Abstract
A global isotopic labeling strategy combined with multidimensional liquid chromatographies and tandem mass spectrometry was used for quantitative proteome analysis of a presymptomatic A53T α-synuclein Drosophila model of Parkinson disease (PD). Multiple internal standard proteins at different concentration ratios were spiked into samples from PD-like and control animals to assess quantification accuracy. Two biological replicates isotopically labeled in forward and reverse directions were analyzed. A total of 253 proteins were quantified with a minimum of two identified peptide sequences (for each protein); 180 (∼71%) proteins were detected in both forward and reverse labeling measurements. Twenty-four proteins were differentially expressed in A53T α-synuclein Drosophila; up-regulation of troponin T and down-regulation of fat body protein 1 were confirmed by Western blot analysis. Elevated expressions of heat shock protein 70 cognate 3 and ATP synthase are known to be directly involved in A53T α-synuclein-mediated toxicity and PD; three up-regulated proteins (muscle LIM protein at 60A, manganese-superoxide dismutase, and troponin T) and two down-regulated proteins (chaoptin and retinal degeneration A) have literature-supported associations with cellular malfunctions. That these variations were observed in presymptomatic animals may shed light on the etiology of PD. Protein interaction network analysis indicated that seven proteins belong to a single network, which may provide insight into molecular pathways underlying PD. Gene Ontology analysis indicated that the dysregulated proteins are primarily associated with membrane, endoplasmic reticulum, actin cytoskeleton, mitochondria, and ribosome. These associations support prior findings in studies of the A30P α-synuclein Drosophila model (Xun, Z. Y., Sowell, R. A., Kaufman, T. C., and Clemmer, D. E. (2007) Protein expression in a Drosophila model of Parkinson's disease. J. Proteome Res. 6, 348–357; Xun, Z. Y., Sowell, R. A., Kaufman, T. C., and Clemmer, D. E. (2007) Lifetime proteomic profiling of an A30P α-synuclein Drosophila model of Parkinson's disease. J. Proteome Res. 6, 3729–3738) that defects in cellular components such as actin cytoskeleton and mitochondria may contribute to the development of later symptoms.
Parkinson disease (PD)1 is the second most common age-related neurodegenerative disorder; it is characterized by loss of dopaminergic neurons in the substantia nigra coupled with formation of intracytoplasmic Lewy body (LB) inclusions (1,2). Both environmental perturbations and genetic abnormalities have been proposed to induce the development of PD (1). There is evidence that oxidative stress (3), mitochondrial dysfunction (4), and ubiquitin-proteasome system defects (5) are each associated with PD; however, the molecular pathways that induce dopamine neuron loss remain poorly understood, and early diagnostic tools and neuroprotective therapies are still lacking.
One promising approach to studying the underlying molecular mechanisms associated with PD involves the α-synuclein protein. This is primarily due to two factors: 1) α-Synuclein is linked to the disease as a primary component of LBs found in brains of PD patients (6), and 2) three missense mutations (A30P, A53T, and E46K) in the α-synuclein gene are linked to early onset familial PD (7–9). In addition, α-synuclein is linked to other neurodegenerative diseases. For example, it is a major constituent of insoluble inclusions found in dementias with LBs and multiple system atrophy (6). Additionally, an internal fragment of α-synuclein (residues 60–95, termed the non-Aβ component) is found in amyloid plaques in the brains of patients with Alzheimer disease (10,11).
With these associations in mind, there has been a significant effort to develop α-synuclein animal models for PD (12); models are now available for a number of organisms, including yeast (13,14), flies (15), mice (16,17), rats (18,19), and primates (20). These models express (or overexpress) either wild-type or disease-linked mutant human α-synuclein. At present, no single model fully presents all human PD features; some fail to show dopamine neuron loss (17), whereas others do not develop true LBs (19). Thus, there is a need to investigate many different systems to understand underlying features that are general to disease mechanisms.
For the last decade, our group has been interested in the development of new high throughput analytical technologies for proteome analysis (21–24). We have recently focused on Drosophila melanogaster (fruit flies). Drosophila has a relatively simple central nervous system and lacks an endogenous α-synuclein gene or α-synuclein gene ortholog (12); however, Feany and Bender (15) have shown that flies expressing both wild-type and mutated (A30P or A53T) human α-synuclein genes display symptoms that resemble those found in human PD patients. That is, the flies are viable but gradually exhibit progressive degeneration of dopaminergic neurons, formation of LB-like inclusions, and impairment of climbing ability. In addition, this organism has a relatively short life cycle, making it possible to carry out many studies with relative rapidity, and extensive studies at the level of the genome and proteome have been reported (25–33). Thus, the Drosophila model appears to be a useful tool for addressing issues associated with the underlying molecular mechanisms of α-synuclein-mediated neurotoxicity in PD.
In closely related work, we recently reported a proteome analysis of an A30P α-synuclein Drosophila model of PD at three different disease stages and a comparison of changes in gene expression profiles at the proteome and transcriptome levels (29). The studies indicated that as early as day 1 (presymptomatic stage) actin cytoskeletal and mitochondrial proteins were perturbed. In addition, our proteomics studies covering seven different time points across the adult life span implied that dysregulated proteins appear to be age- or disease stage-specific (30). This suggests that although analyses covering different disease stages help to elucidate disease progression, studies at the presymptomatic stage appear to be crucial for developing diagnostic tools, determining causation of disease, and intervening in neuron degeneration. Because Drosophila expressing A53T α-synuclein develop similar human PD-like symptoms as Drosophila expressing A30P α-synuclein (15), a proteomics study of the A53T mutant α-synuclein flies is expected to provide a more general (and possibly more comprehensive) view of protein changes that may be responsible for PD. Thus, in the present study we performed a quantitative proteomics analysis of an A53T α-synuclein Drosophila model of PD at the presymptomatic stage. This study also used a different experimental approach, an isotopic labeling strategy utilizing global internal standard technology (GIST) (23, 34–36) in combination with multidimensional LC coupled to MS/MS for protein identification and quantification.
EXPERIMENTAL PROCEDURES
Drosophila Stocks and Harvesting—
This study utilized the following control and PD-generating fly genotypes: elav::Gal4 (Pw+mW.hs = GawBelavC155, Bloomington Stock Center, Indiana University) and UAS-A53T α-synuclein (the P(UAS-HsapSNCA.A53T)15.3 line was obtained from Mel Feany, Harvard Medical School), respectively. To obtain the elav::Gal4⇒UAS::A53T α-synuclein experimental flies (PD-like), virgin females from the elav::Gal4 line were crossed to males from the UAS-A53T α-synuclein stock. Control and PD-like flies were cultured on standard cornmeal medium, maintained at identical conditions (25 ± 1 °C), and harvested at the same time. To avoid differences that arise from gender, only male flies were used. A population of 250 adult fly heads was collected for each genotype at day 1 (within 24 h) posteclosion for protein extraction. Fly heads were collected and stored as described previously (29). A new batch of flies was raised for an independent biological replicate measurement.
Fly Head Sample Preparation—
Fly samples were prepared as described elsewhere (29) with minor modifications. Briefly, fly head proteins were extracted using a mortar and electric pestle in a 0.2 m phosphate buffer saline solution (pH 7.0) containing 8.0 m urea and 0.1 mm phenylmethylsulfonyl fluoride. After centrifugation at 13,000 rpm for 10 min the supernatant was collected. A Bradford assay (Pierce) indicated that ∼2.5 mg of proteins were obtained from 250 adult fly heads. Three standard proteins (i.e. human hemoglobin (purity unavailable), human albumin (96–99%), and horse heart myoglobin (≥90%) (Sigma-Aldrich)) were each spiked into equal amounts of control and PD-like fly samples at known concentration ratios of 2:1, 1:1, and 1:2, respectively. Protein mixtures were reduced, alkylated, and tryptically digested as described previously (29). Finally, tryptic peptides were cleaned, dried, and stored at −80 °C until further analysis.
Synthesis of N-Acetoxysuccinimide and N-Acetoxy-d3-succinimide—
N-Hydroxysuccinimide (Sigma-Aldrich), acetic anhydride (>99% purity, Sigma-Aldrich), and acetic anhydride-d6 (>99% purity, Sigma-Aldrich) were used as purchased. N-Acetoxy-d0-succinimide (light) and N-acetoxy-d3-succinimide (heavy) were synthesized as described previously (34). Briefly, 8 g of N-hydroxysuccinimide was mixed with either 19.8 ml of acetic anhydride or 18.6 ml of acetic anhydride-d6. Mixtures were stirred under nitrogen at room temperature for 15 h. White crystal products were washed, filtered with hexane, dried under nitrogen, and stored at −20 °C for future use. Nuclear magnetic resonance analysis indicated 100% purity of the synthesized products (1H chemical shift, single peak at 2.915 ppm for N-acetoxy-d3-succinimide; two peaks at 2.907 and 2.361 ppm for N-hydroxy-d0-succinimide).
