or MBP-1, but not CBF1, bind to the c-myc promoter in chromosomal DNA of K562/HA-N1IC cells, whereas N1IC and CBF1, but not YY1 and α-enolase or MBP-1, bind to the Hes-1 promoter.
FIG. 5.
N1IC, YY1, and α-enolase or MBP-1 but not CBF1 bind to the c-myc promoter in cells through associating with the YY1 response element. (A) The hatched box indicates sequences of the c-myc promoter in chromosomal DNA and reporter plasmids, including KXM-Luc, KXdPSM-Luc, and KXM-YY1 (mt)-Luc (left). The arrows indicate positions of primer pairs used to amplify the PCR products of the c-myc promoter. The star indicates the position of the mutated YY1 response element (nt −244 to −240 in relation to the P2 promoter). K562/HA-N1IC cells were harvested for ChIP assay using anti-IgG, anti-Notch1 C-ter, anti-YY1, anti-CBF1, and anti-α-enolase antibodies (middle). The immunoprecipitated DNA was used to amplify the 210-bp PCR products of the c-myc promoter and the 312-bp PCR products of the Hes1 promoter. The percentages of immunoprecipitated DNAs were also quantified by real-time PCR and normalized to total input DNA (right). (B) K562/HA-N1IC cells were transfected with KXM-Luc and KXdPSM-Luc reporter plasmids (left). Twenty-four hours after transfection, cells were harvested for ChIP assay using anti-IgG, anti-Notch1 C-ter, anti-YY1, anti-CBF1, and anti-α-enolase antibodies. The immunoprecipitated DNAs were used to amplify the 210-bp PCR products in the region of the c-myc promoter (upper panel). The immunoprecipitated DNAs were also used to amplify the 473-bp and 225-bp PCR products in the region of the c-myc promoter in the KXM-Luc and KXdPSM-Luc reporter plasmids (lower panel), respectively. K562/HA-N1IC cells were transfected with KXM-Luc and KXM-YY1 (mt)-Luc reporter plasmids (middle and right). Twenty-four hours after transfection, cells were harvested for ChIP assay using anti-IgG, anti-Notch1 C-ter, anti-YY1, and anti-α-enolase antibodies. The immunoprecipitated DNA was used to amplify the 210-bp PCR products in the region of the c-myc promoter. (C) K562/HA-N1IC cells were transfected with pYY1-RE-Luc and pYY1-RE (mt)-Luc reporter plasmids. Twenty-four hours after transfection, cells were harvested for ChIP assay using anti-IgG and anti-α-enolase antibodies. The immunoprecipitated DNA was used to amplify the 334-bp PCR products in the region of the YY1 response elements in the reporter plasmid. Input, 10% of cell lysates. Error bars indicate standard deviations.
To further delineate whether the YY1 response element is essential and sufficient for binding of N1IC, YY1, and α-enolase or MBP-1 to the c-myc promoter, the ChIP assay was applied after transfection of reporter plasmids into K562/HA-N1IC cells. When the KXM-Luc reporter plasmid was transfected, both 210-bp and 473-bp PCR products of the c-myc promoter were present in samples immunoprecipitated with anti-Notch1 C-ter, anti-YY1, and anti-α-enolase antibodies but not in those immunoprecipitated mouse anti-IgG and anti-CBF1 antibodies (Fig. 5B, left). However, the 225-bp PCR products of the c-myc promoter were absent in those immunoprecipitated with anti-Notch1 C-ter, anti-YY1, anti-CBF1, and anti-α-enolase antibodies when the KXdPSM-Luc reporter plasmid was transfected.
After transfection of the KXM-YY1 (mt)-Luc reporter plasmid, the 210-bp PCR products of the c-myc promoter were faintly present in samples immunoprecipitated with anti-Notch1 C-ter, anti-YY1, and anti-α-enolase antibodies but not in those immunoprecipitated with anti-IgG antibody (Fig. 5B, middle and right). These results demonstrated that the YY1 response element is important for binding of N1IC, YY1, and α-enolase or MBP-1 to the c-myc promoter in cells.
