Abstract
PDZ-binding motifs are found in the C-terminal tails of numerous integral membrane proteins where they mediate specific protein-protein interactions by binding to PDZ-containing proteins. Conventional yeast two-hybrid screens have been used to probe protein-protein interactions of these soluble C termini. However, to date no in vivo technology has been available to study interactions between the full-length integral membrane proteins and their cognate PDZ-interacting partners. We previously developed a split-ubiquitin membrane yeast two-hybrid (MYTH) system to test interactions between such integral membrane proteins by using a transcriptional output based on cleavage of a transcription factor from the C terminus of membrane-inserted baits. Here we modified MYTH to permit detection of C-terminal PDZ domain interactions by redirecting the transcription factor moiety from the C to the N terminus of a given integral membrane protein thus liberating their native C termini. We successfully applied this “MYTH 2.0” system to five different mammalian full-length renal transporters and identified novel PDZ domain-containing partners of the phosphate (NaPi-IIa) and sulfate (NaS1) transporters that would have otherwise not been detectable. Furthermore this assay was applied to locate the PDZ-binding domain on the NaS1 protein. We showed that the PDZ-binding domain for PDZK1 on NaS1 is upstream of its C terminus, whereas the two interacting proteins, NHERF-1 and NHERF-2, bind at a location closer to the N terminus of NaS1. Moreover NHERF-1 and NHERF-2 increased functional sulfate uptake in Xenopus oocytes when co-expressed with NaS1. Finally we used MYTH 2.0 to demonstrate that the NaPi-IIa transporter homodimerizes via protein-protein interactions within the lipid bilayer. In summary, our study establishes the MYTH 2.0 system as a novel tool for interactive proteomics studies of membrane protein complexes.
The mammalian renal proximal tubule sustains reabsorption of fluid and solutes of the highest variety and magnitude. Vectorial transport in this epithelium is mediated by the polarized distribution of polytopic transport proteins. Strikingly many membrane proteins located at the apical but not at the basolateral side harbor a C-terminal PDZ-binding motif. The acronym PDZ denotes a conserved modular domain of 90 amino acids that recognizes an octapeptide PDZ-binding motif primarily residing at the C termini of target proteins (1–3). PDZ domains are classified into three groups according to their binding specificity. Class I modules represent PDZ proteins that bind the canonical consensus sequence X(S/T)XΦ where X is unspecified and Φ is a hydrophobic residue (4).
There is an ever growing membership of class I PDZ-binding membrane proteins in the apical brush border membrane of proximal tubular cells (5, 6). The Na+/H+ exchanger (NHE)1-3 (C-terminal STHM) contributes to trans- and paracellular absorption of Na+, Cl−, HCO3−, and water (7). The co-transporters NaPi-IIa (C-terminal ATRL) and NaS1 (C-terminal QTMP) support Na+-coupled uptake of Pi (8) and sulfate (9), respectively. The membrane-associated protein MAP17 (C-terminal STPM) of 17 kDa, which likely has multiple unknown functions other than transport, is classified into this group of apical class I PDZ-binding proteins as well (10, 11).
Conventional yeast two-hybrid (YTH) assays and classical biochemical techniques such as GST pulldowns identified proximal tubule brush border class I PDZ proteins as binding partners to the apical transmembrane proteins NHE-3, NaPi-IIa, and MAP17 (5). The Na+/H+ exchanger-regulating factor NHERF-1 and NHERF-2 were characterized as cofactors that confer cAMP-induced inhibition of NHE-3 (12, 13). Despite encompassing two PDZ modules in tandem (PDZ1 and PDZ2), both NHERF isoforms seem to preferentially utilize PDZ2 to interact with NHE-3 (14). Apart from the canonical C-terminal site, an internal PDZ-binding site in NHE-3 has been proposed (15). Via their PDZ1 domains, NHERF-1 and NHERF-2 can also interact with the C-terminal TRL motif of NaPi-IIa (16). Two other related PDZ proteins called PDZK1 and PDZK2 contain four PDZ modules of which the third PDZ domain (PDZ3) interacts with the NaPi-IIa C-terminal canonical site as the sole site of PDZ recognition (6, 16). In addition, PDZ1 and PDZ4 of PDZK1 but not NHERF-1 or NHERF-2 bind to MAP17 (6, 17, 18). Notably a putative connection between PDZ proteins and the sulfate transporter NaS1 has not been determined so far.
Studies performed in cell models have enhanced the understanding of the physiological role of these PDZ proteins. Unlike the strict microvillar localization of NHERF-1/2 and PDZK1/2 in proximal kidney cells (5), CAL was demonstrated to traffic between the Golgi and the plasma membrane (19, 20). In these cells, apical expression of NaPi-IIa partially depends on NHERF-1 (5), and cAMP-mediated regulation of NHE-3 requires NHERF isoforms (12, 13, 21). PDZK1 was proposed to constitute an apical scaffold for the targeting/retention of diverse renal ion transporters (6, 22). As the PDZK1-interacting protein MAP17 is most abundant at the apical membrane of opossum kidney cells, it is also plausible that MAP17 might operate as an apical adaptor of PDZK1 (17). CAL in turn was reported to facilitate the retrograde trafficking of the mature Cl− channel CFTR resulting in CFTR degradation (19).
Acquiring valuable information about interactions of integral membrane proteins has posed a major challenge because the hydrophobic nature of the membrane-spanning proteins prevents them from being used in the conventional YTH assay where both bait and prey need to be obligatory soluble polypeptides (23). To overcome this hurdle, we previously developed the conventional split-ubiquitin (Ub) membrane YTH (MYTH) technology based on a split-ubiquitin protein complementation readout (24–27). The small protein Ub, once conjugated to the C termini of its targets, destines proteins for proteasomal degradation in eukaryotic cells. The Ub moiety is then salvaged for recycling by Ub-specific proteases (UBPs) resident in the nucleus and cytosol of all eukaryotic cells (28). Split-Ub systems rely on the fact that severed C-terminal (Cub) and N-terminal (Nub) halves of Ub spontaneously reassemble to form a functional Ub molecule (29). The mutation of a single residue near the N terminus of Ub from isoleucine (NubI) to glycine (NubG) renders the two separated Cub and NubG halves incompetent for self-reconstitution. The only way for Cub to associate with NubG and to reconstruct the intact functional Ub is for them to be brought together by an auxiliary mechanism that can be provided by interaction of proteins that are additionally annexed to Cub and NubG (25, 30). The reconstituted Ub, but not its halves alone (Cub or NubG), are recognized by UBPs that cause the release of a transcription factor (TF) strategically placed C-terminal to Cub. The released TF enters the nucleus and activates the reporter cascade in a manner similar to conventional YTH. Therefore, the conventional MYTH system consists of a membrane-inserted bait whose C terminus is fused to a Cub-TF hybrid. The inbound interacting prey (either a membrane or cytosolic protein) is fused to NubG, which will furnish the other half of Ub to permit UBP-mediated release of TF.
In this study, we applied a modified MYTH system to the full-length proximal tubule membrane proteins NaPi-IIa, MAP17, NHE-1, NHE-3, and NaS1 as baits integrated into a plasma membrane environment. We designed a MYTH 2.0 system that redirects the TF and Cub moieties from the C to the N terminus of a transmembrane bait protein (see Fig. 1A) thereby exposing the free C termini of the above polytopic membrane proteins to permit interactions with PDZK1, NHERF-1, NHERF-2, and CAL (see Fig. 2A). Using this approach, we made four important observations. First, we validated the MYTH 2.0 system as a tool to investigate interactions between five distinct mammalian full-length integral membrane proteins and their PDZ domain-containing partners. Second, we discovered novel interacting proteins of the phosphate (NaPi-IIa) and sulfate (NaS1) renal transporters using this MYTH 2.0 approach. Third, we showed that NHERF-1 and NHERF-2, the two novel NaS1 interactors found in this study, lead to increased functional sulfate uptake in Xenopus oocytes when co-injected with NaS1. Fourth, we demonstrated homo-oligomerization of the full-length NaPi-IIa protein within the bilayer by interactions of its transmembrane domains. In summary, our collective results demonstrate that the MYTH 2.0 system provides a useful tool to promote further analyses of integral membrane proteins from any model organism.
Fig. 1.
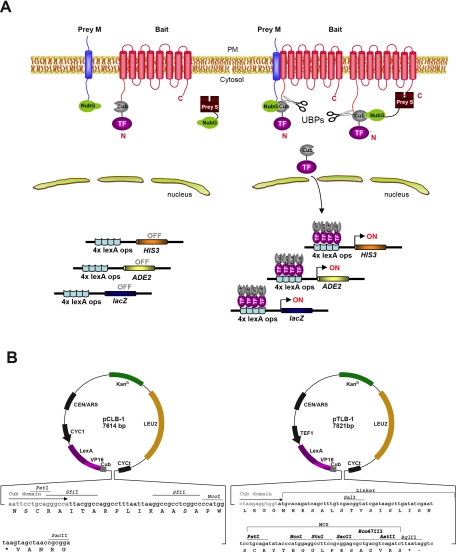
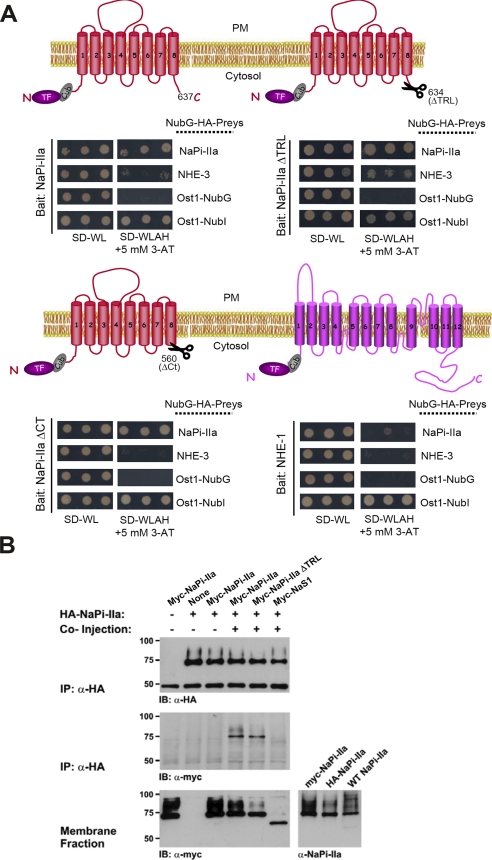
Principle of the MYTH 2.0 system. A, schema of the MYTH 2.0 basic principle to monitor protein-protein interactions. The MYTH 2.0 format differs from its previously described counterpart in that the reporters are placed N-terminal instead of C-terminal to the bait. The bait needs to be firmly attached to the membrane, whereas membrane anchorage of the prey is not mandatory. For a transmembrane prey protein, a default orientation of its termini is not pivotal provided that NubG resides within the cytosol. Upon association of TF-Cub-Bait with NubG-Prey, reconstitution of Ub from C- and N-terminal halves (i.e. Cub and NubG) leads to UBP-dependent proteolytic liberation (open scissors) of the transactivator TF fused to Cub and consequent activation of the reporter genes. TF is a chimera of the LexA DNA-binding domain with the VP16 activation domain. The TF triggers the induction of exogenous HIS3 and lacZ reporter genes that allow for the colorimetric detection of β-galactosidase (lacZ) and prototrophic growth without histidine (HIS3). C, C terminus; N, N terminus; PM, plasma membrane; Prey S, soluble prey; Prey M, membrane-associated prey. B, novel bait vectors (pCLB-1 and pTLB-1) for the MYTH 2.0 system. Both vectors are LEU2-based low copy number (CEN/ARS) vectors bearing either a weak yeast CYC1 (pCLB-1) or a strong TEF1 (pTLB-1) promoter. A TF (LexA-VP16) is followed by the Cub domain and the MCS. The foreign cDNA sequence encoding a transmembrane bait protein of interest is introduced into the MCS in-frame to the TF-Cub portion. Also shown in the MCS are unique PstI, Nco1, Stu1, SacII, Nco47III, AstII, and BglII restriction sites. *, stop codons.