Differential Isotopic Labeling of Tryptic Peptides—
An equal amount of control and PD-like fly tryptic peptides was resuspended in phosphate buffer (pH 7.5) to a 1 mg·ml−1 solution. The isotopic labeling reactions were processed as described by Regnier and co-worker (34). Briefly, a 100-fold molar excess of N-acetoxy-d0-succinimide and N-acetoxy-d3-succinimide was added to control or PD-like samples (In one experiment, the control sample was light labeled, and the PD-like sample was heavy labeled; we refer to this as forward labeling. In a second experiment with an independent biological replicate, the control sample was heavy labeled, and the PD-like sample was light labeled; we refer to this as reverse labeling.). The reactions proceeded for 5 h with constant stirring at room temperature. At the end of the reaction, two equal aliquots of samples were combined and treated with an excess amount of N-hydroxylamine at pH 10–11 for 20 min. Finally, the isotopically labeled peptide mixture was adjusted to pH 7, cleaned, dried, and stored at −80 °C until further analysis.
Strong Cation Exchange (SCX) Chromatography—
Differentially labeled tryptic peptides were dissolved in 5.0 mm potassium phosphate buffer in 75:25 water:acetonitrile at pH 3.0. The peptide solution was injected onto a javelin guard column (10 × 2.1 mm) that preceded a polysulfoethyl aspartamide column (100 × 2.1 mm, 5 μm, 200 Å; PolyLC Inc., Southboro, MA). The gradient was delivered at a flow rate of 0.2 ml·min−1 by a Waters 600 multisolvent delivery system, and peptides were detected at 214 nm by a Waters 2487 dual λ absorbance detector. Mobile phases consisted of 5 mm potassium phosphate in 75:25 water:acetonitrile at pH 3.0 (solvent A) and solvent A with the addition of 350 mm potassium chloride (designated as solvent B). Binary gradients with respect to the percentage of solvent B were as follows: 0–5 min, 0%; 5–45 min, 0–40%; 45–90 min, 40–80%; 90–100 min, 80–100%; 100–110 min, 100%; 110–111 min, 100–0%; and 111–121 min, 0%. One-minute collections into 96-well plates (Corning Inc., Corning, NY) over the 121-min gradient were combined into six fractions as follows: 1) 0–34 min, 2) 34–40 min, 3) 40–44 min, 4) 44–50 min, 5) 50–58 min, and 6) 58–121 min. Pooled fractions were desalted, dried, and stored at −80 °C until further analysis.
Reversed Phase LC-MS/MS Analysis—
Reversed phase LC-MS/MS analysis was performed on an LTQ-FT hybrid linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (ThermoElectron, San Jose, CA) equipped with a Finnigan Nanospray II electrospray ionization source (ThermoElectron) and an UltiMate 3000 HPLC system (Dionex Corp., Sunnyvale, CA). Peptide mixtures were separated on a self-pack PicoFrit column (75-μm inner diameter; New Objective, Woburn, MA) packed with Magic C18AQ (5 μm, 100 Å; Michrom Bioresources Inc., Auburn, CA) in a methanol slurry. A column length of 13.2 cm was used for peptide separation. A sample volume of 6.4 μl was loaded onto a 300-μm-inner diameter × 5-mm precolumn cartridge (packed with C18 PreMap 100, 5 μm, 100 Å; LC Packings, a Dionex Corp. company) at a flow rate of 10 μl·min−1. Mobile phases of 96.95:2.95:0.1 water:acetonitrile:formic acid (solvent A) and 99.9:0.1 acetonitrile:formic acid (solvent B) were delivered by an Ultimate micropump (Dionex Corp.) at a flow rate of 250 nl·min−1. The gradient with respect to the percentage of solvent B was as follows: 0–15 min, 10%; 15–105 min, 10–30%; 105–135 min, 30–60%; 135–140 min, 60–80%; 140–150 min, 80%; 150–151 min, 80–10%; and 151–165 min, 10%.
Eluted peptides were analyzed using a hybrid LTQ-FT mass spectrometer. The instrument was operated to acquire a full FT-MS scan (m/z range of 300–2000) followed by MS/MS scans of the top three most intense ions in the LTQ with data-dependent function enabled. The resolution was set to 100,000 (at m/z 400) for the survey FT-MS scan. The data-dependent acquisition uses a 30-s exclusion duration time and a collision energy of 35%.
Data Analysis—
Raw MS/MS spectra (.RAW) were processed using the Xcalibur software package Bioworks (version 3.3.1, ThermoElectron) to create a peak list (.DTA) with 50-ppm precursor ion tolerance. Individual .DTA files were grouped according to charge states of precursor ions using an in-house written algorithm. Charge-sorted .DTA files were submitted to MASCOT (Matrix Science, Version 2.1) and searched against the National Center for Biotechnology Information nonredundant (NCBInr) Drosophila protein database (28,642 sequence entries) that is correlated with FBgn accession numbers in the FlyBase database for Drosophila protein identification. The same files were also searched against a home-built database containing protein sequences of human albumin, hemoglobin, and horse heart myoglobin for standard protein assignment. Carbamidomethylation of cysteine residues was used as a fixed modification. Variable modifications included acetylation (light or heavy) of lysine residues and the N termini of peptides. Additional parameters included a maximum of two trypsin miscleavages, a precursor tolerance of 15 ppm, and an MS/MS tolerance of 0.8 Da. Spectra that led to scores at or above the MASCOT-assigned homology score (the homology score defines a spectral match at a 95% confidence level) were assigned to specific peptide sequences; peptide sequences matching to multiple FBgn accession numbers were discarded (i.e. only peptides having sequences that were unique to a single protein were considered). To estimate the false positive rate of peptide identifications, we searched one of the most peptide-rich fractions (SCX fraction 3) against the reverse NCBInr Drosophila database (28,642 sequence entries) and the FlyBase database using the same parameters described above; the false positive rate calculated according to the formula by Gygi and co-workers (37) was 2.2% for doubly charged peptides and 3.4% for triply charged peptides.
Abundance ratios of differentially labeled peptides were computed using peak intensities from extracted ion chromatograms of light or heavy labeled peptides with an in-house written algorithm as described previously by our laboratory (38). For peptides identified in multiple SCX fractions or charge states, the intensities of peaks were summed to obtain abundance ratios. Relative quantification of proteins was obtained by averaging the intensity ratios of multiple derived peptides. Only proteins with a minimum of two peptide sequence identifications from the two independent experiments were considered for quantification (this requirement is expected to substantially lower the false positive rate associated with protein identification using the MASCOT algorithm). Abundance ratios for proteins reported as differentially expressed in this study were confirmed by manual inspection. The criteria for passing the manual inspection were as follows: (i) signal-to-noise ratios of both light and heavy labeled peptide pairs ≥10, (ii) extracted ion chromatogram must show approximate Gaussian shape as well as no coeluting ions, and (iii) light and heavy labeled peptide ion spectra must show approximate theoretically predicted isotope patterns and expected mass shift between doublet clusters.
Western Blot Analysis—
For Western blot analysis, a new batch of male control and A53T α-synuclein flies was raised and harvested to obtain a total of 250 adult fly heads each (an independent batch of flies was utilized for Western blot analysis of individual proteins in this study, i.e. troponin T and fat body protein 1). Proteins were extracted (as described above) in 400 μl of ice-cold 50 mm Tris buffer (pH 7.5) containing 10 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 50 mm dithiothreitol, and 4% CHAPS. Protein concentrations were determined with a Bradford assay (Pierce). Equal amounts of proteins were separated by SDS-PAGE with 8% polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes. Membranes were blocked overnight at 4 °C in Odyssey blocking buffer (LI-COR, Lincoln, NE) and simultaneously incubated with anti-β-tubulin antibody (1:2500 dilution; Developmental Studies Hybridoma Bank, Iowa City, IA) and anti-troponin T antibody (1:500 dilution; Babraham Bioscience Technologies, Cambridge, UK) or anti-fat body protein 1 polyclonal antibody (1:500 dilution; kindly provided by Qisheng Song) for 2 h at room temperature with gentle shaking. Next, infrared dye 800-conjugated affinity-purified goat anti-rat IgG (Rockland Inc., Gilbertsville, PA) and infrared dye 700-labeled goat anti-mouse IgG (LI-COR) or anti-rabbit IgG (LI-COR) secondary antibodies were added and allowed to incubate for 2 h at room temperature. Immunoblotting bands were detected and quantified utilizing the LI-COR Odyssey infrared imaging system (simultaneous two-color targeted analysis) and software (LI-COR).