The ChIP assay was also used to examine the specific associations between YY1 response elements and α-enolase or MBP-1 in the context of living cells. K562/HA-N1IC cells were transiently transfected with the pYY1-RE-Luc reporter plasmid containing the wild-type YY1 response elements. At 24 hours after transfection, cells were harvested for the ChIP assay using anti-IgG or anti-α-enolase antibodies. The amplified PCR product of 334 bp was present only in the sample immunoprecipitated with anti-α-enolase antibody (Fig. 5C) and not in that immunoprecipitated by anti-IgG control antibody. However, the amplified 334-bp PCR products were absent in the samples immunoprecipitated by both anti-α-enolase and control antibodies after transfection with the pYY1-RE (mt)-Luc reporter plasmid containing the mutant YY1 response elements.
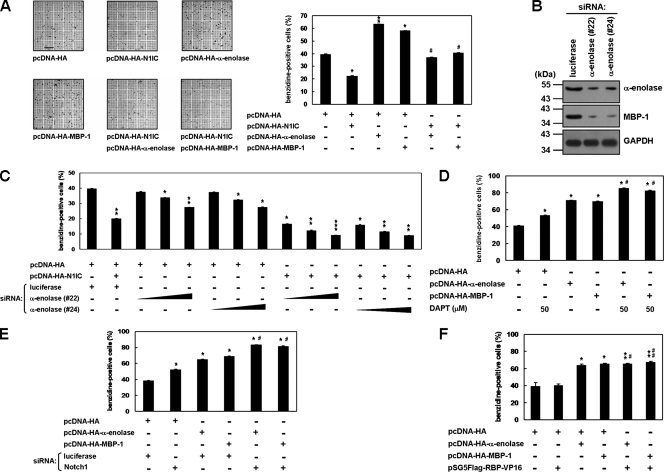
α-Enolase and MBP-1 relieve N1IC-suppressed erythroid differentiation in K562 cells.
It was found that N1IC suppresses erythroid differentiation of K562 cells (21). To elucidate the biological function of the cooperation between N1IC and α-enolase or MBP-1 in tumorigenesis, erythroid differentiation of K562 cells was evaluated in the present study. After the transfection of α-enolase- or MBP-1-expressing constructs, erythroid differentiation of K562 cells was promoted in the presence of hemin (Fig. 6A). Furthermore, the expression of α-enolase and MBP-1 also relieved the N1IC-suppressed erythroid differentiation in K562 cells.
FIG. 6.
α-Enolase and MBP-1 relieve the N1IC-suppressed erythroid differentiation in K562 cells. (A) K562 cells were transfected with plasmids expressing the indicated proteins or their control vector for 2 days. The transfected cells were treated with 40 μM hemin for 2 days to induce erythroid differentiation, and then the benzidine-positive cells were counted after staining. Bar, 0.2 mm. (B) K562 cells were transfected with siRNA vectors against α-enolase and MBP-1 (no. 22 and 24) or luciferase for 2 days. Whole-cell extracts of the transfected cells were used to evaluate knockdown of endogenous α-enolase and MBP-1. Western blot analysis was performed using anti-α-enolase and anti-GAPDH antibodies. (C) K562 cells were cotransfected with the pcDNA-HA-N1IC expression plasmid and various amounts (0.25, 0.5, and 1.0 μg) of siRNA vectors against α-enolase and MBP-1 (no. 22 and 24). (D) MBP-1- or α-enolase-expressing constructs were transfected into K562 cells treated with 50 μM DAPT. (E) K562 cells were cotransfected with expression constructs of the indicated proteins and siRNA vector against Notch1 receptor or its control vector. (F) Expression constructs of the indicated proteins were cotransfected into K562 cells. Two days after transfection, the cells were treated with hemin to induce erythroid differentiation, and then the cells were stained and counted as described above. * and #, P < 0.05; ** and ##, P < 0.01; ***, P < 0.001. Error bars indicate standard deviations.
The siRNA method was also applied to confirm the roles of endogenous α-enolase and MBP-1 in the regulation of N1IC-suppressed erythroid differentiation in K562 cells. As presented in Fig. 6B, transient transfection with siRNA vectors (no. 22 and 24) against α-enolase and MBP-1 could knock down the endogenous expression of these two proteins in K562 cells. The erythroid differentiation of K562 cells was inhibited after transfection with siRNA vectors against α-enolase and MBP-1 in a dose-dependent manner (Fig. 6C). When the siRNA vectors against α-enolase and MBP-1 were transfected into K562 cells, the N1IC-suppressed erythroid differentiation was further downregulated.