Fig. 2.
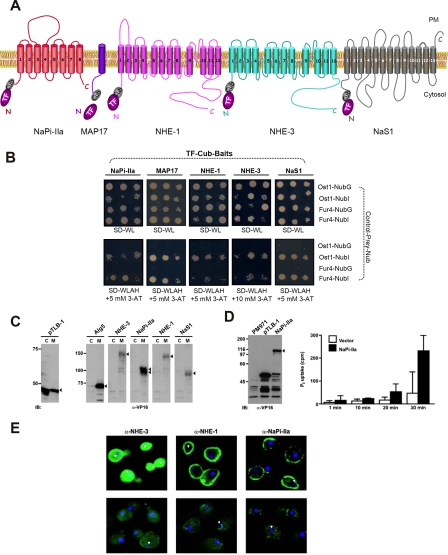
Characterization of various mammalian TF-Cub-Baits in yeast. A, membrane topology of TF-Cub-Baits (NaPi-IIa, MAP17, NHE-1, NHE-3, and NaS1) with N-terminally tagged TF-Cub fusion. B, proper insertion of TF-Cub-Baits into the membrane as exemplified by genetic NubG/NubI experiments. Yeast cells expressing TF-Cub-Baits of NaPi-IIa, MAP17, NHE-1, NHE-3 (pTLB-1 context), and NaS1 (pCLB-1 context) were transformed with control constructs of either yeast Ost1p (ER) or Fur4p (plasma membrane) fused to NubG and NubI. Yeast growth was challenged on minimal SD medium depleted of Trp and Leu (SD−WL) or Trp, Leu, Ade, and His (SD−WLAH) by spotting three independent transformants on the different media. A lack of growth in the presence of the two yeast integral membrane proteins fused to NubG confirms that TF-Cub-tagged mammalian renal baits are not self-activating. In contrast, growth on the same plates in the presence of either NubI construct indicates that the chimeric bait proteins are expressed, and the proteins are properly inserted into the membrane. C, targeting of TF-Cub-Baits to the membrane of yeast. Full-length proteins NaPi-IIa, NHE-1, NHE-3, and NaS1 were expressed in yeast as TF-Cub-Baits from pTLB-1. Lysates thereof were subjected to two consecutive ultracentrifugations to isolate the cytosolic fraction, C, from the detergent-soluble membrane fraction, M. An equal volume from each supernatant was used for the detection of VP16 from the TF-Cub-Bait proteins in immunoblots (IB). The localization of the endogenous ER membrane protein Alg5 to the detergent-soluble membrane fraction validates the centrifugation procedure (n = 2). D, functional phosphate transport activity of TF-Cub-NaPi-IIa in yeast. Yeast strain PM971 with a deficiency of two high affinity Pi transporters was transformed with TF-Cub-NaPi-IIa. In this strain, the bait was properly expressed as a post-translationally modified full-length and single form (left panel, arrowhead). Timed 32Pi uptake was measured at 30 °C on Pi-starved PM971 cells. Relative net Na+-dependent transport of Pi was calculated by subtracting the Na+-independent component (∼40%) from the total Pi uptake (right panel). Values are means of triplicate. The experiment was repeated twice with similar results (n = 3). E, localization of TF-Cub-Baits in yeast. Fixed and acetone-treated spheroplasts of yeast were processed for confocal microscopy using transporter-specific primary and FITC-conjugated secondary antibodies (green). Nonspecific immunofluorescence is shown in the lower panels from vector-transformed cells (n = 2). Confocal planes were based on the strongest 4`,6-diamidino-2-phenylindole stain (blue) of the nuclei, the centers of which are designated by arrows. Secondary antibodies alone yielded no signals (not shown). PM, plasma membrane.
EXPERIMENTAL PROCEDURES
Cloning of the MYTH 2.0 Bait Vectors pTLB-1 and pCLB-1—
The MYTH bait vector pTMBV4 (Dualsystems Biotech AG, Schlieren, Switzerland) allows the fusion of a bait to the N terminus of a Cub-(LexA+VP16) (Cub-TF) reporter module (conventional MYTH system) whose LexA is mutated at R156G to abate the affinity toward the exogenous reporter gene promoters (Dualsystems Biotech AG). As a shuttle vector, pTMBV4 contains a kanamycin resistance gene, a LEU2 gene for auxotrophic selection in yeast, a highly potent TEF1 promoter, and a CEN/ARS origin of replication (Ori) for low copy propagation in yeast (Dualsystems Biotech AG). Related to this vector is pTMBV12 that contains an expanded multiple cloning site (MCS) with among others AatII and Eco47III sites between the SfiI sites of pTMBV4.
The vector pTMBV1 was modified to pTMBV2 by inserting the portion AatII-BglII-Stop codon (TAA) upstream of the stop codon from VP16. For this purpose, pTMBV1 was amplified by means of bidirectional PCR using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), and its parental DNA was digested with DpnI (Stratagene). The extended and nicked vector fragment was religated with T4 DNA ligase (Roche Applied Science) after treatment with T4 polynucleotide kinase (Promega, Madison, WI). To generate pTLB-1 (pTEF-LexMut+Cub-Bait-CEN/ARS), the following amplicons were designed by PCR using pTMBV1 as a template: the TF fragment as XbaI-Kozak (GCCACC)-(LexA+VP16)-T-NotI and the Cub fragment as XbaI-Spacer (GAACTA)-NotI-Cub-TT-SalI. The nucleotide T in the TF fragment brings the reporter in-frame to Cub, whereas nucleotides TT in the Cub fragment bypass a stop codon within the preceding region. A Linker-MCS fragment was obtained directly from pTMBV1 by digestion with SalI and AatII. Linearized pTMBV2 (XbaI and AatII) was complemented with fragments derived from Cub (XbaI and SalI) and Linker-MCS (SalI and AatII). Finally the resulting construct was assembled to pTLB-1 by insertion of the TF fragment between XbaI and NotI sites. Thus, the new MYTH 2.0 bait vector pTLB-1 comprises a very strong TEF1 promoter, a LEU2 gene, a Kozak initiation signal sequence, an inverted cassette of pTMBV1 with the composites (LexA+VP16)-Cub-Linker-MCS, and additional stop codons guaranteeing termination (see Fig. 1B). The vector pCLB-1 contains the backbone of pCMBV4 (Dualsystems Biotech AG) plus the XbaI-FspI fragment of pTLB-1 and thus is similar to pTLB-1, but expression is driven via a weak CYC1 promoter.
DNA Constructs for the MYTH—
All bait plasmids encoded entire transmembrane proteins fused to the C terminus of the TF-Cub reporter. The corresponding ORFs of the baits were abridged at their start ATG codon, terminated by ochre plus opal stop codons at the respective C-terminal positions, and flanked with restriction sites by the Expand High Fidelity PCR kit (Roche Applied Science). After digestion, the inserts were ligated in-frame into pTLB-1. The 5`-Pi of the linearized plasmid was removed by calf intestinal phosphatase (New England Biolabs, Beverly, MA) in the case of single cut assemblies as follows (all accession numbers reported are from GenBank™): mouse NHE-3,3 mouse NaPi-IIa (accession number BC020529) as well as mouse NaS1 (accession number AF199365) in StuI only, truncations of mouse NaPi-IIa (ΔTRL and ΔCT void of 67 residues) and NHE-3 (ΔTHM and ΔCT void of 370 residues) between StuI and SacII, rat NHE-1 (accession number M85299) as an NcoI-EcoRV embedded fragment between NcoI and Eco47III, and mouse MAP17 (accession number NM_026018) between NcoI and SacII. Human NaS1 (accession number NM_022444) was cloned into pCLB-1 between NcoI and SacII. The bait yeast GPR1 was integrated, and the baits yeast Alg5p and human ErbB3 have been described recently (24, 25).
The ORFs of preys were derived from full-length soluble or membrane proteins and integrated in-frame into pNubG-HA-X (Dualsystems Biotech AG). This vector creates a fusion to the HA tag located at the C terminus of the NubG domain and is maintained via a strong constitutive ADH1 promoter, a TRP1 selection marker, and a 2μ Ori for high copy propagation. The same cloning procedure of the baits was applied to the following preys: mouse NHE-3 between NcoI and SmaI; mouse NaPi-IIa as a BglII embedded fragment in BamHI only (compatible ligation of cohesive ends); mouse PDZK1 (accession number AF220100) containing a blunted NcoI site at amino acid position 473 (site-directed mutagenesis; Stratagene); and mouse NHERF-1 (accession number U74079), mouse NHERF-2 (accession number NM_023055), and mouse CAL (accession number BC051171) between NcoI and BamHI. For NubG or NubI constructs of yeast Ost1p and Fur4p, refer to other references (24, 27).
MYTH Analysis—
Host Saccharomyces cerevisiae strain THY AP4 with the genotype MATa, ura3, leu2, lexA::lacZ::trp1, lexA::HIS3, lexA::ADE2 (31) was grown in YPAD medium (1% yeast extract, 2% Bacto peptone, 2% glucose, 0.01% adenine hemisulfate, 2% select agar) and transformed with baits according to Gietz and Woods (32). Yeast cells from a 50-ml overnight culture grown to ∼0.8 A546 were harvested at 700 × g for 5 min at 4 °C, washed in 40 ml of ice-cold sterile double distilled H2O, and resuspended in 1 ml of double distilled H2O. An aliquot of 50 μl of competent cells was treated with TRAFO mixture (240 μl of 50% (w/v) polyethylene glycol, 36 μl of 1 m LiOAc, 25 μl of 2.0 mg/ml single-stranded DNA, 50 μl of distilled H2O, 1 μg of plasmid) for 45 min at 42 °C, then pelleted, and resuspended in 100 μl of 0.9% NaCl. At most, 45 μl was plated on synthetic dropout (SD) agar plates devoid of Leu (TF-Cub-Baits) or Trp (HA-Preys). The SD medium was composed of 0.67% yeast nitrogen base without amino acids (Difco), dropout supplement powder (Clontech), 2% glucose, 0.01% adenine hemisulfate at pH 5.6, 2% select agar for plates. The bait GPR1 was maintained on YPAD medium-G418 via resistance to kanamycin.
Co-transformation of preys was always performed sequentially following the above procedure except that overnight cultures were derived from two colonies grown in 30 ml of adequate SD medium and that the two-hybrid selection was achieved on minimal SD plates (−Trp/−Leu/−His/−Ade). To minimize inherent leaky HIS3 expression, co-transformants of baits with pNubG-HA-X were streaked out on minimal SD plates containing increasing amounts of 3-amino-1,2,4-triazole (3-AT; Sigma). The amount of 3-AT was determined that can abolish growth with the NubG control prey but not with a known interacting prey.