RESULTS
Quantification of Internal Standard Proteins—
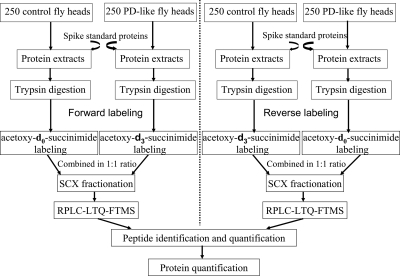
The present study used the GIST coupled to LC-MS/MS approach for protein identification and quantification (Fig. 1). Standard proteins were spiked at an early stage of sample preparation and at known concentration ratios into equal amounts of control and PD-like fly proteins to assess the variability that arises from sample handling and preparation. Human hemoglobin, human albumin, and horse heart myoglobin were chosen as internal standards because BLASTP searches indicate that there are no identical or homologous tryptic peptide sequences in the Drosophila NCBInr genome database. The spiked ratios for hemoglobin, albumin, and myoglobin were 2:1 (down-regulated), 1:1 (not regulated), and 1:2 (up-regulated), respectively, in PD-like samples compared with controls. In the forward labeling measurement, a total of 10, three, and seven peptides were quantified for hemoglobin, albumin, and myoglobin, respectively. The observed average ratio was 0.45 ± 0.07, 0.99 ± 0.16, and 2.35 ± 0.35, respectively, as given in Table I. In the reverse labeling measurement, nine, three, and six peptides were detected for hemoglobin, albumin, and myoglobin with an observed average ratio of 0.45 ± 0.09, 1.31 ± 0.14, and 2.69 ± 0.47, respectively (Table I). Based on these results from internal standard proteins, we expect that this experimental approach accurately yields relative protein amounts in PD-like and control fly samples.
Fig. 1.
Experimental design for quantitative proteomics analysis. Standard proteins spiked into control and A53T α-synuclein-expressing (PD-like) fly samples are human hemoglobin (2:1), human albumin (1:1), and horse heart myoglobin (1:2). Two independent experiments were carried out by isotopically labeling biological replicate samples in forward and reverse directions. RP, reversed phase.
Table I.
Quantification of internal standard proteins spiked into PD-like and control fly samples
| Protein name | Expected ratio (PD-like/control) | Forward labelinga
|
Reverse labelinga
|
||
|---|---|---|---|---|---|
| Observed ratio ± S.D. (PD-like/control) | Error | Observed ratio ± S.D. (PD-like/control) | Error | ||
| % | % | ||||
| Human hemoglobin | 0.50 | 0.45 ± 0.07 (10) | −9.1 | 0.45 ± 0.09 (9) | −10.6 |
| Human albumin | 1.00 | 0.99 ± 0.16 (3) | −1.0 | 1.31 ± 0.14 (3) | 31.3 |
| Horse heart myoglobin | 2.00 | 2.35 ± 0.35 (7) | 17.5 | 2.69 ± 0.47 (6) | 34.4 |
The numbers in parentheses indicate the total number of peptides used for protein quantification. The observed ratio ± S.D. indicates the average ratio and standard deviation of multiple peptides detected for a protein.
Quantitative Proteomics of a Drosophila Model of PD—
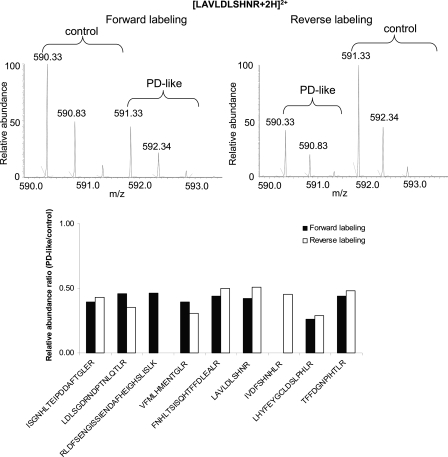
To increase the confidence of protein quantification, we used forward and reverse labeling with two independent biological sets of animals. Fig. 2 shows an example of mass spectra of light and heavy labeled peptide pairs from the forward and reverse labeling measurements of the protein chaoptin (encoded by chaoptic). From the relative intensities of the example peptide observed as [LAVLDLSHNR + 2H]2+ ion, it is apparent that the peptide shows down-regulation in PD-like flies in both the forward and reverse labeling measurements (Fig. 2, top). A total of nine peptides were identified and quantified for chaoptin, comprising ∼11% sequence coverage. Seven of the nine peptides were in common between the forward and reverse labeling experiments, and all nine exhibited down-regulation in PD-like flies in comparison with controls (Fig. 2, bottom), indicating down-regulation of the protein.
Fig. 2.
Example mass spectra for light and heavy labeled peptide (LAVLDLSHNR) pairs from forward and reverse labeling measurements for the protein encoded by chaoptic(top) and column representation of identification and quantification of all nine identified peptides (bottom).
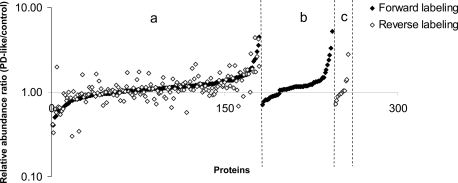
Although a single peptide can be utilized for the quantification of proteins (39), in the present study we required a minimum of two (arbitrarily chosen) peptides for a protein to be considered for quantification from the forward and reverse labeling experiments. In total, 253 proteins (supplemental Table SI) meet this criterion, among which 180 proteins (71%) were quantified in both forward and reverse labeling measurements (Fig. 3). A total of 61 and 12 proteins were uniquely quantified with multiple peptides from the forward and reverse labeling experiments, respectively. For proteins quantified in both forward and reverse labeling experiments, similar -fold-changes were obtained for the majority of the proteins (i.e. 157 of 180 (87%) proteins had relative S.D. values ≤25% between the two experiments). A linear regression analysis of the relative abundance ratios commonly obtained from the two experiments gave a Pearson product moment correlation coefficient of 0.69, and the p value from a Student's t test (two-tailed distribution and two-sample unequal variance) was 0.84 (p ≫ 0.05). These analyses indicate that the two independent experiments were positively correlated and did not differ significantly. However, there were five proteins that differed by a relative S.D. ≥ 50% between the two measurements. Ribosomal protein S3A (FBgn0017545) exhibited fold changes in opposite directions. We speculate that this inconsistency typically arises from the use of different peptides that are identified in the separate experiments that may have varying ionization efficiencies. For example, the ribosomal protein S3A-derived peptide ion [LLELHGDGGGK + 2H]2+ identified only in the forward labeling experiment showed a 0.59-fold change, whereas the peptide ion [IYPLHDVYIR + 2H]2+ identified only in the reverse labeling experiment exhibited a 1.98-fold change. Similar discrepancies in peptide fold-change values have also been observed using an isotope-coded affinity tag approach (40). As discussed previously by others, poor agreement in relative abundance of different peptides from the same protein may also arise due to post-translational modifications (39).
Fig. 3.
Relative quantification of the 253 proteins with a minimum of two peptide sequences from two independent biological experiments on a logarithm scale. Proteins were classified into three groups: a, proteins (i.e. 180) identified in both forward and reverse labeling experiments; b, proteins (i.e. 61) that were uniquely quantified in the forward labeling experiments; and c, proteins (i.e. 12) that were uniquely quantified in the reverse labeling experiments. Each group of proteins was sorted by relative abundance ratios in ascending order.
Similar to many other quantitative proteomics analyses (39–42), the majority of proteins identified in the present study did not change in PD-like flies compared with controls (Fig. 3). Currently, various criteria have been utilized to determine differential expression of proteins based on data quality (39–43). For instance, Griffin et al. (43) applied a 1.5-fold difference criterion from the ICAT method as a threshold indicating significant change, and Chiang et al. (40) considered a protein with a calibrated ratio (ICAT analyses calibrated by Western blot analysis) of either ≥1.23 or ≤0.77 to be differentially expressed. The present study utilized three proteins as internal standards at different concentration ratios at an early stage of sample preparation to estimate experimental variability, e.g. isotopic labeling reagents and sample handling variability. On the basis of the quantitative results of internal standard proteins (discrepancies of −10.6 to 34.4% between the measured and expected values), we considered a 1.5-fold expression difference as a suitable threshold for significant changes (We note that individual peptides from the same protein can differ drastically, causing decent standard deviation among the measured peptide ratios. A protein is considered differentially expressed if it exhibits ≥1.5-fold change from multiple peptides in both forward and reverse labeling experiments despite large standard deviations among different peptides.). Proteins that exhibited ≥1.5-fold changes in only one of the experiments were discarded (these include proteins that did not show change or were not identified in the other experiment) to eliminate suspicious protein candidates. A total of 24 proteins were differentially expressed; they are listed in Table II. Among the dysregulated proteins, nine proteins were down-regulated that are encoded by fat body protein 1, chaoptic, retinin, odorant-binding protein 99b, retinal degeneration A, karst, Rtnl1, calnexin 99A, and cytochrome P450 reductase; the remaining 15 proteins that were up-regulated in PD-like flies are encoded by paramyosin, upheld, ribosomal protein L6, glycerol-3-phosphate dehydrogenase, muscle LIM protein at 60A, ATP synthase-β, superoxide dismutase 2 (manganese), failed axon connections, ribosomal protein L14, ribosomal protein S8, heat shock protein cognate 3, ribosomal protein L23A, and three unnamed genes, i.e. CG32029, CG7675, and CG15006. The proteins encoded by chaoptic, upheld, paramyosin, and ATP synthase β subunit were quantified with a minimum of six peptides from each independent measurement (a minimum of nine peptides from two biological experiments). An increased number of peptides that are used for quantification should increase the reliability of determining protein abundance changes.