The cooperation of N1IC and α-enolase or MBP-1 in modulating erythroid differentiation was also investigated by suppression of endogenous Notch signaling. The erythroid differentiation of K562 cells was enhanced by treatment with DAPT, knock down of the Notch1 receptor, and expression of α-enolase or MBP-1 (Fig. 6D and E). The erythroid differentiation enhanced by treatment with DAPT or knockdown of the Notch1 receptor was further promoted by the expression of α-enolase or MBP-1. To check whether the enhancement of erythroid differentiation by α-enolase and MBP-1 is CBF1 independent, the RBP-Jκ-VP16 fusion protein expression construct was transfected into K562 cells. The results showed that erythroid differentiation promoted by exogenous α-enolase or MBP-1 was not further affected by the expression of RBP-Jκ-VP16 fusion protein (Fig. 6F). Therefore, the regulation of erythroid differentiation in K562 cells by α-enolase and MBP-1 is CBF1 independent.
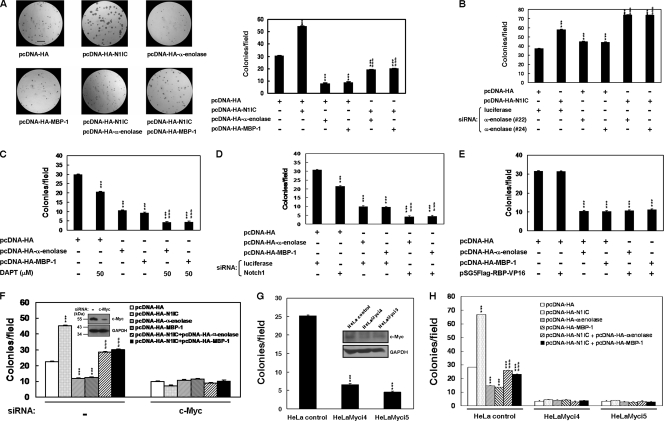
α-Enolase and MBP-1 suppress the colony-forming ability enhanced by N1IC through c-myc.
The role of the cooperation between N1IC and α-enolase or MBP-1 in colony-forming ability was also assessed in soft agar. As shown in Fig. 7A, the colony-forming ability of K562 cells was increased by N1IC and inhibited by α-enolase and MBP-1. Moreover, the increment of colony-forming ability in K562 cells by caused N1IC was suppressed after cotransfection with α-enolase or MBP-1 constructs. We also found that the enhancement of colony-forming ability in K562 cells by N1IC was further increased after cotransfection with siRNA vectors against α-enolase and MBP-1 (Fig. 7B).
FIG. 7.
α-Enolase and MBP-1 suppress the colony-forming ability enhanced by N1IC through c-myc. (A) K562 cells were transfected with plasmids expressing the indicated proteins or their control vector for 2 days. The transfected cells were harvested, seeded in top agar, and incubated for 7 days, and then cells were stained with 0.005% crystal violet for 1 hour and counted under the microscope from 10 random fields. Bar, 1.0 mm. (B) K562 cells were cotransfected with pcDNA-HA-N1IC expression plasmid and siRNA vectors against α-enolase and MBP-1 (no. 22 and 24). Two days after transfection, the colony-forming ability of the cells was determined. (C) The α-enolase or MBP-1 expression plasmids were transfected into K562 cells treated with 50 μM DAPT. (D) K562 cells were cotransfected with expression constructs of the indicated proteins and siRNA vector against Notch1 receptor or its control vector. (E) K562 cells were cotransfected with expression constructs of the indicated proteins or their control vector. (F) K562 cells were cotransfected with expression constructs of the indicated proteins and siRNA vector against c-Myc or its control vector (−). The upper inset shows results of c-Myc knockdown after transfection with siRNA vector in K562 cells by Western blot analysis. (G) The c-Myc-knockdown HeLa cells (HeLaMyci4 and HeLaMyci5) and their control cells (HeLa control) were seeded in top agar and incubated for 14 days. The colony-forming assay was performed as described above. The upper inset shows results of c-Myc expression in HeLaMyci4, HeLaMyci5, and HeLa control cells by Western blot analysis. (H) The HeLaMyci4, HeLaMyci5, and HeLa control cells were transiently transfected with plasmids expressing the indicated proteins or their control vector for 2 days. The cells were then assayed for their colony-forming ability as described above. *** and ###, P < 0.001. Error bars indicate standard deviations.