Construction of a NubG-X Human Kidney cDNA Library and MYTH 2.0 Screen Using the TF-Cub-NaS1 as a Bait—
An oligo(dT)-primed, size-selected (0.6–8 kb; average insert size of 1.8 kb) human kidney cDNA library with 2.6 × 106 independent clones was custom constructed in the prey vector pPR3-N by Dualsystems Biotech AG and was a kind gift of Dr. Reinhart Reithmeier. This cDNA library was transformed into the yeast reporter strain THY AP4 expressing the human TF-Cub-NaS1 bait. Approximately 8 × 106 transformants were screened for colonies that would grow on media lacking Trp, Ade, His, and Leu supplemented with 5 mm 3-AT. Library plasmids were isolated from 192 positive clones, amplified in Escherichia coli, and their sequences were analyzed. The plasmids that contained an insert were further processed by a bait dependence test. For this purpose, the individual prey plasmids were retransformed into THY AP4 yeast cells in which the bait TF-Cub-NaS1 or a human plasma membrane protein, β2-adrenergic receptor (β2AR), had been expressed. Thereby 35 TF-Cub-NaS1-dependent clones were identified of which three encoded PDZK1, NHERF-1, and NHERF-2, respectively. The nature of 28 TF-Cub-NaS1-dependent clones will be described elsewhere. Lastly the remaining four clones interacted with both TF-Cub-NaS1 and TF-Cub-β2AR and were therefore considered as artifacts.
Protein Extraction and Immunoblotting—
Proper expression of baits and preys was confirmed by immunoblotting as described previously (33). Cells were propagated to an A546 of 1.0 in the appropriate SD medium to maintain the plasmid and collected at 700 × g for 5 min at 4 °C. A total lysate was produced from 10 ml of culture by vortexing the cells for 5 min in extraction buffer (1× SDS sample buffer, 100 mm DTT, 1 mm EDTA, complete protease inhibitor (Roche Applied Science)) containing 200 μl of acid-washed glass beads (425–600 μm; Sigma). To obtain a membrane extract, cells from 50 ml of culture were subjected to repetitive vortex pulses (8 × 30 s) in lysis buffer (50 mm Tris-HCl at pH 7.5, 1 mm EDTA, Complete protease inhibitor) containing 300 μl of glass beads on ice. A debris-cleared fraction (700 × g for 20 min at 4 °C) was centrifuged at 150,000 × g for 2 h at 4 °C to yield the cytosolic fraction (supernatant). The pellet was resuspended in 150 μl of lysis buffer supplemented with 1% Triton X-100 by passages through a 25-gauge needle. A subsequent ultracentrifugation as above resulted in the soluble membrane fraction. From both soluble fractions, 150-μl aliquots were taken up in an equal amount of 2× SDS sample buffer (+DTT).
Each sample was denatured at 45 °C for 20 min, and 15 μl was used for SDS-PAGE followed by transfer to nitrocellulose (Schleicher & Schuell). The membrane was blocked (TBS at pH 7.4, 0.25% Triton X-100, 5% milk powder) for 2 h and incubated overnight either with rabbit polyclonal antibodies raised against the C terminus of rat NHE-3 (1:3000; serum 1314), the N terminus of NaPi-IIa (1:6000), and VP16 (1:350; Clontech) or with a mouse monoclonal antibody raised against the HA tag (1:2500; Sigma). After washing (TBS at pH 7.4, 0.1% Triton X-100), secondary horseradish peroxidase-coupled antibodies (1:10,000; Amersham Biosciences) were applied for enhanced chemiluminescence development (Pierce).
Determination of β-Galactosidase Expression—
Activity of the lacZ reporter gene was monitored on X-gal colony lifts (34) and quantified from o-nitrophenyl-β-d-galactopyranoside (ONPG) liquid assays (35) with some modifications to the protocol. For ONPG tests, one single positive colony was picked in triplicate to inoculate 5 ml of each minimal SD medium (devoid of 3-AT). Cells from 1.5 ml of overnight culture grown to A600 0.5–0.8 were harvested at 9000 × g for 3 min at 4 °C, washed in 1.5 ml of Z buffer (40 mm NaH2PO4, 60 mm Na2HPO4 at pH 7.0, 10 mm KCl, 1 mm MgSO4, 50 mm β-mercaptoethanol), and snap frozen in liquid nitrogen. Generally 15% of total cells were permeabilized in 800 μl of Z buffer, 80 μl of chloroform, and 40 μl of 0.1% SDS by vortexing for 10 s. The reaction was initiated after preincubation for 5 min at 28 °C by adding 160 μl of 4 mg/ml ONPG (Sigma) in Z buffer and terminated by adding 400 μl of 1 m Na2CO3. After spinning with a microcentrifuge at top speed for 10 min, the product at A420 and the light scatter at A550 were measured against blanks (no cells) to determine normalized Miller units, which = 1000 × (A420 − (1.75 × A550))/(1.875 × T × V × A600) where T and V represent reaction time (min) and volume of withdrawn culture (ml), respectively (36).
Phosphate Uptake in Yeast—
Phosphate acquisition in yeast is accomplished by a low affinity system (Km of Pi ∼1 mm) and by two high affinity symporters, a H+-coupled Pho84p (Km of Pi 1–15 μm; Vmax at pH 5.0) and a Na+-coupled Pho89p (Km of Pi 0.5 μm; Vmax at pH 9.5) (37). Yeast PM971 (Δpho89::TRP1 Δpho84::HIS3 ade2 leu2 his3 trp1 ura3), a knock-out strain of both high affinity Pi transporters (38), was transformed with TF-Cub-NaPi-IIa in pTLB-1 and grown overnight in 30 ml of SD−Leu. Cells from 10.5 A600 units were collected (700 × g for 5 min) and washed in SD−Leu/−Pi (20 mm Mes at pH 7.4, SD−Leu devoid of KH2PO4 (ForMedium, CYN0801)). After Pi starvation for 4 h in 30 ml of SD−Leu/−Pi, cells from 10 ml of culture were washed (1400 × g for 5 min at 4 °C) twice in uptake buffer ± sodium (10 mm Hepes-Tris at pH 7.4, 1 mm MgSO4, 1 mm CaCl2, 2% glucose, 100 mm NaCl or choline-Cl) and concentrated to a density of 10 A600. For 32Pi uptake experiments (39), cells at a final concentrations of 6 A600 in uptake buffer ± sodium supplemented with 200 μm of KH2PO4 plus 20 μCi/ml 32Pi were incubated at 30 °C. For each time point, a 20-μl aliquot was transferred in 5 ml of ice-cold stop solution (5 mm Tris-HCl at pH 7.4, 100 mm NaCl, 5 mm NaH2PO4). Cells were rapidly filtered on nitrocellulose membranes, washed twice, and counted by scintillation spectrometry. Uptake was normalized to total counts in uptake buffers and performed in triplicate.
Immunofluorescence of Yeast—
Cells were grown overnight to 1 A600 in 30 ml of SD−Leu and fixed for 30 min in 5% formaldehyde at room temperature. Approximately 108 cells (10 ml of culture) were collected (700 × g for 5 min) and incubated for 2 h at room temperature in PF buffer (100 mm KPO4 at pH 6.5, 0.5 mm MgCl2, 5% formaldehyde). After equilibration to SP buffer (40 mm KPO4 at pH 6.5, 0.5 mm MgCl2, 1.2 m sorbitol), cells were resuspended in 1 ml of 1.35 mg/ml Zymolyase-20T (Seikagaku Corp., Tokyo, Japan) dissolved in SP buffer plus 0.2% (v/v) β-mercaptoethanol and converted to spheroplasts with shaking at 100 rpm for 30 min at 30 °C. Cells were gently collected at 100 × g for 5 min at 4 °C, washed three times in SP buffer, spotted on precoated poly-l-lysine slides (O. Kindler GmbH and Co., Freiburg, Germany) and subjected to an acetone/methanol treatment as described by Gotta et al. (40). Washed spots were blocked with PBS-T (PBS at pH 7.4, 2% BSA, 0.1% Triton X-100) and incubated overnight with the following polyclonal antibodies: rabbit anti-rat NHE-3 (1:1000, C-terminal; serum 1568), rabbit anti-rat NaPi-IIa (1:1000, N-terminal), and goat anti-mouse NHE-1 (1:200, C-terminal; Santa Cruz Biotechnology). After rinsing in PBS-T, primary antibodies were detected using goat anti-rabbit FITC (1:50; Dako Cytomation) or donkey anti-goat Alexa Fluor 488 (1:200; Molecular Probes). Spots were intensively washed in PBS-T, stained for 3 min with 1 μg/ml 4`,6-diamidino-2-phenylindole, and mounted using Dako Glycergel. Finally a Leica TCSSP laser scanning microscope (Wetzlar, Germany) equipped with a 63× oil-immersed objective was applied for confocal microscopy.
Overlay of the Class I PDZ Domain Proteomic Array—
The C-terminal 79 (CT) or 76 (ΔTRL) amino acids of mouse NaPi-IIa were expressed between EcoRI and XhoI sites of pGEX-6P-2 (GE Healthcare) and batch-purified on glutathione-agarose beads (∼0.3 ml of bead resin/ml of cleared lysate) following the procedure as delineated previously (16). Upon elution in EB buffer (50 mm Tris-HCl at pH 8.0, 120 mm NaCl, 1 mm DTT, 0.1% Igepal CA-630, 20 mm reduced glutathione) for 30 min at 4 °C, supernatants were dialyzed against SB buffer (EB buffer devoid of glutathione) on Microcon YM-10 Centricon concentrators (Millipore; cutoff, 10 kDa). Proteins concentrated to 10 mg/ml in SB buffer plus 10% glycerol were stored at −80 °C until use. Processing of the overlay derived from a 100 nm concentration of each purified GST fusion protein has been described elsewhere (41).
Transport Measurements in Xenopus Oocytes—
Mature Xenopus laevis females were purchased from the African Xenopus Facility C.C. (Noordhoek, South Africa) and kept under standard conditions. Stage V and VI oocytes from X. laevis were maintained at 18 °C in modified Barth's solution MBS (88 mm NaCl, 1 mm KCl, 0.82 mm MgSO4, 0.4 mm CaCl2, 0.33 mm Ca(NO3)2, 2.4 mm NaHCO3, 20 mg/liter gentamicin sulfate, 10 mm HEPES/Tris at pH 7.4). Human NaS1 was subcloned into plasmid pSPORT, whereas human NHERF-1 (accession number NM_004252) and NHERF-2 (accession number NM_004785) were generated in pGEX-6P-2. For cRNA synthesis, each plasmid was linearized by NotI digestion. The cDNA was in vitro transcribed using the mMessage mMachine (Ambion) T7 RNA polymerase kit, and the resulting capped cRNA was dissolved in Milli-Q H2O. Oocytes were injected with either 50 nl of water (control), 5 ng of human NaS1 cRNA, 2.5 ng of human NHERF-1 cRNA, 2.5 ng of human NHERF-2 cRNA, or combinations using a Nanojet automatic injector (Drummond Scientific Co., Broomall, PA). Transport measurements were performed by a [35S]sulfate uptake on day 3 postinjection. Briefly 10 oocytes (individual data point) were washed at room temperature for 2 min in solution A (100 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 10 mm HEPES/Tris at pH 7.5) and placed for 30 min at room temperature into 100 μl of solution A containing 0.1 mm Na2SO4 and 10 mCi/ml of 35SO42− (PerkinElmer Life Sciences). The oocytes were washed four times with ice-cold solution A, lysed with 1% SDS, and dissolved in scintillant (Emulsifier Safe, Canberra Packard) before counting by liquid scintillation spectrometry.