Table II.
Differentially expressed proteins in A53T α-synuclein flies relative to controls
| Gene name or IDa | FlyBase IDa | Observed ratio ± S.D. (PD-like/control)
|
Total no. peptides | |
|---|---|---|---|---|
| Forward labelingb | Reverse labelingb | |||
| Fat body protein 1 (Fbp1) | FBgn0000639 | 0.41 ± 0.12 (3) | 0.33 ± 0.08 (3) | 4 |
| chaoptic (chp) | FBgn0000313 | 0.41 ± 0.07 (8) | 0.42 ± 0.09 (8) | 9 |
| karst (kst) | FBgn0004167 | 0.51 ± 0.15 (2) | 0.63 (1) | 3 |
| Odorant-binding protein 99b (Obp99b) | FBgn0039685 | 0.53 ± 0.02 (2) | 0.57 ± 0.07 (2) | 2 |
| Cytochrome P450 reductase (Cpr) | FBgn0015623 | 0.60 ± 0.01 (2) | 0.67 (1) | 2 |
| Retinin | FBgn0040074 | 0.61 ± 0.03 (2) | 0.54 (1) | 2 |
| Calnexin 99A (Cnx99A) | FBgn0015622 | 0.61 ± 0.30 (2) | 0.66 (1) | 2 |
| Retinal degeneration A (rdgA) | FBgn0003217 | 0.64 ± 0.03 (2) | 0.60 (1) | 2 |
| Rtnl1 | FBgn0053113 | 0.65 ± 0.08 (4) | 0.64 ± 0.28 (3) | 4 |
| Muscle LIM protein at 60A (Mlp60A) | FBgn0011643 | 1.53 ± 0.11 (2) | 1.72 ± 0.08 (2) | 2 |
| Glycerol-3-phosphate dehydrogenase (Gpdh) | FBgn0001128 | 1.56 ± 0.25 (3) | 1.70 ± 0.26 (3) | 3 |
| Paramyosin (Prm) | FBgn0003149 | 1.57 ± 0.26 (21) | 1.60 ± 0.38 (16) | 23 |
| Superoxide dismutase 2 (manganese) (Sod2) | FBgn0010213 | 1.60 ± 0.26 (2) | 1.84 ± 0.23 (2) | 2 |
| Ribosomal protein L6 (RpL6) | FBgn0039857 | 1.61 ± 0.92 (2) | 1.63 ± 0.76 (2) | 2 |
| Failed axon connections (fax) | FBgn0014163 | 1.66 ± 0.89 (2) | 1.84 ± 1.57 (3) | 3 |
| upheld (up) | FBgn0004169 | 1.82 ± 0.22 (6) | 1.63 ± 0.21 (7) | 9 |
| Heat shock protein cognate 3 (Hsc70-3) | FBgn0001218 | 1.91 ± 1.60 (5) | 2.84 ± 2.72 (3) | 5 |
| Ribosomal protein L14 (RpL14) | FBgn0017579 | 1.94 ± 1.18 (2) | 1.87 ± 1.15 (2) | 3 |
| ATP synthase-β (ATPsyn-β) | FBgn0010217 | 2.30 ± 2.56 (9) | 1.72 ± 1.28 (14) | 15 |
| CG15006 | FBgn0035510 | 2.45 ± 0.05 (2) | 4.42 (1) | 3 |
| CG32029 | FBgn0052029 | 2.52 ± 2.06 (2) | 1.72 ± 0.79 (3) | 3 |
| Ribosomal protein S8 (RpS8) | FBgn0039713 | 3.04 ± 3.26 (3) | 2.64 ± 2.38 (3) | 3 |
| Ribosomal protein L23A (RpL23A) | FBgn0026372 | 3.56 ± 0.52 (2) | 4.21 ± 0.56 (2) | 2 |
| CG7675 | FBgn0038610 | 4.52 (1) | 2.01 ± 0.09 (2) | 2 |
Gene name or IDs and FlyBase IDs were obtained from the FlyBase database. Gene symbols are denoted in parentheses to facilitate understanding of Figs. 7 and 8.
The numbers in parentheses indicate the total number of peptides used for protein quantification. The observed ratio ± S.D. indicates the average ratio and standard deviation of multiple peptides detected for a protein. Detailed information on peptide identification is in supplemental Table SI.
Validation of Protein Expression by Western Blot Analysis—
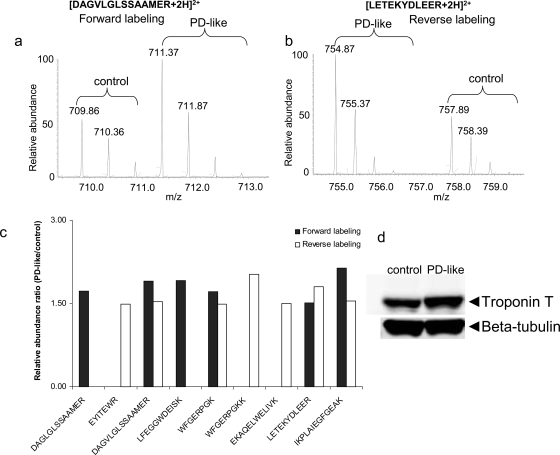
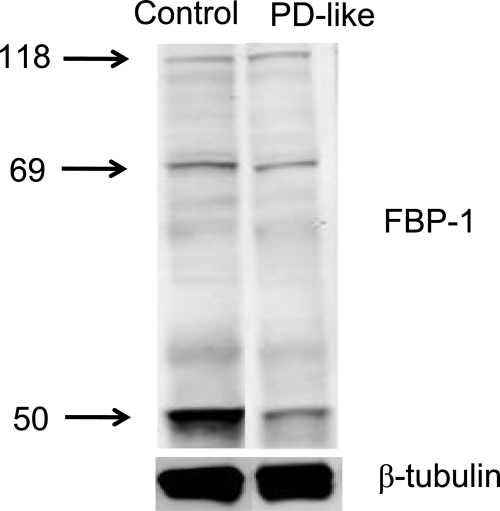
To validate protein changes observed in the present study, we considered Western blot analysis. For instance, Drosophila troponin T (encoded by upheld) was identified with a total of nine peptides from the GIST-coupled LC-MS/MS approach. Example mass spectra for [DAGLGLSSAAMER + 2H]2+ and [LETEKYDLEER + 2H]2+ peptides belonging to troponin T show up-regulation in PD-like flies based on the relative intensities of peaks (Fig. 4, a and b). Seven other peptides identified also display up-regulation in PD-like flies compared with controls (Fig. 4c), suggesting up-regulation of the protein (i.e. 1.82 ± 0.22 from the forward labeling experiment and 1.63 ± 0.21 from the reverse labeling measurement). Consistent with these changes, Western blot analysis also showed up-regulation of the protein (Fig. 4d, top). In the present study, expression of β-tubulin (multiple β-tubulin isoforms) did not change in PD-like flies in comparison with controls (i.e. 0.97 ± 0.28 from the forward labeling experiment and 0.96 ± 0.07 from the reverse labeling experiment) and thus was utilized as a loading control (Fig. 4d, bottom). With normalization of the integrated intensities of the immunoblotting bands of troponin T to β-tubulin, a 1.51-fold up-regulation of troponin T was obtained in PD-like flies with a 13.2 and 5.4% relative S.D. from the forward and reverse labeling measurements, respectively. Another example is fat body protein 1, which exhibits down-regulation in PD-like flies from the proteomics approach (Table II). Western blot analysis detected three main bands (Fig. 5) that correspond to the FBP-1 protein (118 kDa) and two FBP-1 fragments (69 and 50 kDa); this is consistent with previous reports (44,45). Western blot analysis indicated that the native FBP-1 protein levels do not exhibit substantial differences between PD-like flies and controls. However, the two FBP-1 fragments were at lower levels; there was a 0.64- and 0.37-fold down-regulation of these fragments at 69 and 50 kDa, respectively. Overall, there was a 0.43-fold down-regulation for the sum of the three detected bands in PD-like flies in accordance with our proteomics results (i.e. an average of 0.37-fold down-regulation from the forward and reverse labeling experiments). Thus, these Western blot analyses provide evidence, in addition to the internal standards, that the quantitative results obtained with the GIST approach are sufficiently accurate.