The cooperation of N1IC and α-enolase or MBP-1 in modulating colony-forming ability was also assessed by suppression of endogenous Notch signaling. The colony-forming ability of K562 cells was inhibited by treatment with DAPT, knockdown of Notch1 receptor, and expression of α-enolase or MBP-1 (Fig. 7C and D). The colony-forming ability inhibited by treatment with DAPT or knockdown of the Notch1 receptor was further suppressed by α-enolase or MBP-1. To check whether the modulation of colony-forming ability by α-enolase and MBP-1 is CBF1 independent, the RBP-Jκ-VP16 fusion protein expression construct was transfected into K562 cells. The data showed that colony-forming ability inhibited by exogenous α-enolase or MBP-1 was not further affected by the expression of RBP-Jκ-VP16 fusion protein (Fig. 7E). Therefore, the regulation of colony-forming ability in K562 cells by α-enolase and MBP-1 is CBF1 independent.
To check whether c-myc is necessary for the regulation of colony-forming ability by cooperation of N1IC and α-enolase or MBP-1, the colony-forming assay was applied after knockdown of endogenous c-myc in K562 and HeLa cells. The increment of colony-forming ability in K562 cells caused by N1IC was suppressed after cotransfection with α-enolase- or MBP-1-expressing constructs (Fig. 7F). However, the effect elicited by the cooperation of N1IC and α-enolase or MBP-1 on colony-forming ability was abolished after knockdown of c-Myc in K562 cells.
As demonstrated in a previous report (8), the knockdown of c-Myc was also observed in both HeLaMyci4 and HeLaMyci5 cells compared with HeLa control cells (Fig. 7G). The colony-forming ability was suppressed after knockdown of endogenous c-myc in HeLaMyci4 and HeLaMyci5 cells. Furthermore, the increase of colony-forming ability in HeLa control cells by N1IC was suppressed after cotransfection with α-enolase- or MBP-1-expressing constructs (Fig. 7H). However, the effect on colony-forming ability caused by the cooperation of N1IC and α-enolase or MBP-1 was abolished after knockdown of c-Myc in HeLaMyci4 and HeLaMyci5 cells.
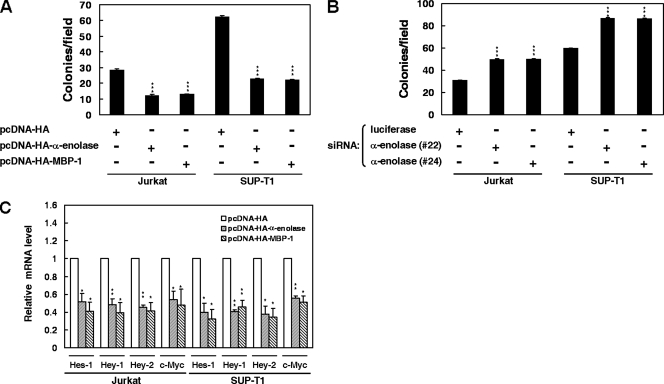
α-Enolase and MBP-1 suppress colony-forming ability and Notch target gene expression in leukemia cells.
Owing to the important roles of the Notch1 receptor and c-Myc activation in T-cell development and leukemia, we surmised that α-enolase and MBP-1 might participate in regulation of tumorigenesis in T-cell leukemia. Therefore, the colony-forming ability of Jurkat and SUP-T1 cells was evaluated to address the role of α-enolase and MBP-1 in T-cell leukemia. The results showed that expression of α-enolase and MBP-1 suppressed the colony-forming ability of these cells (Fig. 8A). Additionally, knockdown of α-enolase and MBP-1 enhanced colony-forming ability in Jurkat and SUP-T1 cells (Fig. 8B). Expression of α-enolase and MBP-1 also inhibited the mRNA expression of Notch target genes, including Hes-1, Hey-1, Hey-2, and c-Myc, in Jurkat and SUP-T1 cells (Fig. 8C).