Immunoprecipitation from Xenopus Oocyte Membranes—
Rat NaPi-IIa (±ΔTRL) and rat NaS1 were cloned in-frame between XhoI and ClaI sites of pSD-HA or pSD-myc, both descendent vectors of pSD-Easy with additional N-terminal tags. The resulting constructs were linearized with BglII and transcribed on SP6 (Ambion). X. laevis oocytes were (co-)injected with 10 ng of each cRNA and incubated for 3 days at 16 °C. Total membranes were isolated from 30 oocytes and solubilized in 0.5% (w/v) sodium deoxycholate for 1 h at room temperature. The solubilized material was used for immunoprecipitations at 4 °C for 16 h in sucrose buffer containing 0.1% (v/v) of Triton X-100 as described by de Jong et al. (42). Modifications from this protocol included the removal of yolk proteins twice by centrifugation at 200 × g (instead of 3000 × g) and the immobilization of 1.25 μl from a mouse anti-HA antibody (clone HA-7; Sigma) on 40 μl of Protein G plus/Protein A-agarose suspension (Calbiochem) in IPP500 (42). After four washes in IPP100 (42), proteins were eluted in 50 μl of 2× SDS sample buffer (+DTT) for 30 min at 37 °C and subjected in equal amounts to immunoblotting as delineated before. Primary antibodies were as follows: mouse anti-HA (clone HA-7, 1:10,000; Sigma), rabbit anti-MYC (2 μg/ml; Upstate, Waltham, MA), or rabbit anti-NaPi-IIa (1:6000).
RESULTS
Construction of MYTH 2.0 Bait Vectors—
The current bait vectors for the conventional MYTH system fuse the Cub-TF reporter to the C terminus of a membrane-associated bait. The appropriate centromeric bait construct pCYC-Bait-Cub-TF permits constitutive low level expression in yeast driven by a weak CYC1 promoter and a low copy origin of replication (CEN/ARS) under auxotrophic selection from a LEU2 nutritional marker (24). Here two MYTH 2.0 vectors (pCLB-1 and pTLB-1) were generated with an inverted cassette that brings a membrane bait N-terminal to the reporters (i.e. TF-Cub-Bait) exposing its free C terminus for interaction with preys (Fig. 1, A and B, and refer to “Experimental Procedures”). From these two vectors, heterologous expression is initiated by an additional Kozak vertebrate consensus signal. Except for NaS1, all required target proteins were not expressed from the pCLB-1 vector in which the expression of the TF-Cub-Bait chimeric protein is under control of a weak CYC1 promoter. Consequently a stronger TEF1 promoter was also placed in front of the cassette resulting in vector pTLB-1. Notably a MYTH 2.0 TF-Cub-Bait in contrast to the conventional MYTH system liberates TF that is still affixed to the Cub domain (Fig. 1A). Although sterically bigger than TF alone, the TF-Cub from pCLB-1 and pTLB-1 showed robust β-galactosidase activity by induction of the lacZ gene in both X-gal colony filter lifts or ONPG liquid assays (data not shown).
Choice, Expression, and Localization of Baits—
The mammalian membrane proteins NaPi-IIa (16), MAP17 (6, 17, 43), and NHE-3 (22, 44), which are all apically localized, require at least their free C termini to bind the previously identified PDZ proteins (6, 14, 16, 17, 45). Along with apical NaS1 (9, 46, 47) and basolateral NHE-1 (7), these particular integral membrane proteins were generated as TF-Cub-Baits in the setting of the MYTH 2.0 system (Fig. 2A) with the goal to verify PDZ protein-membrane protein interactions based herein on full-length membrane proteins. Because only the MYTH 2.0 aligns with the prerequisite of C-terminal unmasked baits, the conventional MYTH system was found to be not applicable in this sense (data not shown).
To illustrate that the five renal epithelial membrane protein baits are (i) properly expressed, (ii) inserted into the yeast membrane, and (iii) not self-activated by triggering reporter genes in the absence of any interacting protein, we conducted genetic, biochemical, and immunofluorescence assays. The five TF-Cub-membrane bait proteins were analyzed in the presence of two non-interacting yeast integral membrane proteins, namely Ost1p as a member of the oligosaccharyltransferase complex in ER and Fur4p as a uracil permease localized to the plasma membrane. They were fused to either NubI (Ost1-NubI and Fur4-NubI) or NubG (Ost1-NubG and Fur4-NubG). Both NubI control fusion constructs are designed to activate the yeast reporter system irrespective of a protein-protein interaction because of the high affinity of NubI to associate with Cub into active ubiquitin (25). Activation of the reporter system was measured as growth on media lacking histidine and adenine in the presence of the optimal concentration of 3-AT (Fig. 2B). All five mammalian TF-Cub-Bait chimeras exhibited growth on the selective medium in the case of co-expression with either NubI fusion, indicating that they are properly inserted into the membrane. On the other hand, co-expression of the five mammalian TF-Cub-Bait fusions with the non-interacting Ost1-NubG and Fur4-NubG protein fusions did not lead to initiation of the reporter system, indicating that the chosen TF-Cub-Bait chimeras are not self-activating (Fig. 2B).
In addition, membrane targeting of TF-Cub-Baits was analyzed from yeast lysates after sequential high speed centrifugations. Upon segregation of the cytosolic fraction, the residual pellet was solubilized in 1% of Triton X-100 and subsequently ultracentrifuged to obtain the membrane fraction (Fig. 2C). The yeast ER membrane protein Alg5p was included to validate this fractionation procedure. Although the soluble TF-Cub moiety alone from vector pTLB-1 was found in the cytosolic fraction, all five mammalian full-length TF-Cub-Bait proteins were present only in the membrane fraction (Fig. 2C). Furthermore immunoblotting of total extracts from yeast against VP16 from the TF module verified the correct expressions of all TF-Cub-Baits (supplemental Fig. 1A). Two similarly sized immunoreactive bands of NHE-3 and NaPi-IIa were detected by anti-VP16, but only the upper band of NHE-3 and both bands of NaPi-IIa were recognized by the appropriate transporter-specific antibodies.
To address whether a chosen mammalian protein can fulfill transport activity as a TF-Cub fusion in yeast, NaPi-IIa bait protein was tested for phosphate uptake in yeast strain PM971, which is devoid of high affinity Pi co-transport systems (38). PM971 was transformed with the bait TF-Cub-NaPi-IIa and deprived of Pi before uptake measurements. In this strain, the type IIa NaPi cotransporter was functionally expressed with concomitant Na+-dependent Pi transport (Fig. 2D). As shown in the bar graph, the amount of Pi uptake rose relative to time of incubation indicating that the NaPi-IIa transporter is functionally active in the yeast cell membrane.
Lastly we conducted immunofluorescence analysis to localize NHE-3, NHE-1, and NaPi-IIa in yeast. In spheroplasts from yeast cells treated with zymolyase, transporter-specific immunofluorescence of TF-Cub-Bait from NaPi-IIa was strictly confined to the plasma membrane. The analog bait of NHE-1 was expressed exclusively in the plasma membrane, whereas that of NHE-3 was distributed in both intracellular and cellular membrane pools (Fig. 2E). In summary, the results of these genetic, biochemical, and cell biological experiments strongly suggest that the N-terminal TF-Cub fusions of NaPi-IIa, MAP17, NHE-3, NaS1, and NHE-1 are properly inserted into the yeast membrane and are not self-activating and that at least the TF-Cub-NaPi-IIa bait is functionally active in yeast.
Choice of PDZ Targets as Preys—
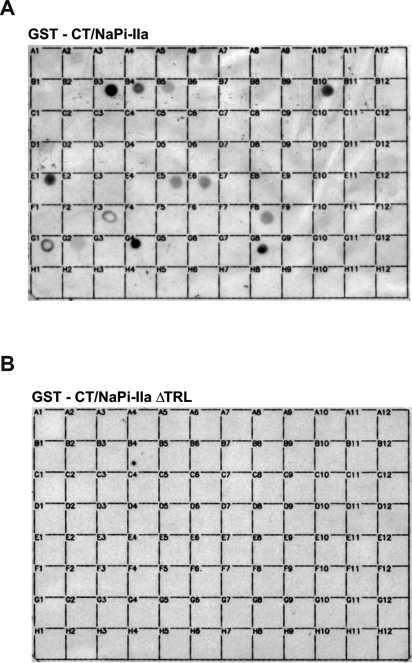
The interactions of the C termini derived from NHE-3, NaPi-IIa, and MAP17 with the PDZ proteins PDZK1, PDZK2, NHERF-1, NHERF-2, and CAL have been demonstrated using genetic as well as biochemical techniques (6, 16, 17). Recently a proteomic PDZ array was constructed that comprised almost 100 class I PDZ domains from proteins of different tissues (Table I) (41). The interaction between NaPi-IIa and PDZ domains is exemplified in Fig. 3. An overlay with a GST-fused C-terminal tail of NaPi-IIa identified PDZ3 of PDZK1 (bin G4), PDZ3 of PDZK2 (bin G8), and PDZ1 of NHERF-1 (bin B3) with high affinity as well as PDZ2 of NHERF-1 (bin B4), PDZ1 of NHERF-2 (bin B5), CAL (bin B10), and Densin-180 (bin E1) with moderate affinity (Fig. 3A). These interactions depend on TRL from the extreme C-terminal end of NaPi-IIa because removal of this motif resulted in no detectable binding (Fig. 3B). Thus, this array identified only NaPi-IIa-interacting PDZ proteins that had already been published thereby supporting the strikingly high sensitivity and selectivity of NaPi-IIa toward these PDZ proteins.
Table I.