Fig. 4.
Data from peptides belonging to troponin T. a, example mass spectra for light and heavy labeled peptide (DAGVLGLSSAAMER) pairs from the forward labeling measurement. b, example mass spectra for light and heavy labeled peptide (LETEKYDLEER) pairs from the reverse labeling measurement. c, column representation of identification and quantification of all nine identified peptides. d, Western blot analysis of troponin T with β-tubulin as the loading control.
Fig. 5.
Western blot analysis of FBP-1 using polyclonal antibodies (top) with β-tubulin (bottom) as the loading control. Three bands corresponding to the FBP-1 protein (118 kDa) and two FBP-1 fragments (69 and 50 kDa) were detected.
DISCUSSION
Overview—
The results presented above describe a quantitative proteome analysis of an A53T α-synuclein Drosophila model of PD utilizing forward and reverse isotopic labeling coupled with an LC-MS/MS approach. Two independent biological replicates (PD model and control animals) were used. Quantification was aided by additional incorporation of internal standards, i.e. tryptic peptides from proteins at defined concentration ratios. These internal standards allowed us to assess the experimental variability associated with isotopic labeling reagents, such as preferential labeling, inefficient labeling, and variation in ionization efficiency, sample handling, and preparation. The percent errors obtained in our study ranged from −10.6 to 34.4%. This provides confidence that a 1.5-fold difference in abundance of light and heavy labeled peptide pairs is a suitable criterion to determine that a protein is significantly differentially expressed.
A total of 253 proteins were quantified with a minimum of two peptides. Of these, 24 proteins (Table II) met our criteria for differential expression in the PD-like flies relative to the controls (We note that the relatively high percentage (i.e. 10%) of dysregulated proteins is closely associated with a relatively small number of proteins (i.e. 253) that were considered for quantification (described in detail below), and this percentage may not be applicable to the remaining of the fly proteome. Thus, we caution that it may be inappropriate to deduce that a global disruption of the proteome was induced at the presymptomatic stage in the PD-like animals from these data.). Up-regulation of troponin T and down-regulation of fat body protein 1 were confirmed by Western blot analysis, providing confidence of our experimental design. Collectively, we expect that the present quantitative proteomics approach provides valuable information about protein alterations that appear to be associated with PD at the presymptomatic stage.
The relatively low proteome coverage found in the present study primarily arises from several factors. First, high abundance proteins (e.g. myosin and actin) were not removed during sample preparation. These proteins may mask detection of many low abundance proteins, e.g. myosin heavy chain contributes to ∼6% of the total identified peptides (see supplemental Table SI). Second, the isotopic labeling approach used in the study acetylates the primary amines (N termini and lysine residues), removing positive charge and thus making the lysine-containing peptides harder to detect. Third, most of the proteins (i.e. 572) identified in our study were assigned with a single peptide; however, we required a minimum of two peptides for quantification to be more confident in changes. Other factors that may be associated with low proteome coverage include database searching parameters used (e.g. a stringent precursor mass tolerance (15 ppm) was utilized, and variable protein post-translational modifications were not considered when running MASCOT searches); thus, this approach may miss some protein identifications. Overall, the 253 proteins assigned correspond to abundant proteins in flies based on our data-dependent acquisition method (i.e. generally, the number of peptides detected positively correlate with the protein). The cellular compartments associated with the 253 proteins can be obtained from Gene Ontology analysis (46); however, these are not described in this study. Rather we focused our attention on the differentially expressed proteins, and the cellular compartments of the 24 proteins are presented in Fig. 6, which shows that five have no available cellular component annotation, four are ribosomal proteins, four are membrane proteins, four are from the endoplasmic reticulum (ER), three are actin cytoskeletal proteins, two are mitochondria-associated proteins, and the last two are assigned to the extracellular and cytoplasmic compartments. Below we provide a brief discussion of the relevance of individual proteins.
Fig. 6.
Pie chart representation of cellular localization for the 24 dysregulated proteins in A53T α-synuclein flies in comparison with controls. A protein was grouped into a single well known category if it was associated with several cellular components.
Membrane Proteins—
We identified four membrane-associated proteins whose expression was perturbed in A53T α-synuclein-expressing flies in comparison with controls. Chaoptin, a glycoprotein that localizes to the extracellular surface of the plasma membrane by covalent interaction with glycosylphosphatidylinositol, plays a key role in photoreceptor cell morphogenesis by serving as a photoreceptor-specific cell adhesion molecule (47–49). Drosophila chaoptin mutants display remarkable membrane abnormalities (49). The protein encoded by retinal degeneration A (rdgA) is associated with the transport of phospholipids to the photoreceptive membranes, and the Drosophila rdgA mutant induces degeneration in photoreceptor cells within a week posteclosion (50). Down-regulation of both chaoptic and rdgA at the protein level suggests disruption of the structural integrity of neuronal cell membranes that could lead to failure in cell communications and subsequent malfunction of cellular processes. This hypothesis appears to be supported by a previous report that suggests that α-synuclein plays a role in membrane transport and synaptic membrane biogenesis, and A53T mutant α-synuclein likely disrupts the plasma lipid bilayers (51). Because the plasma membrane serves as the attachment point for intracellular cytoskeletal structure, dysregulation of membrane-associated proteins could be linked to cytoskeletal defects.
Cytoskeletal Proteins—
We observed three actin cytoskeleton-associated proteins that were disturbed. Troponin T, encoded by upheld, is a member of the actin filament-associated troponin complex (52). Drosophila troponin T has sequences similar to vertebrate troponin T (53), and the importance of troponin T has been demonstrated in both vertebrates and invertebrates (52–54). For example, mutations in cardiac troponin T causes familial hypertrophic cardiomyopathy in humans (54); similarly, mutations in Drosophila troponin T induce muscle abnormalities (53). Muscle LIM protein at 60A (Mlp60A) was up-regulated in A53T α-synuclein-expressing flies relative to controls (Table II). Mlp60A is a member of the cysteine-rich protein family that plays pivotal roles in cell differentiation (55). Mlp60A is associated with the actin cytoskeleton and plays a role in myogenic differentiation (56,57). Arber et al. (57) found that muscle LIM protein was strongly up-regulated in denervated (or paralyzed skeletal muscle) in adult rats; substantially high levels of muscle LIM protein appear to be toxic or to interfere with cell differentiation. Thus, up-regulation of Mlp60A at the presymptomatic stage in PD-like flies appears to corroborate the notion that expression of A53T α-synuclein may be deleterious to actin cytoskeletal muscles. In addition to up-regulation of troponin T and Mlp60A, the thick filament structure protein paramyosin was also up-regulated in A53T α-synuclein flies. The observation of functional perturbations in the actin cytoskeleton agrees with our previous finding in an A30P α-synuclein Drosophila model of PD (29,30). Collectively, our studies suggest that actin cytoskeletal defects may play a role in the onset of PD-like symptoms in flies.
ER Proteins—
Of the four proteins associated with ER, the change in heat shock protein cognate 3 (Hsc3p) is especially intriguing. Hsc3p is a member of the Hsp70 family, a class of molecular chaperones that are associated with a diversity of important cellular activities, such as protein folding and protein degradation (58). Increased levels of Hsc3p alleviate fly lethality caused by D231S mutant Hsc3p (59). Overexpression of Hsp70 has been found to rescue α-synuclein-medicated toxicity in Drosophila (60). We hypothesize that increased Hsc3p levels in A53T α-synuclein-expressing flies may be an indication that cells are encountering an unfavorable environment (e.g. increased concentrations of misfolded proteins). Because Hsc3p is the only ER member in the Hsp70 family (59), overexpression of the ER Hsp70 as a result of expression of A53T α-synuclein may induce ER stress (in contrast, HSC4 protein, the most abundant cytoplasmic Hsp70 member, did not exhibit change in A53T α-synuclein flies, i.e. 1.07 ± 0.07 from the forward labeling experiment and 1.14 ± 0.19 from the reverse labeling experiment). It is worth noting that increased ER stress was also observed in rat PC12 lines as a consequence of induction of A53T α-synuclein (61). The remaining three ER proteins (encoded by Rtnl1, calnexin 99A, and cytochrome P450 reductase) are involved in proton and intracellular protein transport (62) and are down-regulated in PD-like flies.