FIG. 8.
α-Enolase and MBP-1 suppress the colony-forming ability and Notch target gene expression in leukemia cells. (A) Jurkat and SUP-T1 cells were transfected with α-enolase or MBP-1 expression plasmids for colony-forming assay as described above. (B) The siRNA vectors against α-enolase and MBP-1 (no. 22 and 24) or luciferase were transfected into Jurkat or SUP-T1 cells for colony-forming assay. (C) After transfection with α-enolase or MBP-1 expression constructs into Jurkat and SUP-T1 cells for 2 days, the transcript levels of Hes-1, Hey-1, Hey-2, and c-Myc were measured by quantitative real-time PCR. The data were compared after being normalized to GAPDH. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate standard deviations.
DISCUSSION
The role and control of the Notch signal pathway in tumorigenesis are very complicated and still remain unclear. Protein-protein interaction is an important regulatory mechanism of Notch signaling. The Notch receptor intracellular domain elicits its biological function through associating with many cellular factors. Here, we show that endogenous N1IC associates with intrinsic α-enolase and MBP-1 in Jurkat and SUP-T1 cells (Fig. 1A). These associations were also observed in the nuclei (Fig. 1B and C).
N1IC, YY1, α-enolase, and MBP-1 bound on the c-myc promoter to modulate c-myc expression, at least in part, via the YY1 response element (Fig. 5). Therefore, these proteins cooperate in the control of c-myc expression to regulate cellular processes such as colony-forming ability and erythroid differentiation (Fig. 6 and 7). Similarly, Pim1 is recruited to the DNA of the E-box element via associating with c-Myc in tumorigenesis (48). The formation of the Myc-Max-Pim1 ternary complex contributes to the activation of c-Myc target genes.
Our results demonstrated the inhibitory effect of N1IC on erythroid differentiation in K562 cells, which is also consistent with the previously published work of Lam et al. (21). We also demonstrated that the cooperative effect of N1IC and α-enolase or MBP-1 affected the biological function of colony-forming ability in K562 and HeLa cells through c-myc expression (Fig. 7F and H). Additionally, both α-enolase and MBP-1 were involved in the control of tumorigenesis in T-cell leukemia (Fig. 8). This is the first report regarding the role of α-enolase and MBP-1 in the control of c-myc expression modulated by CBF1-independent Notch signaling.
As shown in Fig. 2D and 3D, the activities of reporter genes containing the c-myc promoter with the mutated YY1 response element were slightly enhanced by the expression of N1IC. Actually, YY1 still slightly bound to the mutated YY1 response element after the transfection with the KXM-YY1 (mt)-Luc reporter plasmid (Fig. 5B). Therefore, the activity of the reporter gene containing the c-myc promoter with the mutated YY1 response element was slightly upregulated by the expression of N1IC. However, the mutation of the YY1 response element significantly decreased the binding of N1IC, YY1, and α-enolase to the c-myc promoter (Fig. 5B). Additionally, the other binding sites of transcription factors may also contribute to the transcativation of c-myc by the Notch1 receptor intracellular domain. Many transcription factors, such as NF-κB, are involved in the control of the c-myc promoter (42). The Notch1 receptor intracellular domain can regulate the target genes of the NF-κB pathway (40).
The majority of c-myc RNA in humans is transcribed from the P1 and P2 promoters (4). Here, the results show that the activated Notch1 receptor can significantly enhance the activity of the c-myc promoter, by 30-fold (Fig. 2B). The deregulated Notch1 expression may lead to aberrant c-myc expression to promote tumorigenesis. Therefore, it is important to maintain a moderate level of the Notch signal pathway in cells. Actually, the promotion of c-Myc protein expression is only 3.8-fold in N1IC-expressing K562/HA-N1IC cells (24). It is possible that the Notch1 receptor intracellular domain is unstable (21), and both α-enolase and MBP-1 could also prevent the irregularity of c-myc expression activated by N1IC in two ways. Both α-enolase and MBP-1 suppress the activity of the c-myc promoter via direct binding to its P2 promoter (7, 11, 31, 32, 37). In addition, these two proteins also indirectly bind on the YY1 response element in the c-myc promoter to inhibit c-myc promoter activity (Fig. 3 and 5).