Decryption of bin codes from the class I PDZ domain proteomic array
GEF, guanine nucleotide-exchange factor; nNOS, neuronal nitric-oxide synthase; RA, Ras/Rap1-associating; INADL, INAD-like; GIPC, GAIP-interacting protein C terminus; PAPIN, plakophilin-related armadillo-repeat protein-interacting PDZ protein.
| Bin | PDZ domain |
|---|---|
| A1 | MAGI-1 PDZ1 |
| A2 | MAGI-1 PDZ2 |
| A3 | MAGI-1 PDZ3 |
| A4 | MAGI-1 PDZ4+5 |
| A5 | MAGI-2 PDZ1 |
| A6 | MAGI-2 PDZ2 |
| A7 | MAGI-2 PDZ3 |
| A8 | MAGI-2 PDZ4 |
| A9 | MAGI-2 PDZ5 |
| A10 | MAGI-3 PDZ1 |
| A11 | MAGI-3 PDZ2 |
| A12 | MAGI-3 PDZ3 |
| B1 | MAGI-3 PDZ4 |
| B2 | MAGI-3 PDZ5 |
| B3 | NHERF-1 PDZ1 |
| B4 | NHERF-1 PDZ2 |
| B5 | NHERF-2 PDZ1 |
| B6 | NHERF-2 PDZ2 |
| B7 | PSD-95 PDZ1+2 |
| B8 | PSD-95 PDZ3 |
| B9 | PDZ-GEF-1 PDZ |
| B10 | CAL PDZ |
| B11 | nNOS PDZ |
| B12 | INADL PDZ5 |
| C1 | INADL PDZ6 |
| C2 | INADL PDZ7 |
| C3 | SAP97 PDZ1+2 |
| C4 | SAP97 PDZ3 |
| C5 | SAP102 PDZ1+2 |
| C6 | SAP102 PDZ3 |
| C7 | Chapsyn110 PDZ1+2 |
| C8 | Chapsyn110 PDZ3 |
| C9 | E6TP1 PDZ |
| C10 | ERBIN PDZ |
| C11 | ZO-1 PDZ1 |
| C12 | ZO-1 PDZ2 |
| D1 | ZO-1 PDZ3 |
| D2 | ZO-2 PDZ1 |
| D3 | ZO-2 PDZ2 |
| D4 | ZO-2 PDZ3 |
| D5 | ZO-3 PDZ1 |
| D6 | ZO-3 PDZ2 |
| D7 | ZO-3 PDZ3 |
| D8 | C2PA PDZ |
| D9 | GIPC PDZ |
| D10 | MALS-1 PDZ |
| D11 | MALS-2 PDZ |
| D12 | MALS-3 PDZ |
| E1 | Densin-180 PDZ |
| E2 | Rhophilin PDZ1 |
| E3 | Rhophilin PDZ2 |
| E4 | Harmonin PDZ1 |
| E5 | Harmonin PDZ2 |
| E6 | Neurabin PDZ |
| E7 | Spinophilin PDZ |
| E8 | α1 syntrophin PDZ |
| E9 | β1 syntrophin PDZ |
| E10 | β2 syntrophin PDZ |
| E11 | γ1 syntrophin PDZ |
| E12 | γ2 syntrophin PDZ |
| F1 | PAPIN PDZ1 |
| F2 | MUPP1 PDZ1 |
| F3 | MUPP1 PDZ4 |
| F4 | MUPP1 PDZ6 |
| F5 | MUPP1 PDZ7 |
| F6 | MUPP1 PDZ8 |
| F7 | MUPP1 PDZ10 |
| F8 | MUPP1 PDZ12 |
| F9 | MUPP1 PDZ13 |
| F10 | PTPN13 PDZ1 |
| F11 | PTPN13 PDZ2 |
| F12 | PTPN13 PDZ3 |
| G1 | PTPN13 PDZ4+5 |
| G2 | PDZK1 PDZ1 |
| G3 | PDZK1 PDZ2 |
| G4 | PDZK1 PDZ3 |
| G5 | PDZK1 PDZ4 |
| G6 | PDZK2 PDZ1 |
| G7 | PDZK2 PDZ2 |
| G8 | PDZK2 PDZ3 |
| G9 | PDZK2 PDZ4 |
| G10 | LNX1 PDZ1 |
| G11 | LNX1 PDZ2 |
| G12 | LNX1 PDZ3 |
| H1 | LNX1 PDZ4 |
| H2 | LNX2 PDZ1 |
| H3 | LNX2 PDZ2 |
| H4 | LNX2 PDZ3 |
| H5 | LNX2 PDZ4 |
| H6 | LARG PDZ |
| H7 | MAST-205 PDZ |
| H8 | Tamalin PDZ |
| H9 | Shank PDZ |
| H10 | Rho-GEF PDZ |
| H11 | RA-GEF PDZ |
| H12 | None |
Fig. 3.
Proteomics analysis of NaPi-IIa C terminus binding to PDZ proteins. Arrays of 95 related class I PDZ domains from various proteins were spotted on nylon membranes and overlaid with the GST-fused C terminus of NaPi-IIa (A) or its truncation thereof for the very last amino acids, TRL (B). Interactions were visualized by a horseradish peroxidase-linked antibody directed against the GST moiety as described previously (41). The PDZ domains spotted in each bin are listed in Table I.
PDZ-mediated Protein-Protein Interactions Detected by the MYTH 2.0 System—
Based on the immunoblot against the HA tag from total extracts, fusions of the PDZ proteins PDZK1, NHERF-1, NHERF-2, and CAL to the C terminus of NubG-HA were detected as full-length proteins in yeast (supplemental Fig. 1B). A combination of these NubG-PDZ proteins with various TF-Cub-Baits was then applied to the new MYTH 2.0 system to test whether the entire membrane-inserted mammalian renal baits can still associate with their respective PDZ proteins. Reporter activity was measured by auxotrophic growth selection on media lacking histidine and adenine but containing the optimal concentration of 3-AT, whereas the strength of β-galactosidase expression was determined by ONPG liquid assays after selection in minimal SD medium depleted of histidine and adenine (Fig. 4). Co-expression of full-length NHE-3 in the form of TF-Cub-Bait with NubG-NHERF-1 or NubG-NHERF-2 resulted in a strong MYTH signal (Fig. 4A, left and right panels). In contrast to NHERF isoforms, the MYTH signal for NubG-PDZK1 or NubG-CAL to TF-Cub-NHE-3 was either very weak or negligible, respectively (Fig. 4A, left and right panels).
Fig. 4.
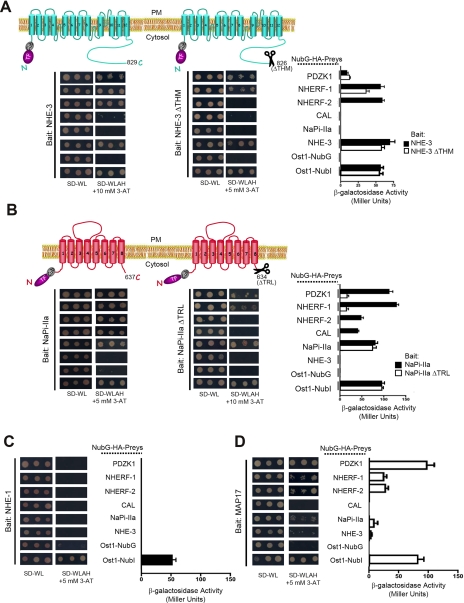
Applicaton of NHE-3, NaPi-IIa, NHE-1, and MAP17 to the MYTH 2.0 system. TF-Cub-Baits (pTLB-1) derived from full-length NHE-3 or its C-terminal truncation for THM (ΔTHM) (A), NaPi-IIa or its C-terminal truncation for TRL (ΔTRL) (B), NHE-1 (C), and MAP17 (D) were co-expressed with the following NubG-HA-Preys: PDZK1, NHERF-1, NHERF-2, CAL, NaPi-IIa, NHE-3, Ost1-NubG (negative control), and Ost1-NubI (positive control). Reconstitution of split Ub was determined by yeast growth on minimal SD−Trp/−Leu (SD−WL) medium. The interactions of baits with preys were analyzed by growth on selective SD−Trp/−Leu/−Ade/−His (SD−WLAH) medium supplemented with 3-AT (left and middle panels) as described in Fig. 2B. Quantification of MYTH reporter activity stemmed from ONPG liquid tests on permeabilized cells and was depicted against Miller units to normalize cell densities (right panels). The error bars show the standard deviations determined from triplicates. PM, plasma membrane.
When the C-terminal PDZ-binding motif of TF-Cub-NHE-3 was removed (NHE-3 ΔTHM), β-galactosidase activity was lost partially for NubG-NHERF-1 and completely for NubG-NHERF-2 (Fig. 4A, middle and right panels) suggesting that there are non-canonical secondary binding determinants for NHERF-1. With respect to NubG-PDZK1, truncation for THM from the NHE-3 MYTH 2.0 bait did not abolish the interaction compared with the full-length NHE-3 counterpart (Fig. 4A, middle and right panels) implying that there are internal binding sites for PDZK1 involved. Truncating the entire C terminus (ΔCT) of NHE-3 as a MYTH 2.0 bait disrupted all PDZ interactions (data not shown). Furthermore full-length NHE-3 and its C-terminal PDZ-binding motif deletion ΔTHM did not interact with CAL, but both NHE-3 forms were detected to homodimerize with full-length NHE-3 (Fig. 4A, all panels). A detailed investigation of the NHE-3 homodimerization will be published separately.4 Pertaining to controls, Ost1-NubG and Ost1-NubI represented negative and positive interactions of NHE-3, respectively.
The interaction pattern of TF-Cub-NaPi-IIa was distinct from that of NHE-3 in terms of MYTH assemblies: it recognized all NubG-PDZ proteins although by different relative reporter activities (Fig. 4B, left and right panels). TF-Cub-NaPi-IIa with a C-terminal TRL deletion (NaPi-IIa ΔTRL) either completely lost its interactions with PDZ proteins or decreased colony survival of PDZ cotransformants to background (Fig. 4B, middle and right panels). In addition, deletion of the entire C terminus (ΔCT) from NaPi-IIa in MYTH showed no binding to all tested PDZ proteins (data not shown). This indicates that putative non-canonical accessory sites within all intracellular domains of NaPi-IIa per se are not implicated in interactions with the applied PDZ proteins. Lastly NaPi-IIa homodimerized with both NaPi-IIa full-length and ΔTRL-truncated proteins in yeast as elucidated in details below (see Fig. 6).
Fig. 6.
Homotropic dimerization of NaPi-IIa. A, MYTH 2.0 dot assays. TF-Cub-Baits in pTLB-1 harboring full-length NaPi-IIa, its C-terminal truncation for TRL (ΔTRL) or the entire tail (ΔCT), and full-length NHE-1 as a negative control were co-transformed in a yeast reporter strain with either NubG-HA-Preys of NaPi-IIa, NHE-3, Ost1-NubG, or Ost1-NubI. Prototrophic cells were rescued on minimal SD−Trp/−Leu/−Ade/−His (SD−WLAH) plates supplemented with 3-AT to suppress leakage of the respective baits. Rescued colonies were blue due to induction of lacZ as determined by β-galactosidase replica colony lift experiments (not shown). B, immunoprecipitation from total membranes of X. laevis oocytes. Various cRNAs (MYC-NaPi-IIa, its corresponding TRL ablation, or MYC-NaS1) were co-injected with cRNA of HA-NaPi-IIa in oocytes. Total solubilized membranes of oocytes were processed for immunoprecipitation by anti-HA. The immunocomplexes were resolved by SDS-PAGE for immunoblotting with the antibodies shown. Results from two other experiments were identical (n = 3). The detection of type IIa homotropic dimers was abrogated when solubilized membranes of oocytes after a single injection of HA-NaPi-IIa were mixed with those of MYC-NaPi-IIa-injected oocytes and used for immunoprecipitation (IP). PM, plasma membrane; SD−WL, SD−Trp/−Leu.
In bait dependence tests, all PDZ proteins displayed high selectivity toward their target membrane bait proteins. The TF-Cub-Bait of NHE-1 that does not possess any of the specified PDZ-binding motifs (C-terminal PKGQ) was negative for all PDZ proteins (Fig. 4C). Analogous results were obtained with unrelated Bait-Cub-TF chimeras of yeast Gpr1p (the yeast G-protein-coupled receptor) and Ost1p (data not shown) as well as with the human receptor tyrosine kinase ErbB3 (supplemental Fig. 2, left panel). In agreement with recent findings (6, 17), TF-Cub-MAP17 might interact more strongly with PDZK1 than with NHERF-1 or NHERF-2 when merely based on semiquantitative ONPG values (Fig. 4D).
