Abstract
Mdm2, a regulator of the tumor suppressor p53, is frequently overexpressed in human malignancies. Mdm2 also has unresolved, p53-independent functions that contribute to tumorigenesis. Here, we show that increased Mdm2 expression induced chromosome/chromatid breaks and delayed DNA double-strand break repair in cells lacking p53 but not in cells with a mutant form of Nbs1, a component of the Mre11/Rad50/Nbs1 DNA repair complex. A 31-amino-acid region of Mdm2 was necessary for binding to Nbs1. Mutation of conserved amino acids in the Nbs1 binding domain of Mdm2 inhibited Mdm2-Nbs1 association and prevented Mdm2 from delaying phosphorylation of H2AX and ATM-S/TQ sites, repair of DNA breaks, and resolution of DNA damage foci. Similarly, the mutation of eight amino acids in the Mdm2 binding domain of Nbs1 inhibited Mdm2-Nbs1 interaction and blocked the ability of Mdm2 to delay DNA break repair. Both Nbs1 and ATM, but not the ubiquitin ligase activity of Mdm2, were necessary to inhibit DNA break repair. Only Mdm2 with an intact Nbs1 binding domain was able to increase the frequency of chromosome/chromatid breaks and the transformation efficiency of cells lacking p53. Therefore, the interaction of Mdm2 with Nbs1 inhibited DNA break repair, leading to chromosome instability and subsequent transformation that was independent of p53.
Mdm2 is considered an oncogene, as its overexpression has been demonstrated to be transforming (16). The ability of Mdm2 to transform cells was linked to its regulation of the tumor suppressor p53 (17). p53 is a target of the E3 ubiquitin ligase activity of Mdm2, resulting in proteosomal degradation of p53 (21, 24). Mdm2 also suppresses p53 transcriptional activity and shuttles p53 out of the nucleus (35, 41). Mdm2, in turn, is regulated by p14/p19ARF, which binds to Mdm2 and inhibits the ability of Mdm2 to control p53 (52). Overexpression of Mdm2 is frequently observed in human and murine malignancies (15, 34, 39). In fact, Mdm2 amplification occurs in 10% of all human cancers and approximately 20% of soft tissue sarcomas and osteosarcomas (34), suggesting that maintaining Mdm2 within normal levels is important for controlling cancer development and/or progression. Indeed, studies have shown that altering Mdm2 levels changes the balance of the p53 pathway, thus influencing tumorigenesis (2, 4, 33, 50).
While p53 regulation is the best-characterized function of Mdm2, evidence also supports a role for p53-independent functions of Mdm2, which also appear to influence tumorigenesis (19). Mdm2 overexpression due to amplification or other mechanisms has been detected in patients with a variety of human cancers that also harbor mutant p53 or lack p53 (12, 51). Soft tissue sarcoma and bladder cancer patients with tumors having both mutant p53 and elevated Mdm2 levels had a worse prognosis than patients with tumors with either abnormality alone (12, 26). Mouse studies also support a p53-independent role for Mdm2 in tumorigenesis. For example, a third of the lymphomas arising in Eμ-myc transgenic mice that have mutated p53 or lack p53 also overexpress Mdm2 (2, 15), suggesting that besides inhibiting p53, the tumor may additionally benefit from elevated Mdm2 levels. p53-null mice heterozygous for Mdm2 or overexpressing Mdm2 from a transgene have an altered tumor spectrum compared to that of mice lacking p53 alone (22, 32). Furthermore, mammary-specific-Mdm2-transgenic mice exhibit increased ploidy, a marker of genome instability, in mammary epithelial cells regardless of p53 expression (27). Finally, Mdm2 splice variants lacking the p53 binding domain identified in tumors promote transformation in vitro despite their lack of p53 association (42). Combined, the data support the hypothesis that Mdm2 has p53-independent oncogenic functions, and elucidation of these functions will explain how Mdm2 contributes to transformation.
Recently, we discovered the interaction between Mdm2 and Nbs1, a member of the Mre11/Rad50/Nbs1 complex that functions in DNA double-strand break repair and other functions that are necessary to maintain chromosomal stability (1, 54). Nbs1 is thought to localize the Mre11/Rad50/Nbs1 complex to DNA damaged sites, while Rad50 tethers the DNA ends together and Mre11 processes the DNA breaks with exonuclease and endonuclease activity (11, 13, 37). Following DNA damage, Nbs1 participates in activating ATM, a kinase responsible for signaling DNA damage (25, 47). The importance of the Mre11/Rad50/Nbs1 complex and ATM in the DNA damage pathway is apparent in humans with Nijmegen breakage syndrome, ataxia-telangiectasia (AT)-like disease, and AT, which are caused by mutations in Nbs1, Mre11, and ATM, respectively (46). Cells from Nijmegen breakage syndrome, AT-like disease, and AT patients exhibit genome instability due to increased sensitivity to DNA-damaging agents and reduced ability to repair DNA breaks, which leads to an increased frequency of malignancies (46). Our recent discovery that Mdm2 binds to Nbs1 suggested that Mdm2 may impact the DNA repair response through its association with Nbs1 (1). Here, we show that Mdm2-Nbs1 interaction inhibited the early response to DNA damage, which delayed DNA double-strand break repair, leading to loss of genome stability and transformation that was independent of p53. These data link Mdm2 oncogenic function to a p53-independent, Nbs1-dependent DNA repair pathway.
MATERIALS AND METHODS
Cell culture.
293T and NIH 3T3 cells were cultured as described by the American Type Culture Collection (Manassas, VA). p53−/−, ARF−/−, and wild-type murine embryo fibroblasts (MEFs) were isolated as previously described (55). ATM−/− ARF−/− MEFs were provided by Michael Kastan (St. Jude Children's Research Hospital, Memphis, TN) and simian virus 40 (SV40)-transformed wild-type and Nbs1ΔB/ΔB MEFs were provided by John H. Petrini (Memorial Sloan Kettering Institute, New York, NY). All MEFs were cultured as described previously (55).
Vector construction and retroviral infection.
A murine Mdm2 mutant consisting of amino acids (aa) 1 to 228 (1-228) was generated by PCR and Mdm2 mutant 231-489 by restriction enzyme digest. Both were cloned into the pJ3H vector to generate N-terminal hemagglutinin (HA) protein tags, and then subcloned into pcDNA3. Human Nbs1 mutants 179-542, 396-542, 179-395, and 269-474 were generated by restriction enzyme digest of wild-type Nbs1, and Nbs1 mutants 269-512 and 513-754 were generated by PCR. Nbs1 mutants were FLAG tagged by being cloned into the pCMV Tag vectors (Stratagene). Mdm2 and Nbs1 point mutants were generated by site-directed mutagenesis. All Mdm2 deletion and point mutants and wild-type and Nbs1 with eight point mutations were subcloned into murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-green fluorescent protein (GFP) retroviral vector (from Robert Hawley). The MSCV-IRES-GFP retroviral vector encoding wild-type Mdm2 and the 198-400 Mdm2 mutant were gifts from Martine Roussel (St. Jude Children's Research Hospital). Wild-type Mdm2 was also subcloned into an MSCV-IRES-yellow fluorescent protein (YFP) retroviral vector. Retroviruses were produced and used to infect MEFs as previously reported (55). Infection was confirmed by flow-cytometric analysis of GFP and/or YFP.
Transient transfection, immunoprecipitation, and Western blotting.
HA-tagged Mdm2 constructs or FLAG-tagged Nbs1 constructs were transfected into 293T cells and harvested 36 h later. ARF- and ubiquitin-encoding vectors from Martine Roussel (St. Jude Children's Research Hospital) and Dirk Bohmann (University of Rochester), respectively, were cotransfected with HA-tagged Mdm2 constructs (see Fig. 2E and F, respectively). MEFs infected with retroviruses (see above), were subjected to 5 Gy of gamma irradiation (137Cs source) and harvested at intervals. Cells were lysed and protein was measured as previously described (1). For phosphorylated H2AX (γ-H2AX) detection, cells were sonicated in 50 mM Tris, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 5 mM sodium pyrophosphate, 10 mM sodium 2-glycerolphosphate, 1 mM phenylmethylsulfonyl fluoride, 0.4 units/ml aprotinin, 1 mM sodium fluoride, and 0.1 mM sodium vanadate. Equal amounts of whole-cell protein lysates were immunoprecipitated with antibodies specific to HA (HA probe F-7; Santa Cruz Biotechnology) and Mdm2 (SMP14; Santa Cruz Biotechnology). Following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to nitrocellulose (Protran; Schleicher & Schuell) or polyvinylidene difluoride (Bio-Rad), proteins from whole-cell lysates and immunoprecipitations were subjected to Western blot analyses with antibodies specific for HA (F-7 from Santa Cruz Biotechnology or HA1.1 from Covance), FLAG (M2; Sigma), Mdm2 (SMP14 from Santa Cruz Biotechnology or 2A10 from Calbiochem), Nbs1 (BD Transduction Laboratories), p53 (DO-1; Santa Cruz Biotechnology), ARF (ab80; Abcam), Mre11 and Rad50 (both from Novus), γ-H2AX (JBW 301; Upstate), and α-tubulin and β-actin (both from Sigma). Immunocomplexes were detected as previously reported (1).