MBP-1 is the product of internal translation initiation from the α-enolase gene (11, 37). Here, it is not excluded that the transfection of the α-enolase-expressing construct could simultaneously express α-enolase and MBP-1 to suppress the c-myc promoter. So far, many genes have been found to possess the ability for internal translation initiation. Apparently, there must be an efficient, rapid, and conserved means for regulating the activity of these genes (7, 11, 31, 32, 37). The regulation of α-enolase and MBP-1 expression by internal translation initiation could participate in the proper control of c-myc activity to protect cells from tumorigenesis.
As presented in Fig. 6C and 7B, the knockdown of α-enolase and MBP-1 suppressed erythroid differentiation and promoted the colony-forming ability of K562 cells. However, these two biological effects were not dramatic. This may be due to the partial knockdown after transient transfection of siRNA vectors (Fig. 6B). Additionally, we found that the effect of N1IC and α-enolase or MBP-1 on the colony-forming ability of K562 and HeLa cells was c-Myc dependent (Fig. 7F and H). The knockdown of MBP-1 also delays cell cycle progression by inhibiting cyclin A and cyclin B1 expression in prostate cancer cells (15, 16). Therefore, MBP-1 could regulate tumorigenesis through different pathways.
The DNA sequence between the PvuII and SmaI sites in the c-myc promoter is necessary for its activation by N1IC (Fig. 2C, right, and 3C, right). There is no CBF1 binding site in the DNA sequence of the 210-bp PCR product of the c-myc promoter between the PvuII and SmaI sites. These results showed that N1IC can activate c-myc expression in a CBF1-independent manner, which is consistent with our previous study (24). Four Notch receptors regulate their target genes in both CBF1-dependent and -independent pathways (29). Here, this activation by N1IC was proven to be suppressed by the expression of α-enolase or MBP-1 (Fig. 3A, right). Whether the other Notch receptors are also involved in the regulation of c-myc expression remains unknown. Further studies are still needed to explain the c-myc expression regulated by Notch signaling.
Acknowledgments
We thank L. M. Boxer for the kind gift of reporter plasmid pLB1530 containing the c-myc promoter and E. Manet for providing the expression construct pSG5Flag-RBP-VP16. RNAi reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica.
The National RNAi Core Facility is supported by National Research Program for Genomic Medicine NSC grants (NSC 94-3112-B-001-003 and NSC 94-3112-B-001-018-Y). This work was supported by the National Science Council (grants NSC 96-3112-B-010-019 and NSC 95-2320-B-010-067-MY2) and in part by a grant from the Ministry of Education, Aim for the Top University Plan.
Footnotes
Published ahead of print on 19 May 2008.
REFERENCES
- 1.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nature Rev. Mol. Cell. Biol. 6635-645. [DOI] [PubMed] [Google Scholar]
- 2.Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front. Biosci. 3d250-d268. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284770-776. [DOI] [PubMed] [Google Scholar]
- 4.Battey, J., C. Moulding, R. Taub, W. Murphy, T. Stewart, H. Potter, G. Lenoir, and P. Leder. 1983. The human c-myc oncogene: structural consequences of translocation into the Igh locus in Burkitt lymphoma. Cell 34779-787. [DOI] [PubMed] [Google Scholar]
- 5.Bolos, V., J. Grego-Bessa, and J. L. de la Pompa. 2007. Notch signaling in development and cancer. Endocrinol. Rev. 28339-363. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. S., W. Wu, G. Walsh, W. K. Hong, and L. Mao. 2003. Enolase-α is frequently down-regulated in non-small cell lung cancer and predicts aggressive biological behavior. Clin. Cancer Res. 93641-3644. [PubMed] [Google Scholar]
- 7.Chaudhary, D., and D. M. Miller. 1995. The c-myc promoter binding protein (MBP-1) and TBP bind simultaneously in the minor groove of the c-myc P2 promoter. Biochemistry 343438-3445. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, Y. C., S. C. Teng, Y. N. Su, F. J. Hsieh, and K. J. Wu. 2003. c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J. Biol. Chem. 27819286-19291. [DOI] [PubMed] [Google Scholar]