Mitochondrial Proteins—
Two key mitochondrial proteins, manganese-superoxide dismutase (Mn-SOD) and ATP synthase β subunit (β-ATPase), were observed at higher levels in A53T α-synuclein flies. The Mn-SOD enzyme is an antioxidant that catalyzes the conversion of reactive oxygen species generated by mitochondrial superoxide to hydrogen peroxide (63) and thus is implicated in cellular defense (preventing oxidative damage). Overexpression of Mn-SOD attenuates 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-mediated toxicity (64) and protects against adriamycin-induced cardiomyopathy in transgenic mice (65). Yeast Flp recombinase-mediated overexpression of Mn-SOD extends life span in Drosophila (63). Increased levels of Mn-SOD in A53T α-synuclein flies suggest that mitochondria may encounter increased oxidative stress. Additionally, mitochondrial disruption can also be seen from the up-regulation of the nuclearly encoded mitochondrial β-ATPase, which plays an essential role in the function of the enzyme ATP synthase (66). This protein was also up-regulated in A30P α-synuclein flies at early disease stage (29), and ATP synthase D was up-regulated in human substantia nigra of PD patients compared with controls (67). From these studies, it seems reasonable to hypothesize that alterations in ATP synthase may be responsible for the malfunction of mitochondria that is widely accepted as a pathological feature of human PD (5).
Ribosomal Proteins—
Four ribosomal proteins were up-regulated in A53T α-synuclein flies: ribosomal proteins L6, L14, L23A, and S8. These ribosomal proteins are structural components of the ribosome, the cellular machinery that synthesizes proteins. Abundance changes in structural constituents of the ribosome suggest that cellular metabolism may be disturbed due to the expression of A53T α-synuclein. Elevated expressions of the ribosomal proteins in the A53T α-synuclein flies may be associated with perturbed abundances of other proteins.
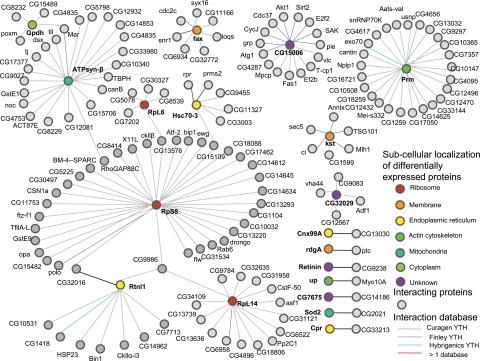
To test the hypothesis that the dysregulated proteins may be connected by intermolecular interactions, we performed a Drosophila protein interaction network analysis using a Web-based tool, Interaction Map Browser, developed by Finley and co-workers (68). By searching the 24 dysregulated proteins against a comprehensive Drosophila protein interaction database combined from the Finley laboratory (69), Curagen (70), and Hybrigenics (71), we found that five proteins have no available protein interactions, seven proteins have one single interaction, and the remaining 12 proteins have multiple interactions (Fig. 7). The top three most abundant interacting proteins are ribosomal protein S8 (35 interactions), paramyosin (25 interactions), and ATP synthase β subunit (22 interactions). A detailed view of the protein interaction network analysis map is shown in Fig. 8. It is interesting to mention that of the 12 proteins that have multiple interactions seven form a single interaction network bridged by ribosomal protein S8 (Fig. 8), including six up-regulated proteins (glycerol-3-phosphate dehydrogenase, β-ATPase, ribosomal protein L6, ribosomal protein S8, Hsc3p, and ribosomal protein L14) and one down-regulated protein encoded by Rtnl1. These potential protein associations may provide insights into molecular pathways underlying PD.
Fig. 7.
Protein interaction profile of differentially expressed proteins. The number of interactions for these 24 differentially expressed proteins was obtained from the Web-based Interaction Map Browser developed by Finley and co-workers (see Ref. 68) by searching against a comprehensive Drosophila protein interaction database generated at the Finley laboratory, Curagen, and Hybrigenics.
Fig. 8.
Protein interaction map of differentially expressed proteins constructed based on the Web-based Interaction Map Browser (see Ref. 68). The Drosophila protein interaction database utilized contains protein interaction data generated using the yeast two-hybrid system (YTH) at Curagen (gray line), the Finley laboratory (blue line), and Hybrigenics (cyan line). Differentially expressed proteins are color-coded according to their subcellular localizations. Proteins (only direct target-interacting proteins are shown) interacting with the differentially expressed proteins are uniformly represented by a gray circle irrespective of their subcellular localizations. This map consists of seven distinct networks: one large network contains 96 proteins, one has 26 proteins, one has 16 proteins, one has eight proteins, one has six proteins, one has five proteins, and seven have two proteins. TBPH, TAR DNA-binding protein homolog; see Table II for other gene symbols.
Finally because transgenic Drosophila expressing human A53T α-synuclein genes develop human PD-like symptoms after day 10 (15), dysregulation of proteins observed at day 1, especially those that were known to be A53T α-synuclein toxicity- or PD-associated, may shed light on potential diagnostic strategies at the presymptomatic stage. Comprehensive studies of these proteins and their involved protein interactions (e.g. the single large protein network that contains seven of the 24 dysregulated proteins) and cellular pathways may provide insight into the molecular mechanisms underlying the causes of neurodegeneration in PD. Although several cellular malfunctions (e.g. mitochondrial dysfunction, oxidative stress, and impairment of the ubiquitin-proteasome system) have been proposed to be closely related to PD (5), the cause and effect relationships remain unknown. Altered proteins in the presymptomatic A53T α-synuclein Drosophila PD model were primarily associated with membrane, actin cytoskeleton, ER, mitochondria, and ribosome. These associations provide further evidence, in addition to our previous studies of the A30P α-synuclein Drosophila model, that defects in cellular components such as actin cytoskeleton and mitochondria may contribute to late onset of PD-like symptoms. Nevertheless, further studies are warranted to understand the relationships among the cellular defects that may induce dopaminergic neuron degeneration. Targeted studies involving dysregulated proteins identified at the presymptomatic stage may facilitate the elucidation of major cellular defects underlying PD.
Supplementary Material
Acknowledgments
We acknowledge Dr. Qisheng Song (Department of Chemistry, University of Missouri, Columbia, MO) for providing the fat body protein 1 antibody and Dr. Kenneth P. Nephew (Department of Medical Sciences, Indiana University, Bloomington, IN) and Xinghua Long (Department of Medical Sciences, Indiana University, Bloomington, IN) for providing assistance with Western blot analysis. The anti-β-tubulin antibody developed by Michael Klymkowsky was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA.
Footnotes
Published, MCP Papers in Press, March 18, 2008, DOI 10.1074/mcp.M700467-MCP200
The abbreviations used are: PD, Parkinson disease; LB, Lewy body; SCX, strong cation exchange; GIST, global internal standard technology; ER, endoplasmic reticulum; rdgA, retinal degeneration A; Mlp60A, muscle LIM protein at 60A; Mn-SOD, manganese-superoxide dismutase; β-ATPase, ATP synthase β subunit; Hsc3p, heat shock protein cognate 3; LIM, Caenorhabditis elegans Lin-1, rat ISI-1, and C. elegans Mec-3.