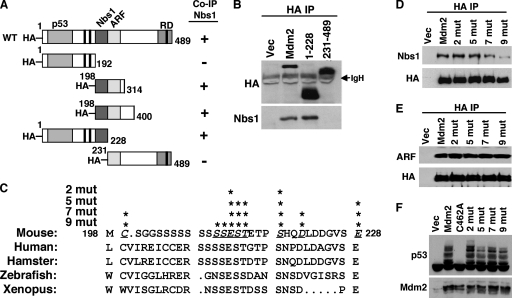
FIG. 2.
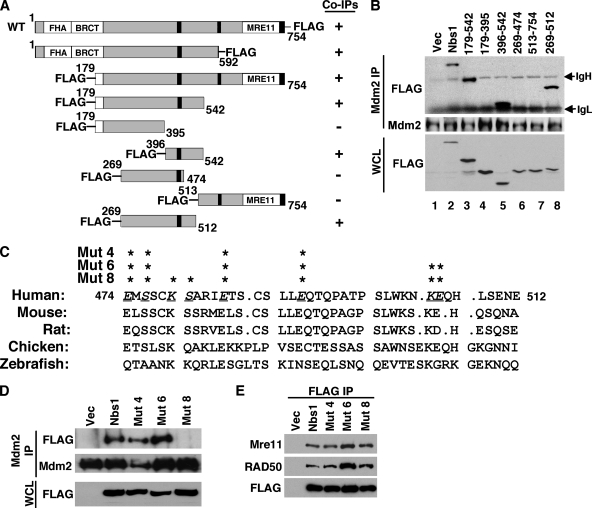
Localization of the Nbs1 binding domain in Mdm2. (A) Schematic diagrams of HA-tagged wild-type (WT) Mdm2 and deletion mutants (amino acids indicated). Boxed areas represent domains that bind p53, Nbs1, and ARF or represent the E3 ubiquitin ligase ring domain (RD). The black boxes represent the locations of the nuclear localization signal, nuclear export signal, and nucleolar localization signal from the N terminus to the C terminus, respectively. Mdm2 mutants that did (+) or did not (−) coimmunoprecipitate (Co-IP) Nbs1 are indicated. (B) Whole-cell lysates from 293T cells expressing empty vector (Vec) or vectors encoding HA-tagged wild-type Mdm2 (Mdm2) or the indicated Mdm2 deletion mutants were immunoprecipitated with anti-HA (HA IP) and Western blotted with antibodies specific for HA and Nbs1. The location of immunoglobulin heavy chain (IgH) is indicated. (C) Alignment of the amino acids of the Nbs1 binding region of Mdm2 from the organisms indicated. Underlined, italic letters and asterisks indicate amino acid positions where alanine substitutions were made. Xenopus indicates Xenopus laevis. (D to F) 293T cells were transiently transfected with empty vector, a vector encoding HA-tagged wild-type Mdm2 (Mdm2) or the indicated Mdm2 point mutants (having two [2 mut], five [5 mut], seven [7 mut], or nine [9 mut] point mutations) and cotransfected with a vector encoding p19ARF (E) or ubiquitin (F). (D and E) Whole-cell lysates were immunoprecipitated with anti-HA (HA IP) and Western blotted with antibodies specific for proteins indicated to the left of each panel. (F) Whole-cell lysates were Western blotted for p53 and Mdm2.
Metaphase analysis.
MEFs and NIH 3T3 cells were infected with empty MSCV-IRES-GFP retrovirus vector or retrovirus encoding wild-type Mdm2 or Mdm2 with nine point mutations in its Nbs1 binding domain. Thirty-six hours after infection, a portion of cells were analyzed by flow cytometry for GFP expression and a portion of cells were treated with colcemid for 4 h, harvested, prepared for blind testing, processed, and analyzed as previously described (49).
Comet assays.
MEFs were infected with retroviruses (see above), exposed to 5 Gy of gamma irradiation (137Cs source), and harvested at specific intervals. Blind neutral comet assays were performed to detect DNA double-strand breaks as directed by the manufacturer's protocol (Trevigen) and as previously described (1). Tail moment (tail length multiplied by percent DNA in tail) is a measurement of DNA breaks (36) and was calculated by CometScore software (TriTek Corp.).
Immunofluorescence.
MEFs infected with retroviruses (see above) were gamma irradiated (5 Gy for γ-H2AX and p-ATM or 12 Gy for Nbs1), and at specific intervals, fixed in 4% paraformaldehyde-4% sucrose for γ-H2AX or 1:1 methanol-acetone for Nbs1 and phosphorylated serine 1981-ATM. Following incubation with antibodies against γ-H2AX (JBW 301; Upstate), Nbs1 (catalog no. 100-143; Novus), or p-S1981-ATM (Rockland), bound antibodies were detected with anti-mouse or anti-rabbit Alexa 594 (Molecular Probes). For γ-H2AX and p-ATM, foci were quantified for at least 50 cells per sample per interval per experiment by using ImageTool software (University of Texas Health Science Center) from photographs taken by fluorescence microscopy. For Nbs1 foci, cells with more than three foci were considered focus positive, and at least 50 cells per sample per interval per experiment were quantified by visual scoring of photographs from fluorescence microscopy. Experiments were performed a minimum of three times.
Radioresistant-DNA-synthesis assay.
Nbs1+/+ or Nbs1ΔB/ΔB MEFs (SV40 transformed) infected with bicistronic retroviruses (MSCV-IRES-GFP or MSCV-Mdm2-IRES-GFP vector) were incubated with 10 nCi/ml of [methyl-14C]thymidine (Amersham Biosciences) for 24 h. MEFs were exposed to 0, 10, or 20 Gy of gamma irradiation (137Cs source) 24 h after the [methyl-14C]thymidine-containing medium was removed. Thirty minutes postirradiation, MEFs were incubated for 15 min with 2 μCi/ml [methyl-3H]thymidine (Amersham Biosciences). MEFs were harvested, washed, fixed in 70% methanol, and transferred to Whatman paper by vacuum filtration. 3H and 14C were measured by liquid scintillation, and the ratio of 3H to 14C in each sample was calculated.
G2/M checkpoint assay.
Nbs1+/+ or Nbs1ΔB/ΔB MEFs (SV40 transformed) infected with bicistronic retroviruses (MSCV-IRES-GFP or MSCV-Mdm2-IRES-GFP) were exposed to 0, 1, 5, or 10 Gy of gamma irradiation (137Cs source). One hour after irradiation, MEFs were harvested and fixed in 70% ethanol. MEFs were permeabilized with 0.25% Triton X-100-phosphate-buffered saline and then blocked in 1% bovine serum albumin-phosphate-buffered saline. Cells were incubated with an antibody specific for phosphorylated serine 10 of histone H3 (catalog no. 06-570, 0.75 μg/100 μl; Upstate Biotechnology) and then with an anti-rabbit Alexa 633 secondary antibody (A21070; Molecular Probes). Following RNase A (Sigma) treatment, DNA was stained with propidium iodide (catalog no. 105458; Sigma). Fluorescence was measured by flow cytometry (FACSCalibur), and data were evaluated with CellQuest software (BD Immunocytometry Systems).
Transformation assays.
p53−/− MEFs (6 × 104) infected with the retroviruses indicated above were resuspended in 0.6% agarose-containing media, and six identical samples were plated into six-well dishes coated with 0.8% agarose. Following approximately two weeks in culture, colonies were counted.
RESULTS
Mdm2 overexpression leads to genomic instability independent of p53.
