Abstract
Protein expression profiles in rat bladder smooth muscle were compared between animal models of streptozotocin-induced diabetes mellitus (STZ-DM) and age-matched controls at 1 week and 2 months after induction of hyperglycemia with STZ treatment. At each time point, protein samples from four STZ-DM and four age-matched control rat bladder tissues were prepared independently and analyzed together across multiple DIGE gels using a pooled internal standard sample to quantify expression changes with statistical confidence. A total of 100 spots were determined to be significantly changing among the four experimental groups. A subsequent mass spectrometry analysis of the 100 spots identified a total of 56 unique proteins. Of the proteins identified by two-dimensional DIGE/MS, 10 exhibited significant changes 1 week after STZ-induced hyperglycemia, whereas the rest showed differential expression after 2 months. A network analysis of these proteins using MetaCore™ suggested induction of transcriptional factors that are too low to be detected by two-dimensional DIGE and identified an enriched cluster of down-regulated proteins that are involved in cell adhesion, cell shape control, and motility, including vinculin, intermediate filaments, Ppp2r1a, and extracellular matrix proteins. The proteins that were up-regulated include proteins involved in muscle contraction (e.g. Mrlcb and Ly-GDI), in glycolysis (e.g. α-enolase and Taldo1), in mRNA processing (e.g. heterogeneous nuclear ribonucleoprotein A2/B1), in inflammatory response (e.g. S100A9, Annexin 1, and apoA-I), and in chromosome segregation and migration (e.g. Tuba1 and Vil2). Our results suggest that the development of diabetes-related complications in this model involves the down-regulation of structural and extracellular matrix proteins in smooth muscle that are essential for normal muscle contraction and relaxation but also induces proteins that are associated with cell proliferation and inflammation that may account for some of the functional deficits known to occur in diabetic complications of bladder.
Diabetic bladder dysfunctions are among the most common complications associated with diabetes mellitus (1, 2). Although the disease is not life threatening, it is associated with several debilitating urological symptoms. Such symptoms include impaired bladder sensation, impaired detrusor contractility, and increased bladder capacity leading to significant distress, limitations in daily functioning, and poor quality of life (1, 3). It appears that up to 80% of patients with diabetes may eventually develop some type of bladder dysfunction (4). Unfortunately there are few systems biology studies focusing on the initiation, development, and progression of the bladder dysfunction in diabetic patients or relevant animal models.
Experimental studies in the diabetic rat have identified a variety of alterations and putative mechanisms that describe the multifactorial and progressive nature of the disease. For example, there are several publications suggesting that diabetes-associated bladder pathology is mainly a result of alterations in neuronal components, including deficiency of acetylcholinesterase (5–7), impaired ATPase activity (8, 9), and diminished neurotrophic factor (10, 11). Recently Sasaki et al. (12) and Goins et al. (13) showed the relation between bladder dysfunction and lower nerve growth factor levels and the feasibility of nerve growth factor gene therapy for treating diabetes mellitus (DM)1 cystopathy. In addition, the results of some studies also showed a correlation between bladder dysfunction and alteration of the myogenic components, including progressive increase in total bladder mass, decrease in collagen, and dilation of the bladder (14). More recently a role for the urothelium (15) and nitric oxide (16–18) effects have also been suggested.
The important impact of changes in gene and/or protein expression to hypertrophic remodeling of the diabetes bladder has long been known (19), and recently a study by Hipp et al. (20) using Affymetrix GeneChip arrays characterized specific genome expression changes in bladder smooth muscle from streptozotocin (STZ)-treated rats 1 week after the onset of diabetes. However, as far as we are aware, there are no studies that have attempted to characterize diabetes-related changes in bladder smooth muscle at the level of the proteome. This highlights the fact that there are uncertainties about the time course, magnitude, and mechanism of diabetes-related changes in bladder function both in this animal model and in humans.
Therefore, we focused this investigation on proteomics studies of the detrusor smooth muscle cell. This seems a reasonable research strategy given that smooth muscle cell tone is a critical determinant of bladder function. The overall goal was to delineate the proteomic changes that occur during the initiation, development, and progression of diabetes-related bladder dysfunction. As a first step, differential protein analysis was carried out using two-dimensional (2D) DIGE coupled with mass spectrometry to compare and identify proteins that are changing at 1 week (a time point when there is no significant effect on bladder physiology) and at 2 months (when significant bladder pathophysiology begins to develop) after the induction of hyperglycemia in an STZ-DM rat model compared with age-matched controls (20–22). Although animal models, in general, may recapitulate only some aspects of human disease, this model system is of particular interest as its appears to mimic many relevant aspects of human urologic dysfunction (23–26). Not surprisingly, the STZ-induced diabetic rat is one of the most commonly used animal models for studying the urologic complications of diabetes.