Analysis of the Human NaS1 Transporter Using the MYTH 2.0 System—
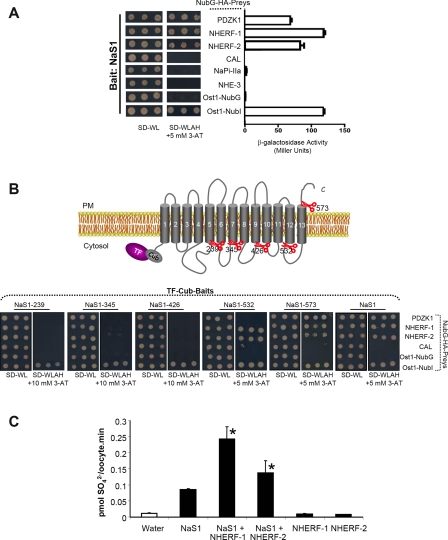
Previous findings of hyposulfatemia and hypersulfaturia in NaS1-null mice (48) evinced the apical NaS1 membrane transporter to play a major role in mediating proximal tubule sulfate reabsorption in kidney. Despite its importance, the investigation of NaS1 interactions has not been tackled by any of the available interactive proteomics technologies so far. To counteract this fact, we conducted a MYTH 2.0 screen using a NubG-tagged human kidney cDNA library to find NaS1 protein interactors. To that end, cDNA fragments originating from human kidney of ∼0.6–8 kb in length were inserted C-terminally to the NubG sequence generating the prey library in the NubG-X orientation where X stands for any cDNA insert. This library was introduced by transformation into the yeast strain expressing the human TF-Cub-NaS1 bait. A MYTH 2.0 screen of ∼8 × 106 yeast transformants revealed 31 novel NaS1-interacting proteins; among them were NHERF-1, NHERF-2, and PDZK1 (Table II). Although this study focused attention on the aforementioned PDZ proteins, the characterization of the remaining 28 newly identified NaS1 interactors will be described elsewhere. When tested in the MYTH 2.0 system, reporter activity was strong from TF-Cub-NaS1 NHERF-1 and weaker with PDZK1 or NHERF-2 (Fig. 5A). These interactions were specific for the N-terminally TF-Cub-tagged NaS1 protein because none of them were detected using an NaS1 bait (NaS1-Cub-TF) in the conventional MYTH format (supplemental Fig. 2, right panel).
Table II.
NaS1 interactors from a MYTH 2.0 screen on a human kidney library
| Accession no. | Description of identified human proteins | Process |
|---|---|---|
| NM_000146 | Ferritin, light polypeptide (FTL) | Phosphorylation |
| NM_018245 | Oxoglutarate dehydrogenase-like (OGDHL) | Metabolism |
| NM_000611 | CD59 molecule, complement-regulatory protein (CD59) | Signal transduction |
| NM_000295 | Serpin peptidase inhibitor, clade A, member 1 (SERPINA1) | Unknown |
| NM_020312 | Coenzyme Q9 homolog (S. cerevisiae) (COQ9) | Metabolism |
| NM_003002 | Succinate dehydrogenase complex, subunit D (SDHD) | Transporter/channel |
| NM_002614 | PDZ domain-containing 1 (PDZK1) | Signal transduction |
| NM_018477 | Actin-related protein 10 homolog (S. cerevisiae) (ACTR10) | Cytoskeleton network |
| NM_013446 | Makorin, ring finger protein, 1 (MKRN1) | Metabolism |
| NM_006821 | Acyl-CoA thioesterase 2 (ACOT2) | Metabolism |
| NM_005345 | Heat shock 70-kDa protein 1A (HSPA1A) | Protein transport |
| NM_013336 | Sec61 α1 subunit (S. cerevisiae) (SEC61A1) | Protein transport |
| NM_004252 | Na+/H+ exchanger, member 3, regulator 1 (SLC9A3R1) | Signal transduction |
| NM_001664 | Ras homolog gene family, member A (RHOA) | Cell organization/biosynthesis |
| NM_006034 | Tumor protein p53-inducible protein 11 (TP53I11) | Protein degradation |
| NM_003006 | Selectin P ligand (SELPLG) | Unknown |
| NM_006827 | Transmembrane emp24-like trafficking protein 10 (TMED10) | Protein transport |
| NM_000050 | Argininosuccinate synthetase | Metabolism |
| NM_002773 | Protease, serine, 8 (prostasin) (PRSS8) | Protein biosynthesis |
| NM_032811 | Transforming growth factor β regulator 1 (TBRG1) | |
| NM_015161 | ADP-ribosylation factor-like 6-interacting protein 1 (ARL6IP1) | Protein biosynthesis |
| NM_014220 | Transmembrane 4 L six family member 1 (TM4SF1) | Unknown |
| NM_001642 | Amyloid β (A4) precursor-like protein 2 (APLP2) | Signal transduction |
| NM_006702 | Patatin-like phospholipase domain-containing 6 (PNPLA6) | Metabolism |
| NM_032341 | Coenzyme Q5 homolog, methyltransferase (COQ5) | Metabolism |
| NM_001037494 | Dynein, light chain, LC8-type 1 (DYNLL1) | Cell organization/biosynthesis |
| NM_002087 | Granulin (GRN) | Signal transduction |
| NM_004552 | NADH dehydrogenase (ubiquinone) Fe-S protein (NDUFS5) | |
| NM_001885 | Crystallin, αB (CRYAB) | Signal transduction |
| NM_005724 | Tetraspanin 3 (TSPAN3) | Unknown |
| NM_004785 | Na+/H+ exchanger, member 3 regulator 1 (SLC9A3R1) | Signal transduction |
Fig. 5.
Characterization of NaS1 using the MYTH 2.0 assay. A, analysis of interactions between TF-Cub-NaS1 (in pCLB-1 backbone) and NubG-HA-Preys. Reporter gene activity of co-transformants was detected in triplicates by growth on minimal selective SD-Trp/-Leu/-Ade/-His (SD−WLAH) medium (left panel) and by ONPG liquid tests (right panel) as delineated in Fig. 4. B, mapping the PDZ-binding domain of NaS1. Five deletion constructs of TF-Cub-NaS1 were generated in pCLB-1 and subjected to yeast dot tests on selective SD-Trp/-Leu/-Ade/-His (SD−WLAH) medium after co-transformation with NubG-HA-Preys PDZK1, NHERF-1, NHERF-2, or CAL. Specificity was corroborated based on positive growth with Ost1-NubI and negative growth with Ost1-NubG. C, impact of NHERF isoforms on NaS1 transport in a Xenopus oocyte expression system. Oocytes were injected with either 50 nl of water (control), 5 ng of human NaS1 cRNA, 2.5 ng of either human NHERF-1/-2 cRNAs, or combinations as depicted. Transport measurements were determined by [35S]sulfate uptake and counted by liquid scintillation spectrometry. Data are shown as mean ± S.E. for 7–10 oocytes per condition and are representative of three similar experiments. S.E. bars not visible were smaller than the symbols. *, p < 0.05 when compared with NaS1-injected oocytes. PM, plasma membrane; SD−WL, SD−Trp/−Leu.
To identify which regions of NaS1 are required for binding to PDZK1, NHERF-1, and NHERF-2, full-length (aa 1–595) TF-Cub-NaS1 bait and several truncations thereof (aa 1–239, 1–345, 1–426, 1–532, and 1–573) were tested for their ability to interact with the newly identified PDZ proteins in MYTH dot assays. The correct expression and localization of all TF-Cub-NaS1 deletion mutants were assessed by NubG and NubI prey tests (Fig. 5B) or by immunoblotting analysis using an anti-VP16 antibody (data not shown). According to our mapping experiments, full-length NaS1 (aa 1–595) possessed strong MYTH activity together with PDZK1, NHERF-1, and NHERF-2 (Fig. 5B). Deletion of the last 22 C-terminal amino acids of NaS1 (aa 1–573) did not abolish the binding of all three PDZ domain proteins. However, deletion of an additional 41 amino acids of NaS1 (aa 1–532) disrupted the binding of PDZK1 suggesting the last cytoplasmic loop to be responsible for PDZK1 recognition. The interaction of NHERF-1 and NHERF-2 was preserved with this NaS1 (aa 1–532) construct but not with the next shorter construct (aa 1–426). This entails an interacting region for NHERF-1/2 isoforms situated in the cytosolic loop between the 10th and 11th transmembrane domains (aa 426–532) on NaS1 (Fig. 5B). Importantly CAL did not associate with any of the above mentioned TF-Cub-NaS1 baits (Fig. 5, A and B) thus further confirming the specificity of the NaS1-PDZK1 and NaS1-NHERF-1/2 interactions. Apart from screening, the MYTH 2.0 is therefore also an efficient means to map internal binding motifs on transmembrane bait proteins as demonstrated herein by the human NaS1 sulfate transporter.
Due to their association with NaS1, NHERF-1 and NHERF-2 might affect NaS1 function. To investigate this assumption, we performed a transport assay in Xenopus oocytes and measured [35S]sulfate uptake. After co-expressing NaS1 with either NHERF-1 or NHERF-2, sulfate uptake was significantly augmented (Fig. 5C), suggesting a functional regulation of NaS1 by NHERF-1/2 isoforms.
Demonstration of NaPi-IIa Oligomerization via the MYTH 2.0—
Besides testing protein-protein interactions between mammalian transporters and their PDZ partners, the MYTH 2.0 system is also suited to investigate the oligo/dimeric state of membrane proteins. As depicted in supplemental Fig. 1B, NHE-3 and NaPi-IIa proteins in a NubG-Prey setting were labeled on immunoblots against the HA tag or against epitopes on the transporters although the undersized band at 70 kDa for NaPi-IIa may imply a deficient N-glycosylation post-translational modification. As shown in Fig. 6A, NubG-NaPi-IIa as a prey strongly formed homomeric dimers with the TF-Cub-Bait of NaPi-IIa or with its C-terminal truncations ΔTRL and ΔCT in dot assays (Fig. 6A). Dimerization was specific because none of the TF-Cub-NaPi-IIa baits induced colony survival in the presence of another renal transporter, NHE-3 (Fig. 6A). These data collectively suggest that the homodimeric property of NaPi-IIa does not involve either the N- or C-terminal ends.
Dimerization of NaPi-IIa was further confirmed in X. laevis oocytes utilizing cRNAs of N-terminal HA- and MYC-tagged NaPi-IIa. The glycosylated and tagged forms of the NaPi-IIa protein were found in the membrane fraction from singly injected oocytes (Fig. 6B, lower panel). An anti-HA immunoprecipitate from membranes of co-injected oocytes contained either MYC-NaPi-IIa or MYC-NaPi-IIa ΔTRL (Fig. 6B). A PDZ scaffold is therefore not required for type IIa NaPi dimerization in oocytes. Homodimerization of NaPi-IIa was specific as neither co-injected MYC-NaS1 nor separately injected MYC-NaPi-IIa from a mixture with separately injected HA-NaPi-IIa was retained (Fig. 6B).
DISCUSSION
The fact that most of the widely used genetic tools for studying protein-protein interactions solely accommodate soluble domains from proteins poses a significant obstacle to our understanding of membrane-anchored or -inserted proteins. Thus far, conventional YTH only accepts the participation of hydrophilic tails from integral membrane proteins. Such soluble tails of transmembrane proteins may acquire partially altered conformations when unhitched from the integrity of their holoproteins begetting either false positive or negative results. Theoretically the previously described conventional MYTH in the split-Ub format can surmount this hurdle. Since its introduction, the MYTH technology indeed has led to the dissection of quaternary structures from protein complexes such as sucrose transporters (49); a yeast protein translocation machinery (25, 50); a receptor tyrosine kinase, ErbB3 (24); physical interactions between plant K+ channels (31); and a yeast ATP-binding cassette transporter Ycf1p (26).