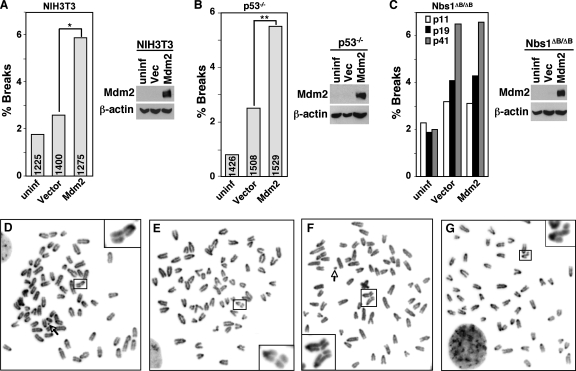
Genome instability manifests itself a number of different ways, including polyploidy and chromosomal breaks (38). Since malfunctions in DNA repair lead to genome instability, we characterized the effects of Mdm2 overexpression on genomic stability. We first analyzed chromosomes in metaphase spreads from NIH 3T3 cells, which do not express ARF but do express wild-type p53, following infection with a bicistronic retrovirus encoding GFP and Mdm2 or a retrovirus vector control. Over 3,000 individual metaphase cells from four independent experiments with GFP+ NIH 3T3 cells were evaluated. Since integration of retroviruses induces DNA breaks (43), NIH 3T3 cells with retroviral infection alone showed a small increase in the number of breaks detected (Fig. 1; Table 1). NIH 3T3 cells overexpressing Mdm2 had twice the frequency of chromosome and chromatide breaks compared to the cells infected with the vector control (P = 0.00004; Fisher's exact test) (Fig. 1A and D). To determine whether the effects of Mdm2 overexpression on the genome were attributable to increased p53 inhibition by Mdm2, we performed similar experiments with p53−/− MEFs. Over 4,000 individual metaphase cells from six independent experiments with GFP+ p53−/− MEFs were analyzed. Notably, Mdm2 overexpression in p53−/− MEFs led to a significant increase in the numbers of chromosome and chromatid breaks compared to the numbers of breaks in control infected p53−/− MEFs (P = 0.00006; Fisher's exact test) (Fig. 1B, E, and F; Table 1). Similar results were observed when wild-type Mdm2 or a ubiquitin ligase-dead mutant of Mdm2 was overexpressed in wild-type MEFs (data not shown). For all samples, chromosome breaks were more frequent than chromatid breaks, and Mdm2 overexpression increased both types of breaks. Therefore, Mdm2 overexpression promoted genomic instability independent of both ARF and p53.
FIG. 1.
Mdm2 overexpression induces chromosome/chromatid breaks. NIH 3T3 cells (A), p53−/− MEFs (B), or Nbs1ΔB/ΔB MEFs (C) were left uninfected (uninf) or infected with an empty bicistronic GFP-encoding retrovirus (vector [Vec]) or a retrovirus encoding GFP and wild-type Mdm2 (Mdm2). Metaphases were examined for chromosome or chromatid breaks (see Table 1 for more details). In panels A and B, the total numbers of metaphases evaluated are indicated in each bar. Significant differences by Fisher's exact test are indicated (*, P = 0.00004; **, P = 0.00006). (C) At least 300 metaphase cells were evaluated for each bar, and the passage numbers of the MEFs are indicated. Representative Western blots for Mdm2 and β-actin are shown. (D to G) Photographs of representative examples of metaphase NIH 3T3 cells (D), p53−/− MEFs (E and F), and Nbs1ΔB/ΔB MEFs (G) overexpressing Mdm2. Chromatid breaks are shown in panels D, F, and G. A chromosome break is shown in panel E. Magnified views of the breaks are displayed in boxes at the corners. Nine normal chromosomes were cropped out of panel G. Arrows point to a detached centromere (D) and a chromosome fragment (F).
TABLE 1.
Mdm2 overexpression increased DNA breaks independent of ARF and p53
| Cell type and experiment no. | No. of breaks/total no. of metaphases analyzeda
|
||
|---|---|---|---|
| Uninfected | Vectorb | Mdm2c | |
| NIH 3T3 | |||
| 1 | 6/346 | 13/348 | 27/327 |
| 2 | 4/233 | 5/344 | 9/268 |
| 3 | 6/299 | 8/350 | 14/311 |
| 4 | 6/347 | 10/358 | 25/369 |
| Totald | 22/1,225 (1.8) | 36/1,400 (2.6) | 75/1,275 (5.9) |
| p53−/− MEFs | |||
| 1 | ND | 3/134 | 13/164 |
| 2 | 0/105 | 4/144 | 7/98 |
| 3 | 2/386 | 4/256 | 10/313 |
| 4 | 5/297 | 15/332 | 27/320 |
| 5 | 2/317 | 8/313 | 10/336 |
| 6 | 2/321 | 4/329 | 17/298 |
| Totale | 11/1,426 (0.8) | 38/1,508 (2.5) | 84/1,529 (5.5) |
The percentages of metaphase cells with breaks are in parentheses. ND, not determined.
Infected with MSCV-IRES-GFP vector.
Infected with MSCV-Mdm2-IRES-GFP vector.
P = 0.00004 (vector in comparison with Mdm2).
P = 0.00006 (vector in comparison with Mdm2).
We previously reported that Mdm2 bound to the DNA repair protein Nbs1 (1). To determine whether Nbs1 was required for the observed Mdm2 effects on the genome, we overexpressed Mdm2 in MEFs that contained a mutant form of Nbs1. MEFs from Nbs1ΔB/ΔB mice express a truncated form of Nbs1 and are defective in DNA repair (53) (see Fig. 6B). Overexpression of Mdm2 in the Nbs1ΔB/ΔB MEFs did not result in an increased number of DNA breaks (Fig. 1C). Similar percentages of cells among control-vector-infected and Mdm2-encoding-retrovirus-infected Nbs1ΔB/ΔB MEFs harbored chromosome and chromatid breaks (Fig. 1G). Since DNA instability increases with increasing passage numbers, we evaluated the impact of Mdm2 overexpression on different passages of Nbs1ΔB/ΔB MEFs. As the numbers of chromosome/chromatid breaks increased over time in the Nbs1ΔB/ΔB MEFs, there was no significant difference between the percentage of breaks in Mdm2-overexpressing cells and the percentage in control cells at any of the passage numbers (Fig. 1C). These results indicate that Nbs1 mediates the effects of Mdm2 on DNA stability.
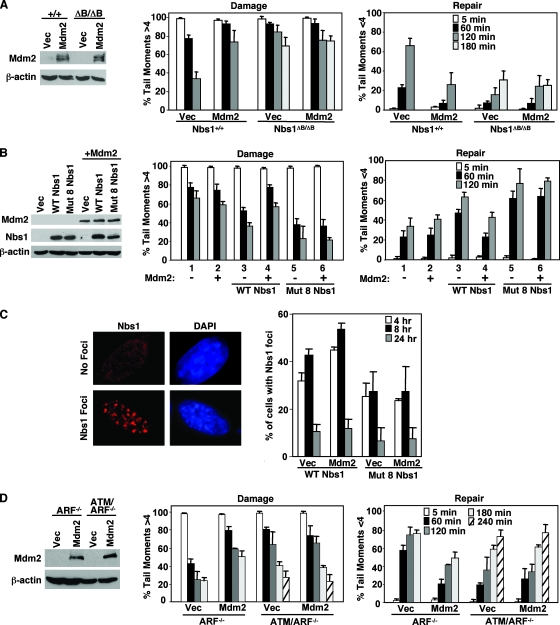
FIG. 6.
Wild-type Nbs1 and ATM are necessary for Mdm2-mediated inhibition of DNA break repair. (A) Wild-type (Nbs1+/+) and Nbs1ΔB/ΔB MEFs were infected with empty bicistronic GFP-encoding retrovirus (Vec) or bicistronic retrovirus encoding GFP and wild-type Mdm2 (Mdm2). Equal concentrations of whole-cell lysates were Western blotted for Mdm2 and β-actin. Comet assays were performed as described in the legend to Fig. 3. (B and C) Mdm2 was overexpressed (+) via a bicistronic YFP-encoding retrovirus in Nbs1ΔB/ΔB MEFs or Nbs1ΔB/ΔB MEFs reconstituted with wild-type (WT) Nbs1 or Nbs1 with eight point mutations (Mut 8) in the Mdm2 binding domain. (B) Equal concentrations of whole-cell lysates were Western blotted to confirm Nbs1 and Mdm2 expression. Comet assays were performed as described in the legend to Fig. 3. (C) At the indicated intervals following exposure to gamma irradiation (12 Gy), Nbs1 foci were detected by immunofluorescence and counted. An example of cells positive and negative for Nbs1 foci eight hours postirradiation is shown. Data are the means of at least three independent experiments, and error bars represent one standard deviation. The DNA stain DAPI (4′,6-diamidino-2-phenylindole) was used to visualize the nucleus. (D) ARF−/− or ATM/ARF−/− MEFs infected with empty bicistronic GFP-encoding retrovirus (Vec) or bicistronic retrovirus encoding GFP and wild-type Mdm2 (Mdm2) were Western blotted with antibodies specific for Mdm2 and β-actin. Comet assays were performed as described in the legend to Fig. 3. All bars show means of at least three independent experiments, and error bars are one standard deviation.
Thirty-one amino acids of Mdm2 are critical for Nbs1 binding.