Our experimental design included four experimental groups (1-week STZ-induced diabetic, 1-week control, 2-month STZ-induced diabetic, and 2-month control). For each experimental group four independent biological replicate samples were prepared and analyzed across multiple 2D DIGE gels using a pooled internal standard sample that ensured precise quantification of expression changes and their detailed statistical analysis (27–29). To probe protein-protein interaction networks that can predict the signaling pathways that are activated or deactivated during the initiation and development of the diabetic bladder dysfunction and also to identify transcriptional factors and relatively low abundance proteins that cannot be identified by 2D DIGE/MS, we further analyzed the 2D DIGE data using MetaCore™ pathway analysis tools (30, 31) along with validation of specific expression changes by Western blotting. With this integrated approach we identified a number of differentially expressed proteins that may provide valuable mechanistic insight into the molecular changes that are precursors for and are associated with DM-induced bladder dysfunction.
MATERIALS AND METHODS
Most of the chemicals used in this study were received from GE Healthcare (formerly Amersham Biosciences), Pierce, and Invitrogen and used without further purification unless otherwise stated. Goat polyclonal anti-Ly-GDI, anti-α-enolase, anti-hnRNP K, mouse monoclonal anti-c-Myc, and anti-p53 antibodies specific for human, rat, and mouse were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Animal Preparations—
Diabetes was induced in male Fisher 344 rats (Taconic Farms, Germantown, NY; 8–10 weeks old and weighing 200–240 g) with a single intraperitoneal injection of STZ (35 mg/kg) dissolved in citrate buffer (0.6 m citric acid, 0.08 m Na2HPO4, pH 4.6). The age-matched control rats received an injection of vehicle only. All procedures were approved by the Animal Institute Care and Use Committee at Albert Einstein College of Medicine. Initiation of the diabetic state was confirmed by the presence of high blood glucose levels for the STZ-treated animals only (control, 132 ± 14 mg/dl glucose; STZ-treated rats at 1 week, 537 ± 22 mg/dl; and STZ-treated rats at 2 months, 562 ± 18 mg/dl); these levels were maintained throughout the study. The animals were then sacrificed 1 week or 2 months following confirmation of STZ-induced diabetes. The bladder from each animal was excised and denuded of the urothelial and suburothelial layers; this provided a tissue preparation that had >90% smooth muscle origin. The bladder strips were flash frozen in liquid nitrogen and stored at −80 °C until used for proteome analysis.
Protein Extraction and Fluorescence Dye Labeling—
The experiments were designed to monitor changes in protein abundance as a function of disease progression, to generate statistically significant results, and to minimize in-gel and gel-to-gel systematic variations. Thus, the bladder tissue samples examined consisted of control Fisher 344 rats sacrificed 1 week (n = 4) and 2 months (n = 4) following vehicle injection and STZ-induced diabetic Fisher 344 rats sacrificed 1 week (n = 4) and 2 months (n = 4) following STZ-induced diabetes. Overall this study design was expected to provide a power of 0.8 to detect a 50% or greater change in protein expression in at least 75% of the detected spot entities using the DIGE method (see below).
The frozen tissue samples were disrupted by grinding in liquid nitrogen with a mortar and pestle. The tissue powder was then homogenized in lysis buffer (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 30 mm Tris) with five cycles of freeze, thaw, and sonication. The protein in the supernatant was separated from the remaining tissue debris by centrifugation at 12,000 × g for 10 min. The proteins samples were then further cleaned (GE Healthcare Clean-Up kit), and the protein concentration was determined using the 2D-Quant kit as described by the manufacturer (GE Healthcare). The lysate was further diluted with lysis buffer to give stock solutions with final concentrations of 5 mg/ml that were used to prepare the experimental samples for 2D DIGE. Sixteen aliquots, each with 25 μg of protein, were collected from the control and STZ-induced diabetic samples and pooled to prepare internal standard samples. Thus, samples from either control or STZ-induced diabetic bladder were labeled with Cy3 or Cy5 cyanine dyes, and internal standard sample was labeled with Cy2 dye by the addition of 400 pmol of CyDye in 1 μl of anhydrous N,N-dimethylformamide/50 μg of protein. A dye-swapping scheme, as shown in Table I, was used such that the four samples for any condition were variously labeled with Cy3 or Cy5 to control for any dye-specific labeling artifacts. Labeling was performed for 30 min of incubation on ice in the dark; the reaction was then quenched with 10 mm lysine and additionally incubated for 10 min.
Table I.