- 9.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejeskar, K., C. Krona, H. Caren, F. Zaibak, L. Li, T. Martinsson, and P. Ioannou. 2005. Introduction of in vitro transcribed ENO1 mRNA into neuroblastoma cells induces cell death. BMC Cancer 5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feo, S., D. Arcuri, E. Piddini, R. Passantino, and A. Giallongo. 2000. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 47347-52. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 171115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garte, S. J. 1993. The c-myc oncogene in tumor progression. Crit. Rev. Oncog. 4435-449. [PubMed] [Google Scholar]
- 14.Ghosh, A., R. Steele, J. Ryerse, and R. Ray. 2006. Tumor-suppressive effects of MBP-1 in non-small cell lung cancer cells. Cancer Res. 6611907-11912. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, A. K., R. Steele, and R. B. Ray. 2005. c-myc promoter-binding protein 1 (MBP-1) regulates prostate cancer cell growth by inhibiting MAPK pathway. J. Biol. Chem. 28014325-14330. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, A. K., R. Steele, and R. B. Ray. 2005. Carboxyl-terminal repressor domain of MBP-1 is sufficient for regression of prostate tumor growth in nude mice. Cancer Res. 65718-721. [PubMed] [Google Scholar]
- 17.Harris, R. A. 2006. Carbohydrate metabolism 1: major metabolic pathways and their control, p. 581-635. In T. M. Devlin (ed.), Textbook of biochemistry with clinical correlations, 6th ed. Wiley-Liss Press, New York, NY.
- 18.Ito, S., T. Honma, K. Ishida, N. Wada, S. Sasaoka, M. Hosoda, and T. Nohno. 2007. Differential expression of the human α-enolase gene in oral epithelium and squamous cell carcinoma. Cancer Sci. 98499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klinakis, A., M. Szabolcs, K. Politi, H. Kiaris, S. Artavanis-Tsakonas, and A. Efstratiadis. 2006. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 1039262-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopan, R. 2002. Notch: a membrane-bound transcription factor. J. Cell Sci. 1151095-1097. [DOI] [PubMed] [Google Scholar]
- 21.Lam, L. T., C. Ronchini, J. Norton, A. J. Capobianco, and E. H. Bresnick. 2000. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by Notch-1. J. Biol. Chem. 27519676-19684. [DOI] [PubMed] [Google Scholar]
- 22.Leong, K. G., and A. Karsan. 2006. Recent insights into the role of Notch signaling in tumorigenesis. Blood 1072223-2233. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 1008164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao, W. R., R. H. Hsieh, K. W. Hsu, M. Z. Wu, M. J. Tseng, R. T. Mai, Y. H. W. Lee, and T. S. Yeh. 2007. The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis 281867-1876. [DOI] [PubMed] [Google Scholar]
- 25.Mai, R. T., T. S. Yeh, C. F. Kao, S. K. Sun, H. H. Huang, and Y. H. W. Lee. 2006. Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene 25448-462. [DOI] [PubMed] [Google Scholar]
- 26.Miele, L., and B. Osborne. 1999. Arbiter of differentiation and death: Notch signaling meets apoptosis. J. Cell. Physiol. 181393-409. [DOI] [PubMed] [Google Scholar]
- 27.Orian, A., B. van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. M. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 171101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palomero, T., W. K. Lim, D. T. Odom, M. L. Sulis, P. J. Real, A. Margolin, K. C. Barnes, J. O'Neil, D. Neuberg, A. P. Weng, J. C. Aster, F. Sigaux, J. Soulier, A. T. Look, R. A. Young, A. Califano, and A. A. Ferrando. 2006. NOTCH1 directly regulates c-Myc and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 10318261-18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radtke, F., and K. Raj. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3756-767. [DOI] [PubMed] [Google Scholar]