Previously, we reported that a region of the central domain of Mdm2 (aa 198 to 314) was necessary for Mdm2 to associate with Nbs1 (1). To identify the Nbs1 binding domain in Mdm2, we generated HA-tagged Mdm2 deletion mutants containing aa 1 to 228 or 231 to 489, essentially splitting the central domain of Mdm2 into two pieces (Fig. 2A). Immunoprecipitation of wild-type Mdm2 transiently expressed in 293T cells showed that Nbs1 coimmunoprecipitated with Mdm2 (Fig. 2B). Although both the truncated forms of Mdm2, aa 1 to 228 and aa 231 to 489, were expressed at higher levels than wild-type Mdm2, only the aa 1 to 228 form of Mdm2 was able to coimmunoprecipitate Nbs1. The aa 1 to 228 form of Mdm2 appeared to coimmunoprecipitate an amount of Nbs1 similar to that which coimmunoprecipitated with wild-type Mdm2 (Fig. 2B). This result, in addition to our previous data, which are summarized in Fig. 1A (1), indicated the Nbs1 binding domain was located within the 31 aa from 198 to 228 of Mdm2.
Amino acid sequence alignment of Mdm2 proteins from multiple species revealed that the Nbs1 binding domain (aa 198 to 228) was highly conserved (Fig. 2C). To determine the critical amino acids for Mdm2-Nbs1 binding, conserved amino acids in the Nbs1 binding domain of Mdm2 were mutated to alanine by site-directed mutagenesis. The Mdm2 point mutants were immunoprecipitated from 293T cells, and Nbs1 association was evaluated by Western blot analysis. Mdm2 point mutants with two (aa 212 and 218) or five (aa 212 to 214, 218, and 228) alanine substitutions in the Nbs1 binding region did not appear to affect Mdm2-Nbs1 association (Fig. 2D). In contrast, Mdm2 point mutants with seven (aa 199, 212 to 214, 218, 221, and 228) or certainly nine (aa 199, 210 to 214, 218, 221, and 228) alanine substitutions showed a reduced amount of Nbs1 coimmunoprecipitating with Mdm2 (Fig. 2D). Since mutations could alter the overall structure of Mdm2, we evaluated whether point mutations in the Nbs1 binding domain of Mdm2 impacted association with known binding partners, such as ARF and p53. ARF binds to Mdm2 directly adjacent to the Nbs1 binding site (5) and therefore may be the most affected by mutations in this region of Mdm2. However, all of the Mdm2 point mutants coimmunoprecipitated amounts of ARF similar to the amounts that coimmunoprecipitated with wild-type Mdm2 (Fig. 2E). Moreover, each of the Mdm2 point mutants was able to effectively ubiquitinate p53 (Fig. 2F). Specifically, expression of wild-type Mdm2 or Mdm2 with point mutations in the Nbs1 binding domain showed a similar characteristic laddering pattern that is indicative of ubiquitination, whereas the Mdm2 E3 ubiquitin ligase mutant (with the C462A mutation) did not ubiquitinate p53 (Fig. 2F). Therefore, the point mutations in the Nbs1 binding domain that inhibited Mdm2-Nbs1 association did not disrupt the interaction of Mdm2 with ARF or the ability of Mdm2 to ubiquitinate p53.
The Nbs1 binding domain of Mdm2 is sufficient to inhibit DNA repair.
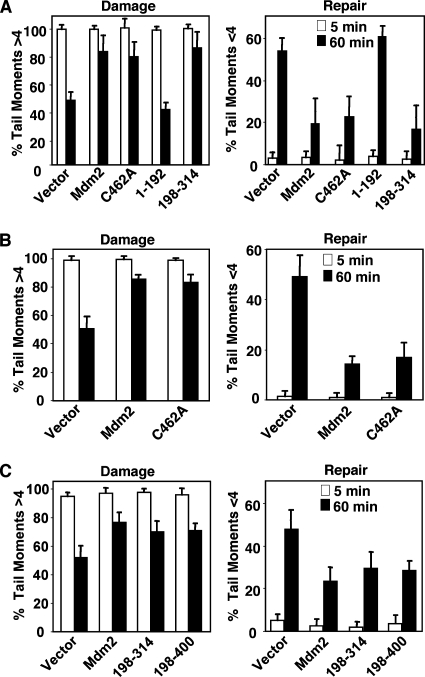
The Mre11/Rad50/Nbs1 complex is involved in the repair of DNA double-strand breaks (54). Initial data from neutral comet assays, which measure DNA double-strand breaks on a single-cell basis and reflect the amount of DNA damage present (36), indicated that overexpression of Mdm2 inhibited DNA double-strand break repair independent of its N or C terminus (1). To begin to determine whether the association of Mdm2 with Nbs1 was responsible for the inhibition of DNA break repair caused by Mdm2, we performed neutral comet assays with p53−/− MEFs. The percentage of broken DNA in each MEF was represented by the tail moment, which was measured by computer software. Tail moments of 0 to 4 corresponded to repaired or undamaged DNA, and tail moments of >4 indicated cells with DNA damage. p53−/− MEFs infected with empty bicistronic GFP-encoding retrovirus or retrovirus encoding GFP and wild-type Mdm2 or mutants (deletion or point) of Mdm2 were subjected to gamma irradiation, which causes DNA double-strand breaks. As expected, all p53−/− MEFs had similar amounts of DNA damage immediately following irradiation (Fig. 3A). However, 60 min following irradiation, p53−/− MEFs with elevated expression of wild-type Mdm2 had an increased percentage of cells with damaged DNA and a reduced percentage of cells with repaired DNA compared to the vector control. Cell cycle analysis showed that the increase in DNA breaks one hour postirradiation was not due to a change in the percentage of cells in S phase when Mdm2 was overexpressed, as there were similar numbers of cells in S phase when Mdm2 was overexpressed and when it was not (data not shown). Analysis of full-length Mdm2 containing a single point mutation in the E3 ubiquitin ligase domain (C462A), which inactivates the ubiquitin ligase function of Mdm2, also showed a delay in DNA break repair that was comparable to that observed for wild-type Mdm2 (Fig. 3A). This result indicated the ubiquitin ligase activity of Mdm2 was dispensable for Mdm2 to inhibit DNA repair. We also performed comet assays on early-passage wild-type MEFs to determine whether the presence of p53 would affect the ability of Mdm2 to inhibit DNA repair. Expression of wild-type Mdm2 or the C462A ubiquitin ligase-dead Mdm2 mutant in wild-type MEFs delayed DNA repair after gamma irradiation to a similar extent as that observed in p53−/− MEFs (Fig. 3B). Thus, Mdm2 inhibited DNA break repair irrespective of the presence of p53 or its ability to ubiquitinate p53. Moreover, expression of only the central region of Mdm2 (aa 198 to 314), containing the Nbs1 binding domain, was sufficient to delay DNA break repair in p53−/− MEFs, whereas a deletion mutant of Mdm2 containing the p53 binding domain (aa 1 to 192) was unable to suppress DNA repair (Fig. 3A). Since the central domain of Mdm2 is also able to bind to ARF and since ARF is expressed in p53-null MEFs, we evaluated whether the inhibition of DNA break repair from these mutants was independent of ARF. In ARF-null MEFs, both Mdm2 deletion mutants containing the central domain of Mdm2 (mutants consisting of aa 198 to 400 and aa 198 to 314) suppressed DNA double-strand break repair as effectively as wild-type Mdm2 following irradiation (Fig. 3C). Expression of Mdm2 deletion mutants that lacked the central domain of Mdm2 were unable to inhibit DNA repair in any of the MEFs analyzed (1; data not shown). Therefore, the Nbs1 binding region of Mdm2 was required and sufficient for Mdm2 to inhibit DNA break repair, and this was independent of p53, ARF, and the ubiquitin ligase function of Mdm2.
FIG. 3.
The Nbs1 binding domain of Mdm2 is required for inhibition of DNA break repair. p53−/− (A), wild-type (B), or ARF−/− (C) MEFs were infected with an empty bicistronic GFP-encoding retrovirus (vector) or a bicistronic retrovirus encoding GFP and wild-type Mdm2 (Mdm2), the indicated Mdm2 deletion mutants, or the ubiquitin ligase-dead Mdm2 (C462A). Following exposure to 5 Gy of gamma irradiation, neutral comet assays were performed on cells harvested at the indicated intervals. DNA double-strand breaks were quantified; tail moments greater than 4 represent damaged DNA, and tail moments equal to or less than 4 represent undamaged/repaired DNA. Data are the means of at least three separate experiments, with error bars representing one standard deviation.
Mdm2 inhibits DNA break repair through interaction with Nbs1.
The data described above suggested that the Mdm2-Nbs1 interaction may be responsible for the delay in DNA break repair. To test this, we performed neutral comet assays with p53−/− MEFs expressing Mdm2 mutants containing seven or nine point mutations in the Nbs1 binding domain, which have reduced Nbs1 association (Fig. 2D). Notably, 60 min after gamma irradiation, MEFs expressing Mdm2 with seven or nine point mutations had amounts of damaged and repaired DNA similar to those in MEFs infected with the retroviral vector control. In contrast, wild-type Mdm2 overexpressing p53−/− MEFs had a significantly greater percentage of cells with damaged DNA and significantly less repaired DNA than MEFs expressing Mdm2 with seven or nine point mutations in the Nbs1 binding domain or vector control (Fig. 4A).