This work was supported, in whole or in part, by National Institutes of Heath Grant RO1-AG-024547. This work was also supported by the Analytical Node of the Indiana University Metabolomics and Cytomics Initiative (funded by the Lilly Endowment). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Olanow, C. W., and Tatton, W. G. ( 1999) Etiology and pathogenesis of Parkinson's disease. Annu. Rev. Neurosci. 22, 123–144 [DOI] [PubMed] [Google Scholar]
- 2.Fearnley, J. M., and Lees, A. J. ( 1991) Aging and Parkinson's disease: substantia nigra regional selectivity. Brain 114, 2283–2301 [DOI] [PubMed] [Google Scholar]
- 3.Sohmiya, M., Tanaka, M., Tak, N. W., Yanagisawa, M., Tanino, Y., Suzuki, Y., Okamoto, K., and Yamamoto, Y. ( 2004) Redox status of plasma coenzyme Q10 indicates elevated systemic oxidative stress in Parkinson's disease. J. Neurol. Sci. 223, 161–166 [DOI] [PubMed] [Google Scholar]
- 4.Keeney, P. M., Xie, J., Capaldi, R. A., and Bennett, J. P. ( 2006) Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 26, 5256–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, T. M., and Dawson, V. L. ( 2003) Molecular pathways of neurodegeneration in Parkinson's disease. Science 302, 819–822 [DOI] [PubMed] [Google Scholar]
- 6.Spillantini, M. G., Schmidt, M. L., Lee, V. M. Y., Trojanowski, J. Q., Jakes, R., and Goedert, M. ( 1997) α-Synuclein in Lewy bodies. Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 7.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L., and Riess, O. ( 1998) Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 8.Spira, P. J., Sharpe, D. M., Halliday, G., Cavanagh, J., and Nicholson, G. A. ( 2001) Clinical and pathological features of a parkinsonian syndrome in a family with an Ala53Thr α-synuclein mutation. Ann. Neurol. 49, 313–319 [PubMed] [Google Scholar]
- 9.Zarranz, J. J., Alegre, J., Gomez-Esteban, J. C., Lezcano, E., Ros, R., Ampuero, I., Vidal, L., Hoenicka, J., Rodriguez, O., Atares, B., Llorens, V., Tortosa, E. G., del Ser, T., Munoz, D. G., and de Yebenes, J. G. ( 2004) The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 [DOI] [PubMed] [Google Scholar]
- 10.Ueda, K., Fukushima, H., Masliah, E., Xia, Y., Iwai, A., Yoshimoto, M., Otero, D. A. C., Kondo, J., Ihara, Y., and Saitoh, T. ( 1993) Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 90, 11282–11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwai, A., Yoshimoto, M., Masliah, E., and Saitoh, T. ( 1995) Non-A β component of Alzheimer's disease amyloid (Nac) is amyloidogenic. Biochemistry 34, 10139–10145 [DOI] [PubMed] [Google Scholar]
- 12.Maries, E., Dass, B., Collier, T. J., Kordower, J. H., and Steece-Collier, K. ( 2003) The role of α-synuclein in Parkinson's disease: insights from animal models. Nat. Rev. Neurosci. 4, 727–738 [DOI] [PubMed] [Google Scholar]
- 13.Scherzer, C. R., and Feany, M. B. ( 2004) Yeast genetics targets lipids in Parkinson's disease. Trends Genet. 20, 273–277 [DOI] [PubMed] [Google Scholar]
- 14.Outeiro, T. F., and Lindquist, S. ( 2003) Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feany, M. B., and Bender, W. W. ( 2000) A Drosophila model of Parkinson's disease. Nature 404, 394–398 [DOI] [PubMed] [Google Scholar]
- 16.Klein, R. L., King, M. A., Hamby, M. E., and Meyer, E. M. ( 2002) Dopaminergic cell loss induced by human A30P α-synuclein gene transfer to the rat substantia nigra. Hum. Gene Ther. 13, 605–612 [DOI] [PubMed] [Google Scholar]
- 17.Giasson, B. I., Duda, J. E., Quinn, S. M., Zhang, B., Trojanowski, J. Q., and Lee, V. M. Y. ( 2002) Neuronal α-synucleinopathy with severe movement disorder in mice expressing A53T human α-synuclein. Neuron 34, 521–533 [DOI] [PubMed] [Google Scholar]
- 18.Yamada, M., Iwatsubo, T., Mizuno, Y., and Mochizuki, H. ( 2004) Overexpression of α-synuclein in rat substantia nigra results in loss of dopaminergic neurons, phosphorylation of α-synuclein and activation of caspase-9: resemblance to pathogenetic changes in Parkinson's disease. J. Neurochem. 91, 451–461 [DOI] [PubMed] [Google Scholar]
- 19.Lo Bianco, C., Ridet, J. L., Schneider, B. L., Deglon, N., and Aebischer, P. ( 2002) α-Synucleinopathy and selective dopaminergic neuron loss in a rat lentiviral-based model of Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 99, 10813–10818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirik, D., Annett, L. E., Burger, C., Muzyczka, N., Mandel, R. J., and Bjorklund, A. ( 2003) Nigrostriatal α-synucleinopathy induced by viral vector-mediated overexpression of human α-synuclein: a new primate model of Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 100, 2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, X. Y., Valentine, S. J., Plasencia, M. D., Trimpin, S., Naylor, S., and Clemmer, D. E. ( 2007) Mapping the human plasma proteome by SCX-LC-IMS-MS. J. Am. Soc. Mass Spectrom. 18, 1249–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valentine, S. J., Plasencia, M. D., Liu, X. Y., Krishnan, M., Naylor, S., Udseth, H. R., Smith, R. D., and Clemmer, D. E. ( 2006) Toward plasma proteome profiling with ion mobility-mass spectrometry. J. Proteome Res. 5, 2977–2984 [DOI] [PubMed] [Google Scholar]
- 23.Kindy, J. M., Taraszka, J. A., Regnier, F. E., and Clemmer, D. E. ( 2002) Quantifying peptides in isotopically labeled protease digests by ion mobility/time-of-flight mass spectrometry. Anal. Chem. 74, 950–958 [DOI] [PubMed] [Google Scholar]
- 24.Hoaglund-Hyzer, C. S., and Clemmer, D. E. ( 2001) Ion trap/ion mobility/quadrupole/time of flight mass spectrometry for peptide mixture analysis. Anal. Chem. 73, 177–184 [DOI] [PubMed] [Google Scholar]
- 25.Taraszka, J. A., Kurulugama, R., Sowell, R. A., Valentine, S. J., Koeniger, S. L., Arnold, R. J., Miller, D. F., Kaufman, T. C., and Clemmer, D. E. ( 2005) Mapping the proteome of Drosophila melanogaster: analysis of embryos and adult heads by LC-IMS-MS methods. J. Proteome Res. 4, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 26.Scherzer, C. R., Jensen, R. V., Gullans, S. R., and Feany, M. B. ( 2003) Gene expression changes presage neurodegeneration in a Drosophila model of Parkinson's disease. Hum. Mol. Genet. 12, 2457–2466 [DOI] [PubMed] [Google Scholar]
- 27.Chen, L., and Feany, M. B. ( 2005) α-Synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat. Neurosci. 8, 657–663 [DOI] [PubMed] [Google Scholar]
- 28.Taraszka, J. A., Gao, X. F., Valentine, S. J., Sowell, R. A., Koeniger, S. L., Miller, D. F., Kaufman, T. C., and Clemmer, D. E. ( 2005) Proteome profiling for assessing diversity: analysis of individual heads of Drosophila melanogaster using LC-ion mobility-MS. J. Proteome Res. 4, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 29.Xun, Z. Y., Sowell, R. A., Kaufman, T. C., and Clemmer, D. E. ( 2007) Protein expression in a Drosophila model of Parkinson's disease. J. Proteome Res. 6, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xun, Z. Y., Sowell, R. A., Kaufman, T. C., and Clemmer, D. E. ( 2007) Lifetime proteomic profiling of an A30P α-synuclein Drosophila model of Parkinson's disease. J. Proteome Res. 6, 3729–3738 [DOI] [PubMed] [Google Scholar]
- 31.Sowell, R. A., Hersberger, K. E., Kaufman, T. C., and Clemmer, D. E. ( 2007) Examining the proteome of Drosophila across organism lifespan. J. Proteome Res. 6, 3637–3647 [DOI] [PubMed] [Google Scholar]
- 32.Feany, M. B. ( 2006) Defining pathways controlling α-synuclein neurotoxicity in Drosophila. Movement Disord. 21, S17–S17 [Google Scholar]
- 33.Kontopoulos, E., Parvin, J. D., and Feany, M. B. ( 2006) α-Synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 15, 3012–3023 [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty, A., and Regnier, F. E. ( 2002) Global internal standard technology for comparative proteomics. J. Chromatogr. A 949, 173–184 [DOI] [PubMed] [Google Scholar]
- 35.Riggs, L., Seeley, E. H., and Regnier, F. E. ( 2005) Quantification of phosphoproteins with global internal standard technology. J. Chromatogr. B 817, 89–96 [DOI] [PubMed] [Google Scholar]
- 36.Ji, J. Y., Chakraborty, A., Geng, M., Zhang, X., Amini, A., Bina, M., and Regnier, F. ( 2000) Strategy for qualitative and quantitative analysis in proteomics based on signature peptides. J. Chromatogr. B 745, 197–210 [DOI] [PubMed] [Google Scholar]