Experimental design for 2D DIGE proteome profiling
Four biological replicate samples for each group (C_1W, control sacrificed at 1 week after vehicle injection; D_1W, 1 week after the confirmation of STZ-induced diabetes; C_2M, 2 months after vehicle injection; and D_2 M, 2 months after the confirmation of STZ-induced diabetes) were used and labeled with Cy3 or Cy5. Each gel contained the pooled standard (equal aliquots of all the samples in all groups) and two other subject samples. Thus, the 16 samples were analyzed in triplicate by running eight gels. (For detailed descriptions refer to “Materials and Methods.”)
| Gel | Cy3 | Cy5 | Cy2 |
|---|---|---|---|
| 1 | C1_1W | D1_1W | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 2 | D2_1W | C2_1W | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 3 | C3_1W | D3_1W | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 4 | D4_1W | C4_1W | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 5 | C1_2M | D1_2M | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 6 | D2_2M | C2_2M | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 7 | C3_2M | D3_2M | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
| 8 | D4_2M | C4_2M | Pooled internal standard sample (C1–4_1W + D1–4_1W + C1–4_2 M + D1–4_2M) |
Gel Electrophoresis—
The quenched Cy3- and Cy5-labeled samples, to be partitioned in the same gel according to the experimental design in Table I, were then combined and mixed with an aliquot of Cy2-labeled standard and an equal volume of 2× sample buffer (8 m urea, 4% (w/v) CHAPS, 2% (w/v) DTT, 2% (v/v) Pharmalyte pH 3–10 non-linear) was added. Prior to IEF an additional 350 μg of unlabeled protein was added (for later spot picking), and the mixtures were brought up to 450 μl with 1× rehydration buffer. Immobiline DryStrips (pH 3–10 non-linear, 24 cm) were rehydrated overnight in 450 μl of sample plus rehydration buffer (8 m urea, 4% (w/v) CHAPS, 1% (v/v) Pharmalyte (pH 3–10), 2% (w/v) DTT) overlaid with 2.0 ml of DryStrip cover fluid in an Immobiline DryStrip holder. Strips were then focused using an IPGphor IEF system for a total of 60.2 kV-h at 20 °C. Prior to SDS-PAGE, each strip was equilibrated with 10 ml of equilibration buffer A (8 m urea, 100 mm Tris-HCl, pH 6.8, 30% (v/v) glycerol, 1% (w/v) SDS, 5 mg/ml DTT) on a rocking shaker for 10 min followed by addition of 10 ml of equilibration buffer B (8 m urea, 100 mm Tris-HCl, pH 6.8, 30% (v/v) glycerol, 1% (w/v) SDS, 45 mg/ml iodoacetamide) for a further 10 min. The second equilibration step in this protocol is included only to help prevent point streaking; it does not help to prevent the regeneration of disulfide bridge formation (32, 33). The strips were then loaded and run on 12.5% acrylamide isocratic Laemmli gels (34) using the Ettan DALT Twelve apparatus. Gels were run at 1 watt/gel constant power at 14 °C for 2 h followed by 2 watts/gel constant power at 14 °C until the bromphenol blue dye front had run off the bottom of the gels. All electrophoresis procedures were performed in the dark.
Image Acquisition and Spot Quantification—
The labeled protein spots in the gel were visualized using the Typhoon 9400 imager (GE Healthcare). The gels were scanned using a 488 nm laser and an emission filter of 520 nm band pass (BP) 40, a 532 nm laser and an emission filter of 580 nm BP30, and a 633 nm laser and a 670-nm BP30 emission filter to acquire the Cy2, Cy3, and Cy5 images, respectively. The narrow BP emission filters ensure that there is negligible cross-talk between fluorescence channels. All gels were prescanned at 1000-μm resolution while adjusting the photomultiplier tube voltage to obtain images with a maximum intensity of 40,000–60,000 units, and then final scans were done at 100-μm resolution. Images were then cropped to remove areas extraneous to the gel image using ImageQuant v5.0 (GE Healthcare) prior to analysis. One of the eight gels was poststained using a deep purple staining protocol, scanned using a 457 nm laser and an emission filter of 610 nm BP30, and used as preparative gel for downstream protein identification.
Data analysis was carried out using a total of 24 gel images consisting of four biological replicate images from 1-week control, four replicates from 1-week treated, four replicates from 2-month control, four replicates from the 2-month treated, and eight replicates from the internal standards, which were pooled mixtures of equal aliquots of each experimental sample. The pick gel image was also processed with the rest of the gel images as a pick gel image but not included in the analysis. To compare protein spots across the eight gels, image analyses were conducted into two steps using DeCyder v6.5 2D Differential Analysis Software (GE Healthcare). In the first step, the set of three images from a single gel was loaded into the differential in-gel analysis (DIA) algorithm within the DeCyder software. Intragel spot detection and quantification was performed using the DeCyder software DIA module. For the subsequent intergel differential analysis, the DIA work spaces for all the gels were saved and loaded into the biological variation analysis (BVA) module. In the BVA module, the image with the largest number of protein spots was assigned as the master image. Sample (Cy3- or Cy5-labeled) spot maps were assigned into four groups (group 1, control, 1 week; group 2, STZ-induced diabetic, 1 week; group 3, control, 2 months; and group 4, STZ-induced diabetic, 2 months), and all internal standard and the pick spot maps were assigned into standard and pick folders, respectively, in the experimental design view of the BVA modules. Once the spots from the common standards were matched across the eight gel images and with the pick gel image, the standardized volume ratio for each standard image from the different gels is set to the value 1.0 to compare ratios between matched protein spots in the different gels (groups). Thus, the ratios of the log-standardized protein spot abundances (differences in expression) between the groups were computed.