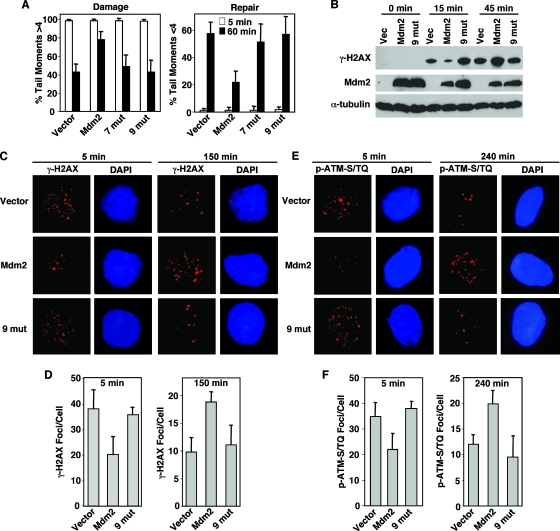
FIG. 4.
Mutations in the Nbs1 binding domain of Mdm2 abrogate its ability to inhibit DNA repair. (A to F) Following exposure to 5 Gy of gamma irradiation, p53−/− MEFs infected with empty bicistronic GFP-encoding retrovirus or bicistronic retrovirus encoding GFP and wild-type Mdm2 (Mdm2) or Mdm2 with seven (7 mut) or nine (9 mut) point mutations in the Nbs1 binding domain were harvested at the indicated intervals. (A) Comet assays, as described in the legend to Fig. 3, were performed. (B) Equal concentrations of whole-cell lysates were Western blotted with antibodies specific for the proteins indicated to the left of each panel. Phosphorylated H2AX (γ-H2AX) (C and D) and phosphorylated ATM-S/TQ sites (p-ATM-S/TQ) (E and F) were detected in fixed cells with antibodies specific for each and a fluorescently labeled secondary antibody. The DNA stain DAPI (4′,6-diamidino-2-phenylindole) was used to visualize the nucleus. Fluorescence was detected by microscopy, and the number of foci per cell was quantified on captured images. Photographs show representative cells. (D and F) Data are the means of three separate experiments, with error bars representing one standard deviation.
For a second independent measure of the effects of Mdm2 on DNA repair, we analyzed phosphorylation of histone H2AX. Histone H2AX is rapidly phosphorylated (becoming γ-H2AX) after DNA has been damaged at and near the damaged site, and as DNA is repaired, levels of γ-H2AX decrease (40). Therefore, levels of γ-H2AX serve as measures of the response to DNA damage and of DNA damage and repair (44). Immediately following gamma irradiation, Mdm2-overexpressing p53−/− MEFs had less γ-H2AX protein than vector control or Nbs1 binding domain mutant Mdm2-expressing MEFs (Fig. 4B). However, within one hour after irradiation, Mdm2-overexpressing p53−/− MEFs had more γ-H2AX protein compared to vector control or MEFs expressing Mdm2 with mutations in the Nbs1 binding domain, indicating that the Mdm2-overexpressing cells had more DNA damage remaining at this time (Fig. 4B). These results are consistent with the comet assay data and indicate that overexpression of wild-type Mdm2 led to a delay in the DNA damage response and DNA break repair following gamma irradiation.
H2AX phosphorylation is one of the earliest events in the DNA double-strand break repair response, and the kinase ATM is primarily responsible for this phosphorylation (6). ATM itself is activated by autophosphorylation of serine 1981 (1987 in mice) in response to DNA damage (3). Phosphorylated ATM localizes to sites of DNA damage, termed nuclear foci, presumably to phosphorylate H2AX and many other targets (3). The commercially available antibody against phosphophorylated serine 1987 of ATM recognizes phosphorylated ATM as well as multiple targets of ATM and ATM family members that are phosphorylated at S/TQ motifs following gamma irradiation (31). Just as with γ-H2AX foci, the number of phosphorylated ATM-S/TQ foci correlates with the amount of DNA damage present (45). Quantification of γ-H2AX foci and phospho-ATM-S/TQ foci immediately following gamma irradiation (5 min later) revealed that Mdm2-overexpressing p53−/− MEFs had significantly fewer of both foci than vector control or Nbs1 binding domain mutant Mdm2-expressing MEFs (Fig. 4C to F). Only a small portion of cells exhibited γ-H2AX foci in the unirradiated samples, and this was not dependent on Mdm2 overexpression (data not shown). By 150 min and as long as 240 min postirradiation, there were significantly greater numbers of γ-H2AX and phospho-ATM-S/TQ foci, respectively, in the Mdm2-overexpressing p53−/− MEFs than in the vector control or the MEFs expressing Mdm2 with nine point mutations in the Nbs1 binding domain (Fig. 4C to F). Since H2AX is phosphorylated during S phase (28), we also evaluated the cell cycle to determine whether this could account for the differences observed. There were similar numbers of cells in S phase for all samples 5 min to 24 h postirradiation (data not shown); therefore, the increase in γ-H2AX in cells that overexpressed wild-type Mdm2 was not due to an increase the percentage of cells in S phase. Combined, these data indicated that overexpression of Mdm2 induced an initial delay in the DNA damage response, resulting in prolonged DNA damage signaling and more DNA damage persisting over time, which were independent of p53. The data also show that loss of Nbs1 binding by Mdm2 abrogated the ability of Mdm2 to inhibit DNA repair, demonstrating that an intact Nbs1 binding domain in Mdm2 was critical for this Mdm2 function.
Eight amino acids of Nbs1 are necessary to mediate binding to Mdm2.
We previously reported that the N-terminal forkhead-associated (FHA) and BRCA1-associated C-terminal (BRCT) domains in the N terminus and the Mre11 binding domain in the C terminus of Nbs1 were dispensable for Mdm2 association (1). Therefore, to assess which region of Nbs1 mediates the interaction with Mdm2, we generated a series of FLAG-tagged Nbs1 deletion mutants (Fig. 5A). Not surprisingly, an Nbs1 mutant (aa 179 to 542) lacking both the N and C termini coimmunoprecipitated with Mdm2 (Fig. 5B, lane 3). Next, mutants that split this central domain of Nbs1 into two pieces were made. The two Nbs1 mutants were expressed at similar levels, but only the Nbs1 mutant consisting of aa 396 to 542, and not the mutant consisting of aa 179 to 395, coimmunoprecipitated with Mdm2 (Fig. 5B, lanes 4 and 5). Three additional Nbs1 deletion mutants were generated to further narrow the Mdm2 binding domain. aa 269 to 474 and 513 to 754 of Nbs1 did not associate with Mdm2 (Fig. 5B, lanes 6 and 7), whereas aa 269 to 512 of Nbs1 did coimmunoprecipitate with Mdm2 (lane 8). These results, together with our previous data (1), indicated that Mdm2 bound to Nbs1 within aa 474 to 512.
FIG. 5.
Localization of the Mdm2 binding domain in Nbs1. (A) Schematic diagrams of FLAG-tagged wild-type (WT) Nbs1 and Nbs1 deletion mutants (amino acids indicated). The FHA domain, the BRCT domain, and the Mre11 binding domain (Mre11) are labeled. The black boxes indicate nuclear localization signals. FLAG-Nbs1 mutants that did (+) or did not (−) coimmunoprecipitate with Mdm2 are indicated. (B) Whole-cell lysates (WCL) and Mdm2 immunoprecipitations (Mdm2 IP) from 293T cells expressing empty vector (Vec) or vectors encoding FLAG-tagged wild-type Nbs1 (Nbs1) or Nbs1 deletion mutants containing the indicated amino acids were Western blotted with antibodies specific for FLAG and Mdm2. The locations of immunoglobulin heavy (IgH) and light (IgL) chains are indicated. (C) Alignment of the Mdm2 binding regions of the Nbs1 proteins from the species indicated. Underlined, italic letters and asterisks indicate amino acid positions where alanine substitutions were made. (D and E) 293T cells with empty vector or vectors encoding FLAG-tagged wild-type Nbs1 (Nbs1) or Nbs1 with four (Mut 4), six (Mut 6), or eight (Mut 8) alanine substitutions in the Mdm2 binding domain. Whole-cell lysates (WCL), Mdm2 immunoprecipitations (Mdm2 IP), and FLAG immunoprecipitations (FLAG IP) were Western blotted with antibodies specified to the left of each panel.