- 37.Peng, J. M., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 38.Liu, X. Y., Miller, B. R., Rebec, G. W., and Clemmer, D. E. ( 2007) Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington's disease. J. Proteome Res. 6, 3134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian, W. J., Monroe, M. E., Liu, T., Jacobs, J. M., Anderson, G. A., Shen, Y. F., Moore, R. J., Anderson, D. J., Zhang, R., Calvano, S. E., Lowry, S. F., Xiao, W. Z., Moldawer, L. L., Davis, R. W., Tompkins, R. G., Camp, D. G., and Smith, R. D. ( 2005) Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics 4, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang, M. C., Juo, C. G., Chang, H. H., Chen, H. M., Yi, E. C., and Chern, Y. ( 2007) Systematic uncovering of multiple pathways underlying the pathology of Huntington disease by an acid-cleavable isotope-coded affinity tag approach. Mol. Cell. Proteomics 6, 781–797 [DOI] [PubMed] [Google Scholar]
- 41.DeSouza, L., Diehl, G., Rodrigues, M. J., Guo, J. Z., Romaschin, A. D., Colgan, T. J., and Siu, K. W. M. ( 2005) Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 4, 377–386 [DOI] [PubMed] [Google Scholar]
- 42.McClatchy, D. B., Liao, L. J., Park, S. K., Venable, J. D., and Yates, J. R. ( 2007) Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 17, 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffin, T. J., Gygi, S. P., Ideker, T., Rist, B., Eng, J., Hood, L., and Aebersold, R. ( 2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 323–333 [DOI] [PubMed] [Google Scholar]
- 44.Burmester, T., Antoniewski, C., and Lepesant, J. A. ( 1999) Ecdysone-regulation of synthesis and processing of Fat Body Protein 1, the larval serum protein receptor of Drosophila melanogaster. Eur. J. Biochem. 262, 49–55 [DOI] [PubMed] [Google Scholar]
- 45.Sun, Y., An, S., Henrich, V. C., Sun, X., and Song, Q. ( 2007) Proteomic identification of PKC-mediated expression of 20E-induced protein in Drosophila melanogaster. J. Proteome Res. 6, 4478–4488 [DOI] [PubMed] [Google Scholar]
- 46.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. ( 2000) Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krantz, D. E., and Zipursky, S. L. ( 1990) Drosophila chaoptin, a member of the leucine-rich repeat family, is a photoreceptor cell-specific adhesion molecule. EMBO J. 9, 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinke, R., Krantz, D. E., Yen, D., and Zipursky, S. L. ( 1988) Chaoptin, a cell-surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell 52, 291–301 [DOI] [PubMed] [Google Scholar]
- 49.Vanvactor, D., Krantz, D. E., Reinke, R., and Zipursky, S. L. ( 1988) Analysis of mutants in chaoptin, a photoreceptor cell specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell 52, 281–290 [DOI] [PubMed] [Google Scholar]
- 50.Masai, I., Suzuki, E., Yoon, C. S., Kohyama, A., and Hotta, Y. ( 1997) Immunolocalization of Drosophila eye-specific diacylglycerol kinase, rdgA, which is essential for the maintenance of the photoreceptor. J. Neurobiol. 32, 695–706 [DOI] [PubMed] [Google Scholar]
- 51.Jo, E. J., McLaurin, J., Yip, C. M., St George-Hyslop, P., and Fraser, P. E. ( 2000) α-Synuclein membrane interactions and lipid specificity. J. Biol. Chem. 275, 34328–34334 [DOI] [PubMed] [Google Scholar]
- 52.Royuela, M., GarciaAnchuelo, R., deMiguel, M. P., Arenas, M. I., Fraile, B., and Paniagua, R. ( 1996) Immunocytochemical electron microscopic study and Western blot analysis of troponin in striated muscle of the fruit fly Drosophila melanogaster and in several muscle cell types of the earthworm Eisenia foetida. Anat. Rec. 244, 148–154 [DOI] [PubMed] [Google Scholar]
- 53.Fyrberg, E., Fyrberg, C. C., Beall, C., and Saville, D. L. ( 1990) Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J. Mol. Biol. 216, 657–675 [DOI] [PubMed] [Google Scholar]
- 54.Watkins, H., Mckenna, W. J., Thierfelder, L., Suk, H. J., Anan, R., Odonoghue, A., Spirito, P., Matsumori, A., Moravec, C. S., Seidman, J. G., and Seidman, C. E. ( 1995) Mutations in the genes for cardiac troponin-T and α-tropomyosin in hypertrophic cardiomyopathy. New Engl. J. Med. 332, 1058–1064 [DOI] [PubMed] [Google Scholar]
- 55.Weiskirchen, R., and Gunther, K. ( 2003) The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays 25, 152–162 [DOI] [PubMed] [Google Scholar]
- 56.Stronach, B. E., Siegrist, S. E., and Beckerle, M. C. ( 1996) Two muscle-specific LIM proteins in Drosophila. J. Cell Biol. 134, 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arber, S., Halder, G., and Caroni, P. ( 1994) Muscle Lim protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 79, 221–231 [DOI] [PubMed] [Google Scholar]
- 58.Hartl, F. U. ( 1996) Molecular chaperones in cellular protein folding. Nature 381, 571–580 [DOI] [PubMed] [Google Scholar]
- 59.Elefant, F., and Palter, K. B. ( 1999) Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol. Biol. Cell 10, 2101–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Outeiro, T. F., Kontopoulos, E., Altmann, S. M., Kufareva, I., Strathearn, K. E., Amore, A. M., Volk, C. B., Maxwell, M. M., Rochet, J. C., McLean, P. J., Young, A. B., Abagyan, R., Feany, M. B., Hyman, B. T., and Kazantsev, A. G. ( 2007) Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519 [DOI] [PubMed] [Google Scholar]
- 61.Smith, W. W., Jiang, H. B., Pei, Z., Tanaka, Y., Morita, H., Sawa, A., Dawson, V. L., Dawson, T. M., and Ross, C. A. ( 2005) Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant α-synuclein-induced toxicity. Hum. Mol. Genet. 14, 3801–3811 [DOI] [PubMed] [Google Scholar]
- 62.Drysdale, R. A., Crosby, M. A., and Consortium, F. ( 2005) FlyBase: genes and gene models. Nucleic Acids Res. 33, D390–D395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun, J. T., Folk, D., Bradley, T. J., and Tower, J. ( 2002) Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics 161, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klivenyi, P., St Clair, D., Wermer, M., Yen, H. C., Oberley, T., Yang, L. C., and Beal, M. F. ( 1998) Manganese superoxide dismutase overexpression attenuates MPTP toxicity. Neurobiol. Dis. 5, 253–258 [DOI] [PubMed] [Google Scholar]
- 65.Yen, H. C., Oberley, T. D., Gairola, C. G., Szweda, L. I., and St Clair, D. K. ( 1999) Manganese superoxide dismutase protects mitochondrial complex I against adriamycin-induced cardiomyopathy in transgenic mice. Arch. Biochem. Biophys. 362, 59–66 [DOI] [PubMed] [Google Scholar]
- 66.Pena, P., Ugalde, C., Calleja, M., and Garesse, R. ( 1995) Analysis of the mitochondrial ATP synthase β-subunit gene in Drosophilidae: structure, transcriptional regulatory features and developmental pattern of expression in Drosophila melanogaster. Biochem. J. 312, 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corpillo, D., Gardini, G., Vaira, A. M., Basso, M., Aime, S., Accotto, G. R., and Fasano, M. ( 2004) Proteomics as a tool to improve investigation of substantial equivalence in genetically modified organisms: the case of a virus-resistant tomato. Proteomics 4, 193–200 [DOI] [PubMed] [Google Scholar]
- 68.Pacifico, S., Liu, G. Z., Guest, S., Parrish, J. R., Fotouhi, F., and Finley, R. L. ( 2006) A database and tool, IM Browser, for exploring and integrating emerging gene and protein interaction data for Drosophila. BMC Bioinformatics 7, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanyon, C. A., Liu, G. Z., Mangiola, B. A., Patel, N., Giot, L., Kuang, B., Zhang, H., Zhong, J. H., and Finley, R. L. ( 2004) A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Res. 5, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., Vijayadamodar, G., Pochart, P., Machineni, H., Welsh, M., Kong, Y., Zerhusen, B., Malcolm, R., Varrone, Z., Collis, A., Minto, M., Burgess, S., McDaniel, L., Stimpson, E., Spriggs, F., Williams, J., Neurath, K., Ioime, N., Agee, M., Voss, E., Furtak, K., Renzulli, R., Aanensen, N., Carrolla, S., Bickelhaupt, E., Lazovatsky, Y., DaSilva, A., Zhong, J., Stanyon, C. A., Finley, R. L., White, K. P., Braverman, M., Jarvie, T., Gold, S., Leach, M., Knight, J., Shimkets, R. A., McKenna, M. P., Chant, J., and Rothberg, J. M. ( 2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 71.Formstecher, E., Aresta, S., Collura, V., Hamburger, A., Meil, A., Trehin, A., Reverdy, C., Betin, V., Maire, S., Brun, C., Jacq, B., Arpin, M., Bellaiche, Y., Bellusci, S., Benaroch, P., Bornens, M., Chanet, R., Chavrier, P., Delattre, O., Doye, V., Fehon, R., Faye, G., Galli, T., Girault, J. A., Goud, B., de Gunzburg, J., Johannes, L., Junier, M. P., Mirouse, V., Mukherjee, A., Papadopoulo, D., Perez, F., Plessis, A., Rosse, C., Saule, S., Stoppa-Lyonnet, D., Vincent, A., White, M., Legrain, P., Wojcik, J., Camonis, J., and Daviet, L. ( 2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.