Scaffolding PDZ proteins usually recognize a few amino acids at the very ends of soluble C-terminal tails from transmembrane proteins (1). Recent studies also showed that PDZ domain-containing proteins recognize and bind non-C-terminal motifs (2, 51, 52). However, the impact of a full-length membrane-inserted protein on a PDZ-mediated association and the possibility of binding sites beyond the C-terminal domain have thus far not been addressed with any interactive proteomics technology. To this end, we modified a conventional MYTH to a new format where the C-terminal tail of a polytopic membrane bait protein is unhindered to interact for instance with PDZ prey proteins (Fig. 1A). This work illustrates five salient features. First, all transmembrane bait proteins examined were targeted to the appropriate location and were processed properly in yeast. Second, we verified interactions of PDZ proteins from proximal tubular cells, some of which were initially identified in conventional YTH traps, but we applied herein full-length membrane proteins instead of only their cytosolic termini as baits. The newly developed MYTH 2.0 system reproduced the PDZ-binding profile with high fidelity. Third, we discovered new interacting candidates such as CAL for NaPi-IIa or PDZK1, NHERF-1, and NHERF-2 for NaS1 by either screening a human kidney NubG-tagged cDNA library or by applying direct trap assays. In this regard, it is worth pointing out the unprecedented and successful subjection of NaS1 to proteomics, which is the merit of the modified MYTH 2.0 system. Fourth, we used the MYTH 2.0 assay to map and confine the regions on NaS1 required for binding to the above PDZ proteins. Finally the modified system disclosed homodimerization of NaPi-IIa that was accomplished by domains other than its cytoplasmic tails. Thus, the MYTH 2.0 represents a useful tool to divulge physiological binding partners/regulators of membrane proteins and establishes a novel horizon for interactive proteomics studies of membrane protein complexes.
When the proteins NHE-3, NHE-1, NaPi-IIa, MAP17, and NaS1 were inserted in the MYTH 2.0 assembly with their C termini uncoupled from reporters, full length and processed TF-Cub-Baits were obtained in yeast (Fig. 2). Several findings imply that the fusions of TF-Cub to the transporter proteins were probably embedded in a natural membranous setting of yeast: (i) lack of transporters in a non-solubilized cell fraction; (ii) distribution of NHE-3, NaPi-IIa, and NHE-1 as TF-Cub-Baits in a pattern similar to their native distribution in mammalian cells; and (iii) Na+-dependent Pi entry after transformation with TF-Cub-NaPi-IIa (Fig. 2, C–E). We were not able to show transformation-dependent Na+/H+ exchange activity in this strain because of a multitude of endogenous alkali-metal-cation transporters that are present in yeast as noted previously (53).
The PDZ proteins PDZK1, NHERF-1, NHERF-2, and CAL were first isolated by conventional YTH using soluble C termini derived from NHE-3, NaPi-IIa, and MAP17 as baits (6, 16, 17, 21). These interactions have been mapped to cover C-terminal class I PDZ motifs of the transporters (6, 16, 17). No further interacting PDZ proteins have been unveiled for NHE-3, NaPi-IIa, and MAP17. This is especially true for NaPi-IIa whose C terminus was overlaid on an array of up to 100 different PDZ proteins resulting in the detection of only PDZK1/2, NHERF-1/2, and CAL (Fig. 3).
In the MYTH 2.0 system, relative quantification of β-galactosidase activity was indicative for strong interaction of NHERF-1 and NHERF-2 with NHE-3 (Fig. 4A). Remarkably truncating NHE-3 of the C-terminal PDZ motif (ΔTHM) or of the entire C terminus either marginally or totally abrogated split-Ub formation along with NHERF-1. Hence internal binding sites within the C-terminal domain of NHE-3 must exist for NHERF-1 but not for NHERF-2 whose target binding site on NHE-3 is completely C-terminal (Fig. 4A). Such deviation from the prototypic C-terminal PDZ recognition was initially attributed to both cofactors by in vitro assays (15).
NaPi-IIa exhibited a broader binding spectrum than NHE-3 because all tested PDZ proteins, PDZK1, NHERF-1/2, and CAL, were able to at least moderately induce the reporter genes in the MYTH 2.0 assay. The finding that binary complexes of NaPi-IIa with NHERF-1 and -2 might have binding affinities similar to that of NHE-3, when judged by Miller units from MYTH complementations, may have consequences to infer Na+-, H+- and Pi-coupled homeostasis. The modality of internal binding sequences probably does not pertain to NaPi-IIa because the last three C-terminal amino acids TRL were mandatory for all PDZ interactions in the MYTH 2.0 (Fig. 4B) and in the proteomic PDZ array (Fig. 3). Interestingly CAL did not associate with any other bait protein used in this study except NaPi-IIa. Therefore, CAL might be involved in specific regulation of NaPi-IIa in a manner similar to that described for the retrograde trafficking of CFTR (19). Furthermore NaPi-IIa was found to form homodimers. This dimerization was confirmed by immunoprecipitation from an oocyte expression system and was achieved by membrane-inserted domains without any engagement of cytosolic termini or a submembranous PDZ scaffold (Fig. 6A).
MAP17 activated the MYTH system much more strongly in the presence of PDZK1 compared with NHERF-1 and NHERF-2 (Fig. 4D). It may be that MAP17 represents a specific auxiliary adaptor for the scaffolding protein PDZK1 from proximal tubular cells that concomitantly regulates the localization or turnover of PDZK1-coupled proteins such as NaPi-IIa (10, 11, 54). Moreover MYTH provided a valuable implement to identify the ability of intact NaS1 to interact with PDZK1, NHERF-1, and NHERF-2 (Fig. 5), a feature that was not achieved by any other interactive proteomics-based methods until now. The putative physiological relevance of NaS1·NHERF-1 and NaS1·NHERF-2 complexes was further validated by a functional assay showing that NHERF-1 and to a lesser extent NHERF-2 stimulate sulfate uptake in Xenopus oocytes when co-injected with NaS1. It is conceivable that NHERF-1 and/or NHERF-2 increase the protein expression, stability, or substrate turnover rate of NaS1 on the plasma membrane; however, the precise mechanism(s) still needs to be clarified. Of particular significance are our findings from the MYTH 2.0 mapping experiments using serial truncations of NaS1 as baits. The existence of internal PDZ-binding determinants on this sulfate transporter beyond the C-terminal motif, i.e. upstream of the C terminus, was revealed for PDZK1 and NHERF-1/2 isoforms comprising residues 532–555 or 426–532 of the transporters, respectively. Only rare cases of such internal PDZ-binding ligands on integral membrane proteins have been reported previously (3, 55).
Our data also demonstrate compelling evidence that NHE-3 and NaPi-IIa must contain N and C termini both facing the cytosol, whereas at least the N termini of MAP17 and NaS1 must be cytosolic. Otherwise it cannot be explained why co-expression of the corresponding TF-Cub-Baits with Ost1-NubI or Fur4-NubI in yeast resulted in a positive MYTH readout and why co-expression with Ost1-NubG or Fur4-NubG was the opposite. In other words, the MYTH 2.0 can also be applied to partially characterize the topology of a particular integral membrane protein.
In conclusion, we have developed an altered membrane YTH system that will allow us to study intact polytopic membrane proteins, which are embedded in or anchored to the lipid bilayer as either bait or prey, for the detection of protein interactions involving this difficult class of hydrophobic proteins. This system can reproduce interactions that have been identified previously and can add additional information about new binding partners that were not detectable by any other protein-protein interaction method. Thus, we anticipate that our approach will greatly enhance the ability to disentangle complexes of membrane proteins and their interacting partners.
Supplementary Material
Acknowledgments
We acknowledge the kind contributions from the following individuals: Bengt Persson for the yeast strain PM971; Alexandru Bobulescu for technical assistance and helpful discussions; Daniel Auerbach, Safia Thaminy, Dawn Edmonds, and Corey Nislow for scores of control constructs and for helpful comments; Reinhart Reithmeier for the human kidney NubG-X cDNA library; and Paul Stehberger for fluorescence microscopy.
Footnotes
Published, MCP Papers in Press, April 11, 2008, DOI 10.1074/mcp.M800079-MCP200
The abbreviations used are: NHE, Na+/H+ exchanger; Ade, adenine; 3-AT, 3-amino-1,2,4-triazole; CAL, CFTR-associated ligand; CT, C terminus; Cub, C-terminal part of ubiquitin; lacZ, gene for β-galactosidase; MAP17, membrane-associated protein of 17 kDa; MYTH, membrane yeast two-hybrid; MYTH conventional format, reporters fused to the CT of the bait; MYTH 2.0 format, reporters fused to the N terminus of the bait; MCS, multiple cloning site; minimal SD, minimal synthetic medium consisting of a dropout for the nutritional requirement of MYTH interaction; NaPi-IIa, type IIa Na+-dependent Pi transporter; NaS1, Na+-coupled sulfate transporter; NHERF, Na+/H+ exchanger-regulating factor; Nub, N-terminal part of ubiquitin; ONPG, o-nitrophenyl-β-d-galactopyranoside; PDZ, domain present in PSD-95, Dlg, and ZO-1/2; PDZK, PDZ protein from kidney; SD, synthetic dropout; TF, transcription factor; Ub, ubiquitin; UBP, Ub-specific protease; YTH, yeast two-hybrid; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactoside; CFTR, cystic fibrosis transmembrane conductance regulator; HA, hemagglutinin; ER, endoplasmic reticulum; aa, amino acids.