Sequence alignment of the Mdm2 binding domain in Nbs1 proteins from different species revealed that this region was highly conserved in mammals (Fig. 5C). To determine the amino acids critical for Mdm2 association, alanines were substituted for four, six, or eight of the conserved amino acids in the Mdm2 binding domain of Nbs1 by site-directed mutagenesis. Nbs1 with four (aa 474, 476, 484, and 491) or six (aa 474, 476, 484, 491, 504, and 505) point mutations in the Mdm2 binding domain were able to associate with Mdm2 as well as wild-type Nbs1 (Fig. 5D). In contrast, Nbs1 with eight point mutations (aa 474, 476, 479, 480, 484, 491, 504, and 505) had severely reduced Mdm2 association (Fig. 5D), indicating that mutation of these amino acids compromised binding to Mdm2. Notably, all of the Nbs1 point mutants coimmunoprecipitated amounts of Mre11 and Rad50 similar to those that coimmunoprecipitated with wild-type Nbs1, demonstrating that mutation of these sites did not alter association with Mre11 (Fig. 5E). Moreover, mutations in the Mdm2 binding domain did not impact the DNA repair activity of Nbs1 (described below). Therefore, mutation of as few as eight aa in Nbs1 disrupted Mdm2-Nbs1 association.
Wild-type Nbs1 is necessary for the Mdm2-mediated delay in DNA repair.
To determine if Nbs1 mediated the Mdm2-induced delay in DNA break repair, we analyzed Nbs1ΔB/ΔB MEFs that contained a mutated form of Nbs1. Nbs1ΔB/ΔB MEFs are sensitive to gamma irradiation and show a slower kinetics of DNA repair (Fig. 6A) (53). Overexpression of Mdm2 in wild-type control MEFs (Nbs1+/+) inhibited DNA repair, as measured by comet assay. In contrast, Mdm2 overexpression did not further inhibit the already delayed repair response of Nbs1ΔB/ΔB MEFs. There were similar high percentages of cells with DNA breaks and low percentages of cells with repaired DNA 180 min postirradiation in Nbs1ΔB/ΔB MEFs overexpressing Mdm2 and vector control (Fig. 6A). Therefore, wild-type Nbs1 appears necessary for Mdm2 to inhibit DNA repair. To determine whether Mdm2 influences Nbs1-dependent intra-S and G2/M cell cycle checkpoints, we performed radioresistant-DNA-synthesis assays and evaluated phosphorylated histone H3 levels, respectively, following irradiation. We did not detect a significant difference for either checkpoint when Mdm2 was overexpressed (data not shown). These results indicate that while Mdm2 delayed DNA repair in wild-type Nbs1-expressing cells, Mdm2 does not appear to influence Nbs1 checkpoint functions, nor does Mdm2 further inhibit checkpoint functions in Nbs1 mutant cells.
To address whether the Nbs1-Mdm2 interaction was required for the Mdm2-mediated delay in DNA repair, we expressed wild-type Nbs1 or the mutant Nbs1 with eight point mutations in the Mdm2 binding domain in Nbs1ΔB/ΔB MEFs. Nbs1ΔB/ΔB MEFs expressing wild-type Nbs1 showed a more rapid decrease in broken DNA and an increase in repaired DNA following gamma irradiation, indicating that wild-type Nbs1 restored the kinetics of DNA repair in the Nbs1ΔB/ΔB MEFs (Fig. 6B, bars 1 versus bars 3). Expression of the Nbs1 mutant with eight point mutations in the Mdm2 binding domain, which has reduced association with Mdm2, also rescued DNA repair in Nbs1ΔB/ΔB MEFs (Fig. 6B, bars 1 versus bars 5). Interestingly, in comparison to MEFs expressing wild-type Nbs1, Nbs1ΔB/ΔB MEFs expressing Nbs1 with eight point mutations appeared to have an accelerated rate of repair with less DNA damage and an increased amount of repaired DNA 60 and 120 min after irradiation (Fig. 6B, bars 3 versus bars 5). Coexpression of Mdm2 in Nbs1ΔB/ΔB MEFs rescued with wild-type Nbs1 delayed the kinetics of repair (bars 3 versus bars 4), but Mdm2 overexpression did not delay repair in Nbs1ΔB/ΔB MEFs rescued with Nbs1 with eight point mutations (Fig. 6B, bars 5 versus bars 6). Therefore, an intact Mdm2 binding domain in Nbs1 was critical for Mdm2-mediated inhibition of DNA repair.
Previous studies have demonstrated that 8 h after DNA damage, a portion of cells contain prominent Mre11/Rad50/Nbs1 foci, and these foci are thought to be sites of remaining DNA damage (8, 30). Therefore, to further investigate the effect of Mdm2 on Nbs1, we examined formation of Nbs1 foci. Nbs1ΔB/ΔB MEFs rescued with wild-type Nbs1 or Nbs1 with eight point mutations that coexpressed either vector control or Mdm2 were exposed to gamma irradiation. Examples of cells positive and negative for Nbs1 foci 8 h after irradiation are shown in Fig. 6C. As early as four hours after irradiation, Mdm2-overexpressing Nbs1ΔB/ΔB MEFs expressing wild-type Nbs1 had a significantly greater percentage of cells with Nbs1 foci than those with the vector control (Fig. 6C). At 8 h following irradiation, the percentage of cells containing Nbs1 foci in the Mdm2-overexpressing Nbs1ΔB/ΔB MEFs reconstituted with wild-type Nbs1 remained elevated over vector control, suggesting that increased levels of Mdm2 inhibited resolution of Nbs1 foci. By 24 h, however, all MEFs had similar low percentages of cells positive for Nbs1 foci indicating that the majority of cells had repaired their DNA by this time. The percentages of cells positive for Nbs1 foci detected in Nbs1ΔB/ΔB MEFs expressing Nbs1 with eight point mutations were similar among Mdm2-overexpressing and vector control cells at all times analyzed (Fig. 6C); these data further support the requirement of Mdm2-Nbs1 association for the Mdm2-induced delay in DNA repair. Notably, there was a decreased percentage of cells containing Nbs1 foci in the Nbs1ΔB/ΔB MEFs expressing Nbs1 with eight point mutations compared to the percentage in the wild-type Nbs1-expressing Nbs1ΔB/ΔB MEFs at both four and eight hours postirradiation, suggesting that there were fewer cells containing unrepaired DNA in MEFs expressing Nbs1 with eight point mutations at these times (Fig. 6C). These data are consistent with the comet assay data showing that Nbs1ΔB/ΔB MEFs expressing Nbs1 with eight point mutations appeared to have an accelerated rate of DNA repair (Fig. 6B). Thus, Mdm2-Nbs1 interactions are required for the increase in the number of cells with DNA damage-induced Nbs1 foci and the delay in resolution of Nbs1 foci.
Since Nbs1 regulates and is regulated by ATM and since ATM signals DNA damage (25, 47, 54), we sought to establish whether the delay in DNA repair induced by Mdm2 required ATM. To do this, we evaluated MEFs lacking ATM to determine whether Mdm2 would inhibit DNA repair in the absence of ATM. Since the loss of ARF eliminates the rapid senescence of ATM-null MEFs and thereby allows expansion and retroviral infection, ATM/ARF double-null MEFs were evaluated. Mdm2 inhibited DNA repair following gamma irradiation in control ARF−/− MEFs; however, Mdm2 overexpression in ATM/ARF double-null MEFs did not further inhibit the delayed DNA repair inherent in cells lacking functional ATM (Fig. 6D). Therefore, ATM appears to be necessary for Mdm2 to delay DNA break repair.
Mdm2 contributes to genome instability and transformation independent of p53.
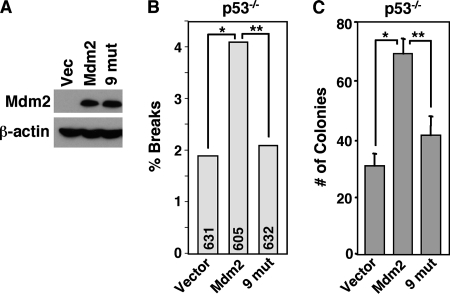
Faithful DNA repair is critical for maintaining genome integrity and preventing transformation (23). Since Mdm2 overexpression led to a delay in DNA break repair and an increase in chromosome and chromatid breaks independent of p53 and ARF but dependent on wild-type Nbs1, we hypothesized that genome instability induced by Mdm2 may be due to its interaction with Nbs1. We infected p53−/− MEFs with bicistronic retroviruses encoding GFP and wild-type Mdm2, Mdm2 with nine point mutations, or empty vector control (Fig. 7A). As previously demonstrated (Fig. 1B), wild-type Mdm2 overexpression in p53−/− MEFs significantly increased the number of chromosome and chromatid breaks over MEFs with retroviral vector control (Fig. 7B). In contrast, overexpression of Mdm2 with nine point mutations that has impaired ability to interact with Nbs1 (Fig. 2D) led to levels of chromosome and chromatid breaks comparable to those obtained from cells with the vector control (Fig. 7B), indicating that an intact Nbs1 binding domain in Mdm2 was critical for Mdm2-promoted genome instability. Therefore, we sought to determine the impact of Mdm2-induced genome instability on transformation. We evaluated the transformation potential of p53−/− MEFs overexpressing Mdm2 by soft agar assays. In repeated experiments, wild-type Mdm2-overexpressing p53−/− MEFs developed an increased number of colonies compared to vector control cells (Fig. 7C). In comparison to p53−/− MEFs overexpressing wild-type Mdm2, a significantly reduced number of colonies formed from MEFs that expressed the Nbs1 binding domain Mdm2 mutant, indicating that decreased association of Mdm2 with Nbs1 compromised its capacity to promote transformation. Since mutation of the Nbs1 binding domain does not completely abrogate association with Nbs1, it was not surprising that there were slightly more colonies that formed in the p53−/− MEFs expressing Mdm2 with nine point mutations than in the vector control (Fig. 7C). Combined, these results demonstrate that Mdm2 induces genomic instability and transformation that is independent of p53 and dependent on Mdm2-Nbs1 interactions.