- 30.Rao, P., and T. Kadesch. 2003. The intracellular form of Notch blocks transforming growth factor β-mediated growth arrest in Mv1Lu epithelial cells. Mol. Cell. Biol. 236694-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray, R. 1995. Induction of cell death in murine fibroblasts by a c-myc promoter binding protein. Cell Growth Differ. 61089-1096. [PubMed] [Google Scholar]
- 32.Ray, R., and D. Miller. 1991. Cloning and characterization of a human c-myc promoter-binding protein. Mol. Cell. Biol. 112154-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray, R. B., R. Steele, E. Seftor, and M. Hendrix. 1995. Human breast carcinoma cells transfected with the gene encoding a c-myc promoter-binding protein (MBP-1) inhibits tumors in nude mice. Cancer Res. 553747-3751. [PubMed] [Google Scholar]
- 34.Roy, M., W. S. Pear, and J. C. Aster. 2007. The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 1752-59. [DOI] [PubMed] [Google Scholar]
- 35.Satoh, Y., I. Matsumura, H. Tanaka, S. Ezoe, H. Sugahara, M. Mizuki, H. Shibayama, E. Ishiko, J. Ishiko, K. Nakajima, and Y. Kanakura. 2004. Roles for c-Myc in self-renewal of hematopoietic stem cells. J. Biol. Chem. 27924986-24993. [DOI] [PubMed] [Google Scholar]
- 36.Sharma, V. M., J. A. Calvo, K. M. Draheim, L. A. Cunningham, N. Hermance, L. Beverly, V. Krishnamoorthy, M. Bhasin, A. J. Capobianco, and M. A. Kelliher. 2006. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 268022-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian, A., and D. M. Miller. 2000. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J. Biol. Chem. 2755958-5965. [DOI] [PubMed] [Google Scholar]
- 38.Takashima, M., Y. Kuramitsu, Y. Yokoyama, N. Iizuka, M. Fujimoto, T. Nishisaka, K. Okita, M. Oka, and K. Nakamura. 2005. Overexpression of alpha enolase in hepatitis C virus-related hepatocellular carcinoma: association with tumor progression as determined by proteomic analysis. Proteomics 51686-1692. [DOI] [PubMed] [Google Scholar]
- 39.Tanigaki, K., and T. Honjo. 2007. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 8451-456. [DOI] [PubMed] [Google Scholar]
- 40.Vilimas, T., J. Mascarenhas, T. Palomero, M. Mandal, S. Buonamici, F. Meng, B. Thompson, C. Spaulding, S. Macaroun, M.-L. Alegre, B. L. Kee, A. Ferrando, L. Miele, and I. Aifantis. 2007. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 1370-77. [DOI] [PubMed] [Google Scholar]
- 41.Wang, W., L. Wang, A. Endoh, G. Hummelke, C. L. Hawks, and P. J. Hornsby. 2005. Identification of α-enolase as a nuclear DNA-binding protein in the zona fasciculata but not the zona reticularis of the human adrenal cortex. J. Endocrinol. 18485-94. [DOI] [PubMed] [Google Scholar]
- 42.Weber, A., J. Liu, I. Collins, and D. Levens. 2005. TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression. Mol. Cell. Biol. 25147-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 2637-47. [DOI] [PubMed] [Google Scholar]
- 44.Weng, A. P., J. M. Millholland, Y. Yashiro-Ohtani, M. L. Arcangeli, A. Lau, C. Wai, C. del Bianco, C. G. Rodriguez, H. Sai, J. Tobias, Y. Li, M. S. Wolfe, C. Shachaf, D. Felsher, S. C. Blacklow, W. S. Pear, and J. C. Aster. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 202096-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeh, T. S., R. H. Hsieh, S. C. Shen, S. H. Wang, M. J. Tseng, C. M. Shih, and J. J. Lin. 2004. Nuclear βII-tubulin associates with the activated Notch receptor to modulate Notch signaling. Cancer Res. 648334-8340. [DOI] [PubMed] [Google Scholar]
- 46.Yeh, T. S., Y. M. Lin, R. H. Hsieh, and M. J. Tseng. 2003. Association of transcription factor YY1 with the high molecular weight Notch complex suppresses the transactivation activity of Notch. J. Biol. Chem. 27841963-41969. [DOI] [PubMed] [Google Scholar]
- 47.Yoo, A. S., C. Bais, and I. Greenwald. 2004. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303663-666. [DOI] [PubMed] [Google Scholar]
- 48.Zippo, A., A. De Robertis, R. Serafini, and S. Oliviero. 2007. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for Myc-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9932-944. [DOI] [PubMed] [Google Scholar]