I. Stagljar, unpublished results.
D. Fuster, unpublished clone.
O. Moe, unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK-48482 and P01-DK-20543 (to the Moe laboratory). This work was also supported by grants from the Canadian Foundation for Innovation, the Canadian Institute for Health Research, the National Cancer Institute of Canada, the Genome Canada and Ontario Genomics Institute, the Gebert Rüf Foundation, Genentech, and Novartis (to the Stagljar laboratory); in part by the Australian Research Council and the National Health and Medical Research Council (to the Markovich laboratory); and by a grant from the Simmons Family Foundation (to the Moe laboratory). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Hung A. Y., and Sheng, M. ( 2002) PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277, 5699–5702 [DOI] [PubMed] [Google Scholar]
- 2.Runyon, S. T., Zhang, Y., Appleton, B. A., Sazinsky, S. L., Wu, P., Pan, B., Wiesmann, C., Skelton, N. J., and Sidhu, S. S. ( 2007) Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3. Protein Sci. 16, 2454–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hillier, B. J., Christopherson, K. S., Prehoda, K. E., Bredt, D. S., and Lim, W. A. ( 1999) Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284, 812–815 [PubMed] [Google Scholar]
- 4.Bezprozvanny, I., and Maximov, A. ( 2001) Classification of PDZ domains. FEBS Lett. 509, 457–462 [DOI] [PubMed] [Google Scholar]
- 5.Hernando, N., Wagner, C. A., Gisler, S. M., Biber, J., and Murer, H. ( 2004) PDZ proteins and proximal ion transport. Curr. Opin. Nephrol. Hypertens. 13, 569–574 [DOI] [PubMed] [Google Scholar]
- 6.Gisler, S. M., Pribanic, S., Bacic, D., Forrer, P., Gantenbein, A., Sabourin, L. A., Tsuji, A., Zhao, Z. S., Manser, E., Biber, J., and Murer, H. ( 2003) PDZK1: I. a major scaffolder in brush borders of proximal tubular cells. Kidney Int. 64, 1733–1745 [DOI] [PubMed] [Google Scholar]
- 7.Burckhardt, G., Di Sole, F., and Helmle-Kolb, C. ( 2002) The Na+/H+ exchanger gene family. J. Nephrol. 15, Suppl. 5, S3–S21 [PubMed] [Google Scholar]
- 8.Murer, H., Hernando, N., Forster, I., and Biber, J. ( 2000) Proximal tubular phosphate reabsorption: molecular mechanisms. Physiol. Rev. 80, 1373–1409 [DOI] [PubMed] [Google Scholar]
- 9.Markovich, D. ( 2001) Physiological roles and regulation of mammalian sulfate transporters. Physiol. Rev. 81, 1499–1533 [DOI] [PubMed] [Google Scholar]
- 10.Kocher, O., Cheresh, P., and Lee, S. W. ( 1996) Identification and partial characterization of a novel membrane-associated protein (MAP17) up-regulated in human carcinomas and modulating cell replication and tumor growth. Am. J. Pathol. 149, 493–500 [PMC free article] [PubMed] [Google Scholar]
- 11.Blasco, T., Aramayona, J. J., Alcalde, A. I., Catalan, J., Sarasa, M., and Sorribas, V. ( 2003) Rat kidney MAP17 induces cotransport of Na-mannose and Na-glucose in Xenopus laevis oocytes. Am. J. Physiol. 285, F799–F810 [DOI] [PubMed] [Google Scholar]
- 12.Weinman, E. J., Steplock, D., and Shenolikar, S. ( 2003) NHERF-1 uniquely transduces the cAMP signals that inhibit sodium-hydrogen exchange in mouse renal apical membranes. FEBS Lett. 536, 141–144 [DOI] [PubMed] [Google Scholar]
- 13.Lamprecht, G., Weinman, E. J., and Yun, C. H. ( 1998) The role of NHERF and E3KARP in the cAMP-mediated inhibition of NHE3. J. Biol. Chem. 273, 29972–29978 [DOI] [PubMed] [Google Scholar]
- 14.Weinman, E. J., Minkoff, C., and Shenolikar, S. ( 2000) Signal complex regulation of renal transport proteins: NHERF and regulation of NHE3 by PKA. Am. J. Physiol. 279, F393–F399 [DOI] [PubMed] [Google Scholar]
- 15.Yun, C. H., Lamprecht, G., Forster, D. V., and Sidor, A. ( 1998) NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J. Biol. Chem. 273, 25856–25863 [DOI] [PubMed] [Google Scholar]
- 16.Gisler, S. M., Stagljar, I., Traebert, M., Bacic, D., Biber, J., and Murer, H. ( 2001) Interaction of the type IIa Na/Pi cotransporter with PDZ proteins. J. Biol. Chem. 276, 9206–9213 [DOI] [PubMed] [Google Scholar]
- 17.Pribanic, S., Gisler, S. M., Bacic, D., Madjdpour, C., Hernando, N., Sorribas, V., Gantenbein, A., Biber, J., and Murer, H. ( 2003) Interactions of MAP17 with the NaPi-IIa/PDZK1 protein complex in renal proximal tubular cells. Am. J. Physiol. 285, F784–F791 [DOI] [PubMed] [Google Scholar]
- 18.Kocher, O., Comella, N., Gilchrist, A., Pal, R., Tognazzi, K., Brown, L. F., and Knoll, J. H. ( 1999) PDZK1, a novel PDZ domain-containing protein up-regulated in carcinomas and mapped to chromosome 1q21, interacts with cMOAT (MRP2), the multidrug resistance-associated protein. Lab. Investig. 79, 1161–1170 [PubMed] [Google Scholar]
- 19.Cheng, J., Wang, H., and Guggino, W. B. ( 2004) Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J. Biol. Chem. 279, 1892–1898 [DOI] [PubMed] [Google Scholar]
- 20.Cheng, J., Moyer, B. D., Milewski, M., Loffing, J., Ikeda, M., Mickle, J. E., Cutting, G. R., Li, M., Stanton, B. A., and Guggino, W. B. ( 2002) A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J. Biol. Chem. 277, 3520–3529 [DOI] [PubMed] [Google Scholar]
- 21.Yun, C. H., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C. M., Weinman, E. J., and Donowitz, M. ( 1997) cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 94, 3010–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson, R. B., Wang, T., Thomson, B. R., Tarrats, L., Girardi, A., Mentone, S., Soleimani, M., Kocher, O., and Aronson, P. S. ( 2005) Role of PDZK1 in membrane expression of renal brush border ion exchangers. Proc. Natl. Acad. Sci. U. S. A. 102, 13331–13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stagljar, I., and Fields, S. ( 2002) Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem. Sci. 27, 559–563 [DOI] [PubMed] [Google Scholar]
- 24.Thaminy, S., Auerbach, D., Arnoldo, A., and Stagljar, I. ( 2003) Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 13, 1744–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. ( 1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 95, 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paumi, C. M., Menendez, J., Arnoldo, A., Engels, K., Iyer, K. R., Thaminy, S., Georgiev, O., Barral, Y., Michaelis, S., and Stagljar, I. ( 2007) Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26, 15–25 [DOI] [PubMed] [Google Scholar]
- 27.Iyer, K., Burkle, L., Auerbach, D., Thaminy, S., Dinkel, M., Engels, K., and Stagljar, I. ( 2005) Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE 2005, pl3. [DOI] [PubMed] [Google Scholar]
- 28.Hershko, A., and Ciechanover, A. ( 1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 [DOI] [PubMed] [Google Scholar]
- 29.Johnsson, N., and Varshavsky, A. ( 1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91, 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields, S., and Song, O. ( 1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 31.Obrdlik, P., El-Bakkoury, M., Hamacher, T., Cappellaro, C., Vilarino, C., Fleischer, C., Ellerbrok, H., Kamuzinzi, R., Ledent, V., Blaudez, D., Sanders, D., Revuelta, J. L., Boles, E., Andre, B., and Frommer, W. B. ( 2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. U. S. A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz, R. D., and Woods, R. A. ( 2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 33.Fetchko, M., and Stagljar, I. ( 2004) Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32, 349–362 [DOI] [PubMed] [Google Scholar]
- 34.Breeden, L., and Nasmyth, K. ( 1985) Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50, 643–650 [DOI] [PubMed] [Google Scholar]
- 35.Guarente, L. ( 1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. ( 1972) Experiments in molecular genetics, in Experiments in Molecular Genetics, pp. 466, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 37.Persson, B. L., Lagerstedt, J. O., Pratt, J. R., Pattison-Granberg, J., Lundh, K., Shokrollahzadeh, S., and Lundh, F. ( 2003) Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43, 225–244 [DOI] [PubMed] [Google Scholar]
- 38.Martinez, P., and Persson, B. L. ( 1998) Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258, 628–638 [DOI] [PubMed] [Google Scholar]
- 39.Daram, P., Brunner, S., Persson, B. L., Amrhein, N., and Bucher, M. ( 1998) Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta 206, 225–233 [DOI] [PubMed] [Google Scholar]
- 40.Gotta, M., Laroche, T., Formenton, A., Maillet, L., Scherthan, H., and Gasser, S. M. ( 1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134, 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fam, S. R., Paquet, M., Castleberry, A. M., Oller, H., Lee, C. J., Traynelis, S. F., Smith, Y., Yun, C. C., and Hall, R. A. ( 2005) P2Y1 receptor signaling is controlled by interaction with the PDZ scaffold NHERF-2. Proc. Natl. Acad. Sci. U. S. A. 102, 8042–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Jong, J. C., Willems, P. H., Mooren, F. J., van den Heuvel, L. P., Knoers, N. V., and Bindels, R. J. ( 2003) The structural unit of the thiazide-sensitive NaCl cotransporter is a homodimer. J. Biol. Chem. 278, 24302–24307 [DOI] [PubMed] [Google Scholar]
- 43.Kocher, O., Comella, N., Tognazzi, K., and Brown, L. F. ( 1998) Identification and partial characterization of PDZK1: a novel protein containing PDZ interaction domains. Lab. Investig. 78, 117–125 [PubMed] [Google Scholar]
- 44.Weinman, E. J., Wang, Y., Wang, F., Greer, C., Steplock, D., and Shenolikar, S. ( 2003) A C-terminal PDZ motif in NHE3 binds NHERF-1 and enhances cAMP inhibition of sodium-hydrogen exchange. Biochemistry 42, 12662–12668 [DOI] [PubMed] [Google Scholar]
- 45.Kocher, O., Pal, R., Roberts, M., Cirovic, C., and Gilchrist, A. ( 2003) Targeted disruption of the PDZK1 gene by homologous recombination. Mol. Cell. Biol. 23, 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck, L., and Markovich, D. ( 2000) The mouse Na+-sulfate cotransporter gene Nas1. Cloning, tissue distribution, gene structure, chromosomal assignment, and transcriptional regulation by vitamin D. J. Biol. Chem. 275, 11880–11890 [DOI] [PubMed] [Google Scholar]
- 47.Beck, L., and Silve, C. ( 2001) Molecular aspects of renal tubular handling and regulation of inorganic sulfate. Kidney Int. 59, 835–845 [DOI] [PubMed] [Google Scholar]
- 48.Dawson, P. A., Beck, L., and Markovich, D. ( 2003) Hyposulfatemia, growth retardation, reduced fertility, and seizures in mice lacking a functional NaSi-1 gene. Proc. Natl. Acad. Sci. U. S. A. 100, 13704–13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinders, A., Schulze, W., Thaminy, S., Stagljar, I., Frommer, W. B., and Ward, J. M. ( 2002) Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure (Camb.) 10, 763–772 [DOI] [PubMed] [Google Scholar]
- 50.Scheper, W., Thaminy, S., Kais, S., Stagljar, I., and Romisch, K. ( 2003) Coordination of N-glycosylation and protein translocation across the endoplasmic reticulum membrane by Sss1 protein. J. Biol. Chem. 278, 37998–38003 [DOI] [PubMed] [Google Scholar]
- 51.Harris, B. Z., and Lim, W. A. ( 2001) Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 52.Wong, H. C., Bourdelas, A., Krauss, A., Lee, H. J., Shao, Y., Wu, D., Mlodzik, M., Shi, D. L., and Zheng, J. ( 2003) Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell 12, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plant, P. J., Manolson, M. F., Grinstein, S., and Demaurex, N. ( 1999) Alternative mechanisms of vacuolar acidification in H+-ATPase-deficient yeast. J. Biol. Chem. 274, 37270–37279 [DOI] [PubMed] [Google Scholar]
- 54.Silver, D. L., Wang, N., and Vogel, S. ( 2003) Identification of small PDZK1-associated protein, DD96/MAP17, as a regulator of PDZK1 and plasma high density lipoprotein levels. J. Biol. Chem. 278, 28528–28532 [DOI] [PubMed] [Google Scholar]
- 55.Christopherson, K. S., Hillier, B. J., Lim, W. A., and Bredt, D. S. ( 1999) PSD-95 assembles a ternary complex with the N-methyl-d-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J. Biol. Chem. 274, 27467–27473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.