FIG. 7.
Mdm2 promotes genome instability and transformation independent of p53. (A to C) p53−/− MEFs were infected with empty bicistronic GFP-encoding retrovirus (Vector) or bicistronic retrovirus encoding GFP and wild-type Mdm2 (Mdm2) or Mdm2 with nine point mutations in the Nbs1 binding domain (9 mut). (A) Equal concentrations of whole-cell lysates were Western blotted for Mdm2 and β-actin. (B) Metaphase cells were evaluated for chromosome and chromatid breaks. The total numbers of metaphases assessed per sample are indicated in each bar. Significant differences by Fisher's exact test are indicated (*, P = 0.030; **, P = 0.048). (C) Soft agar assays were performed with six identical samples of each, and colonies were counted from all six. Means of each are presented, and error bars represent one standard deviation. Data are representative of four independent experiments. Significant differences by the t test are indicated (*, P = 0.00000006; **, P = 0.00001).
DISCUSSION
More-subtle p53-independent functions of Mdm2 that contribute to tumorigenesis have remained elusive, overshadowed by the dominant role of this protein in regulating p53. Our recent discovery of an Mdm2-Nbs1 association (1) provided a new avenue of investigation that, in this study, has revealed a novel p53-independent oncogenic function of Mdm2. Although it is well established that Mdm2 functions to control p53 under conditions of normal cell growth and development (29), cells undergoing transformation acquire a collection of alterations that allow them to bypass the constraints that restrict normal cells (20). For example, there are selective pressures on tumor cells for unregulated proliferation and resistance to apoptosis, and consequently, genetic changes occur to allow tumors to grow and survive. Therefore, it would follow that elevated expression of a protein, such as Mdm2, that would increase the frequency of genetic alterations should be selected for during transformation. Thus, a function of Mdm2 that takes a backseat to p53 regulation under normal circumstances could be utilized by a cancer cell and cooperate with an inactivating mutation of p53. In support of this idea, Mdm2 overexpression, which is frequently selected for during tumorigenesis in mice and humans, has been detected in tumors that also harbor mutant p53 or lack p53 (12, 15, 26, 34, 39, 51). Patients with soft tissue sarcomas or bladder cancers that contained mutant p53 and also overexpressed Mdm2 had a worse prognosis than patients with only one of these defects (12, 26). Moreover, mice lacking p53 and overexpressing Mdm2 had an altered tumor spectrum and frequently developed two primary malignancies (22). Together, these and other observations suggested that there is an advantage for tumor cells to inactivate p53 and overexpress Mdm2, seemingly redundant genetic alterations, unless Mdm2 has functions other than regulating p53. Data shown here that Mdm2 overexpression inhibited Nbs1-mediated DNA double-strand break repair, resulting in increased chromosome/chromatid breaks and transformation of cells lacking p53, represent the first data demonstrating how increased levels of Mdm2 lead to transformation independent of p53. These results also provide an explanation for why tumor cells that have inactivated p53 would also overexpress Mdm2.
Detection and response to DNA damage occurs within minutes of the insult and involves many proteins (23). Phosphorylation of H2AX by the DNA damage sensor ATM, or possibly by ATM family members (ATR and DNA-dependent protein kinase), is one of the earliest events following DNA damage (6). It has been shown that H2AX phosphorylation is necessary for the retention of DNA repair factors at sites of DNA breaks (10). In Mdm2-overexpressing cells, the number of γ-H2AX foci was significantly below normal levels immediately following irradiation. We also observed that the amounts of activated (phosphorylated) ATM and other phosphorylated proteins (ATM/ATR targets) at sites of DNA breaks present minutes following DNA damage were also significantly reduced when Mdm2 was overexpressed. However, overexpression of a mutant Mdm2 that had reduced association with Nbs1 had no effect on rapid phosphorylation of ATM and ATM/ATR targets at foci and the appearance of γ-H2AX foci. These data strongly suggest that elevated levels of Mdm2 and Mdm2-Nbs1 interactions interfere with early events in the DNA damage response, events that are controlled by Nbs1. Nbs1 regulates ATM activation (25, 47). Inactivation of ATM or inhibition of ATM-Nbs1 association results in reduced numbers of γ-H2AX foci and delayed DNA repair (6, 7, 44). In Mdm2-overexpressing cells, we observed an increased percentage of cells with broken DNA, elevated numbers of γ-H2AX foci, and increased levels of phosphorylated ATM and ATM targets at foci hours after cells with normal levels of Mdm2 had repaired their DNA damage. Notably, inhibition of DNA repair by Mdm2 did not require the ubiquitin ligase activity of Mdm2 but only a small region of Mdm2 containing the Nbs1 binding domain. Moreover, we did not detect Mdm2-mediated ubiquitination of Nbs1, and Mdm2 expression did not appear to alter the stability of Nbs1 protein (unpublished data), suggesting that Mdm2 inhibited DNA repair through a mechanism separate from its ubiquitin ligase function. We also determined that Mdm2 did not appear to be impacting the cell cycle checkpoint functions of Nbs1; however, we cannot exclude the possibility that the crude assays used to measure S and G2/M checkpoints may not be sufficiently sensitive to detect small differences in these checkpoints. We did observe that cells expressing Nbs1 with mutations in its Mdm2 binding domain had accelerated rates of DNA repair and resolution of Nbs1 foci, indicating that an inability of Nbs1 to associate with Mdm2 facilitated DNA repair. Therefore, the data are consistent with the notion that Mdm2 inhibits Nbs1 through its interactions with Nbs1, leading to suppression of the early response to DNA damage. Consequently, Nbs1-mediated ATM activation and/or formation of DNA repair protein complexes at sites of DNA damage may be affected by Mdm2 overexpression, but other explanations are also possible. Although exactly how Mdm2 inhibits the early DNA damage response through Nbs1 is the focus of future work, a delay in the DNA damage response would lead to a delay in the repair of broken DNA, resulting in DNA breaks persisting. Unrepaired double-strand breaks progress to chromosomal breaks (18), as we observed. Interestingly, Mdm2 increased the number of both chromosome and chromatid breaks. Therefore, this impact on DNA repair by Mdm2 has negative consequences on the genome.
Preservation of genome integrity is critical to suppress tumorigenesis (23). Cells that have inactivated Nbs1, Mre11, Rad50, ATM, or other proteins involved in detecting and repairing DNA breaks have delayed DNA repair kinetics and higher rates of chromosomal aberrations and transformation (46). Humans with mutations in DNA repair/DNA damage signaling proteins, such as Nbs1 or ATM, have a significantly increased occurrence of malignancies (23, 46). Studies have shown that Mdm2 overexpression can promote certain forms of genome instability, such as aneuploidy and centrosome duplication (9, 50). It was assumed that these effects of Mdm2 on DNA stability were due to Mdm2 inhibiting p53, since inactivation of p53 also negatively impacts chromosomal stability (14). However, one report did show that polyploidy developed in breast epithelial cells that overexpressed Mdm2 regardless of p53 status (27). Here, we provide evidence that Mdm2 promotes chromosomal instability through inhibition of DNA double-strand break repair independent of p53, and instead dependent on Nbs1 and interactions with Nbs1. The delay in DNA break repair following overexpression of Mdm2, but not overexpression of mutant Mdm2 that had reduced association with Nbs1, led to an increased frequency of unrepaired chromosome and chromatid breaks. The consequence of increased genetic instability induced by Mdm2 was increased transformation efficiency of p53-null MEFs. These results should have important implications for cancer treatment, as Mdm2 antagonists, such as Nutlins, were developed to interfere with Mdm2-p53 interactions (48). However, given our data, altering Mdm2-Nbs1 association may increase the effectiveness of treatment of the many cancers that have elevated levels of Mdm2.
Acknowledgments
We thank John Petrini for Nbs1ΔB/ΔB MEFs, Michael Kastan for ATM/ARF-null MEFs, Martine Roussel for the wild-type Mdm2 construct, and Tanya Paull for the wild-type Nbs1 construct. We also thank Jane Kennedy, Jessica Odvody, and Brad Dworak for technical expertise.
This work was supported by NCI grants CA098139 and CA117935 (to C.M.E.) and the University of Nebraska Program of Excellence Graduate Assistantship (to A.B.). C.M.E. is a Leukemia & Lymphoma Society Scholar.
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Alt, J. R., A. Bouska, M. R. Fernandez, R. L. Cerny, H. Xiao, and C. M. Eischen. 2005. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J. Biol. Chem. 28018771-18781. [DOI] [PubMed] [Google Scholar]
- 2.Alt, J. R., T. C. Greiner, J. L. Cleveland, and C. M. Eischen. 2003. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 221442-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 4.Bond, G. L., W. Hu, E. E. Bond, H. Robins, S. G. Lutzker, N. C. Arva, J. Bargonetti, F. Bartel, H. Taubert, P. Wuerl, K. Onel, L. Yip, S. J. Hwang, L. C. Strong, G. Lozano, and A. J. Levine. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119591-602. [DOI] [PubMed] [Google Scholar]
- 5.Bothner, B., W. S. Lewis, E. L. DiGiammarino, J. D. Weber, S. J. Bothner, and R. W. Kriwacki. 2001. Defining the molecular basis of Arf and Hdm2 interactions. J. Mol. Biol. 314263-277. [DOI] [PubMed] [Google Scholar]
- 6.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 27642462-42467. [DOI] [PubMed] [Google Scholar]
- 7.Cariveau, M. J., X. Tang, X. L. Cui, and B. Xu. 2007. Characterization of an NBS1 C-terminal peptide that can inhibit ataxia telangiectasia mutated (ATM)-mediated DNA damage responses and enhance radiosensitivity. Mol. Pharmacol. 72320-326. [DOI] [PubMed] [Google Scholar]
- 8.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93477-486. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, P. E., M. Okuda, H. F. Horn, P. Biddinger, P. J. Stambrook, L. L. Gleich, Y. Q. Li, P. Tarapore, and K. Fukasawa. 1999. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene 181935-1944. [DOI] [PubMed] [Google Scholar]
- 10.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5675-679. [DOI] [PubMed] [Google Scholar]
- 11.Cerosaletti, K. M., and P. Concannon. 2003. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J. Biol. Chem. 27821944-21951. [DOI] [PubMed] [Google Scholar]
- 12.Cordon-Cardo, C., E. Latres, M. Drobnjak, M. R. Oliva, D. Pollack, J. M. Woodruff, V. Marechal, J. Chen, M. F. Brennan, and A. J. Levine. 1994. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 54794-799. [PubMed] [Google Scholar]
- 13.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 81129-1135. [DOI] [PubMed] [Google Scholar]
- 14.Donehower, L. A., L. A. Godley, C. M. Aldaz, R. Pyle, Y. P. Shi, D. Pinkel, J. Gray, A. Bradley, D. Medina, and H. E. Varmus. 1995. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 9882-895. [DOI] [PubMed] [Google Scholar]
- 15.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 132658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fakharzadeh, S. S., S. P. Trusko, and D. L. George. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 101565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlay, C. A. 1993. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol. Cell. Biol. 13301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franco, S., M. Gostissa, S. Zha, D. B. Lombard, M. M. Murphy, A. A. Zarrin, C. Yan, S. Tepsuporn, J. C. Morales, M. M. Adams, Z. Lou, C. H. Bassing, J. P. Manis, J. Chen, P. B. Carpenter, and F. W. Alt. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell 21201-214. [DOI] [PubMed] [Google Scholar]
- 19.Ganguli, G., and B. Wasylyk. 2003. p53-independent functions of MDM2. Mol. Cancer Res. 11027-1035. [PubMed] [Google Scholar]
- 20.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 10057-70. [DOI] [PubMed] [Google Scholar]
- 21.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 42025-27. [DOI] [PubMed] [Google Scholar]
- 22.Jones, S. N., A. R. Hancock, H. Vogel, L. A. Donehower, and A. Bradley. 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 9515608-15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27247-254. [DOI] [PubMed] [Google Scholar]
- 24.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387299-303. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. H., and T. T. Paull. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308551-554. [DOI] [PubMed] [Google Scholar]
- 26.Lu, M. L., F. Wikman, T. F. Orntoft, E. Charytonowicz, F. Rabbani, Z. Zhang, G. Dalbagni, K. S. Pohar, G. Yu, and C. Cordon-Cardo. 2002. Impact of alterations affecting the p53 pathway in bladder cancer on clinical outcome, assessed by conventional and array-based methods. Clin. Cancer Res. 8171-179. [PubMed] [Google Scholar]
- 27.Lundgren, K., R. Montes de Oca Luna, Y. B. McNeill, E. P. Emerick, B. Spencer, C. R. Barfield, G. Lozano, M. P. Rosenberg, and C. A. Finlay. 1997. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 11714-725. [DOI] [PubMed] [Google Scholar]
- 28.MacPhail, S. H., J. P. Banath, Y. Yu, E. Chu, and P. L. Olive. 2003. Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat. Res. 159759-767. [DOI] [PubMed] [Google Scholar]
- 29.Marine, J. C., S. Francoz, M. Maetens, G. Wahl, F. Toledo, and G. Lozano. 2006. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 13927-934. [DOI] [PubMed] [Google Scholar]
- 30.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 176087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuoka, S., B. A. Ballif, A. Smogorzewska, E. R. McDonald III, K. E. Hurov, J. Luo, C. E. Bakalarski, Z. Zhao, N. Solimini, Y. Lerenthal, Y. Shiloh, S. P. Gygi, and S. J. Elledge. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 3161160-1166. [DOI] [PubMed] [Google Scholar]
- 32.McDonnell, T. J., R. Montes de Oca Luna, S. Cho, L. L. Amelse, A. Chavez-Reyes, and G. Lozano. 1999. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J. Pathol. 188322-328. [DOI] [PubMed] [Google Scholar]
- 33.Mendrysa, S. M., K. A. O'Leary, M. K. McElwee, J. Michalowski, R. N. Eisenman, D. A. Powell, and M. E. Perry. 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2016-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Momand, J., D. Jung, S. Wilczynski, and J. Niland. 1998. The MDM2 gene amplification database. Nucleic Acids Res. 263453-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 691237-1245. [DOI] [PubMed] [Google Scholar]
- 36.Olive, P. L. 1999. DNA damage and repair in individual cells: applications of the comet assay in radiobiology. Int. J. Radiat. Biol. 75395-405. [DOI] [PubMed] [Google Scholar]
- 37.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 131276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pihan, G., and S. J. Doxsey. 2003. Mutations and aneuploidy: co-conspirators in cancer? Cancer Cell 489-94. [DOI] [PubMed] [Google Scholar]
- 39.Rayburn, E., R. Zhang, J. He, and H. Wang. 2005. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets 527-41. [DOI] [PubMed] [Google Scholar]
- 40.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigalas, I., A. H. Calvert, J. J. Anderson, D. E. Neal, and J. Lunec. 1996. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat. Med. 2912-917. [DOI] [PubMed] [Google Scholar]
- 43.Skalka, A. M., and R. A. Katz. 2005. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 12(Suppl. 1)971-978. [DOI] [PubMed] [Google Scholar]
- 44.Stucki, M., and S. P. Jackson. 2006. γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amsterdam) 5534-543. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, K., H. Okada, M. Yamauchi, Y. Oka, S. Kodama, and M. Watanabe. 2006. Qualitative and quantitative analysis of phosphorylated ATM foci induced by low-dose ionizing radiation. Radiat. Res. 165499-504. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 50949-78. [DOI] [PubMed] [Google Scholar]
- 47.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 225612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassilev, L. T. 2004. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle 3419-421. [PubMed] [Google Scholar]
- 49.Wang, P., T. C. Greiner, T. Lushnikova, and C. M. Eischen. 2006. Decreased Mdm2 expression inhibits tumor development induced by loss of ARF. Oncogene 253708-3718. [DOI] [PubMed] [Google Scholar]
- 50.Wang, P., T. Lushnikova, J. Odvody, T. C. Greiner, S. N. Jones, and C. M. Eischen. 2008. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 271590-1598. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, T., A. Ichikawa, H. Saito, and T. Hotta. 1996. Overexpression of the MDM2 oncogene in leukemia and lymphoma. Leuk. Lymphoma 21391-397. [DOI] [PubMed] [Google Scholar]
- 52.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 120-26. [DOI] [PubMed] [Google Scholar]
- 53.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12648-653. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., J. Zhou, and C. U. Lim. 2006. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 1645-54. [DOI] [PubMed] [Google Scholar]
- 55.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 122424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]