Statistical Analysis of Protein Expression—
The experiment was designed to minimize both false positive and false negative results for expressed proteins while detecting minimal detectable difference (the smallest difference between the treatments that we wish to be able to detect). Four full replicates (with respect to model animal preparation, protein isolation, sample preparation, and 2D DIGE) were performed for each experimental condition. To estimate the number of samples providing enough power to detect differences in the protein level among experimental groups, sample size and power analysis was performed for several proteomics data sets that we already have in the center. Using a two independent group t test, we calculated the sample size required to detect 1.3-. 1.5-, 2-, and 4-fold changes in each protein spot intensity level as a function of false positive rate, power, and percentage of protein spots that have a standard deviation below a given quartile. Sample sizes providing 80% power to detect a 50% mean difference between groups for 75% of protein spots present in gels, assuming that standard deviation is common for the 75th percentile of all protein spots present in a gel, were 4 at the 0.05 significance level.
To test for significant differences in expression of proteins between the experimental groups, one-way analysis of variance (ANOVA) was performed at a significance level of 0.05; thus, for every hundred spots tested, five false positives would be expected. In addition, two-way ANOVA treatment, two-way ANOVA time, and two-way ANOVA interact were computed to assign statistically significant changes in spot intensity due to the treatment alone, time alone, and due to both treatment and time. The data were filtered using the average volume ratios of 1.5 and above or −1.5 and below -fold differences in expression and with a one-way ANOVA p value of 0.05 or less and assigned to a spot of interest. Thus, of a total of 2508 spots detected on the master gel, 100 spots satisfied these requirements and were highlighted as spots of interest. For the spots that displayed significant differences in expression among the groups, pairwise comparisons of the experimental groups were determined using Tukey's multiple comparison procedure with the extended data analysis modules of the DeCyder software. For these spots a pick list with pick coordinates and pick locations on the pick gel image was generated. The pick list along with the poststained pick gel was transferred to the automated Ettan spot picker, and gel plugs were excised and recovered into 96-well plate for the subsequent in-gel digestion and mass spectrometry analysis of the peptide for protein identification.
In addition to univariate analysis, multivariate analyses were performed to explore categories of differential protein expression. Global relationships among spot maps were visualized by performing a principal component analysis (35) on the spot intensity data for the identified proteins that are listed in supplemental Table 1 and plotting spot maps in two-dimensional space corresponding to the first two principal components, PCA1 and PCA2, which orthogonally divide the spot maps (samples) based on the two largest sources of variation in the data set. The protein expression profile of the spot maps was visualized in two-dimensional Euclidian space by using extended data analysis modules of the DeCyder software.
In-gel Digestion and Protein Identification—
The proteins in the gel plugs were digested with trypsin (Promega) using a modified protocol adapted from Shevchenko et al. (36). Tryptic digests were extracted from the gel matrix and concentrated by SpeedVac avoiding complete drying. For mass spectrometric analysis using MALDI, aliquots of the concentrated peptides were treated and eluted off of the ZipTip (Millipore, Billerica, MA) according to the instructions in the manual and directly deposited onto a MALDI target plate with a matrix solution (50% ACN, 0.1% TFA at a concentration of 5 mg/ml). Mass spectra were acquired on a prOTOF 2000™ MALDI orthogonal TOF mass spectrometer (PerkinElmer Life Sciences) in the positive ion mode using TOFworks™, an integrated work flow-based software platform. The peptide ion masses (M + H) were accurate to within 10 ppm after external calibration using an external calibration mixture, pepmix 1, from the ProXpression MALDI standard kit (PerkinElmer Life Sciences). Monoisotopic peptides peak lists were generated in the mass range m/z 500–5000 with a signal to noise ratio threshold of 3.0 and peak resolution threshold of 10,000 using M/Z peak picking algorithm (Genomic Solutions, Ann Arbor, MI) of the TOFworks software version 1.0.1.798. Trypsin autolysis fragment peaks and peaks from the matrix were not excluded unless otherwise stated. The resulting peptide mass lists were used to search the sequences present in an indexed rat subset database (36,274 sequences) created from the National Center for Biotechnology Information non-redundant (NCBInr) databases containing 3,893,302 sequences (released July 4, 2006) and stored locally by running ProFound™ search engine V2003.6.2.1 (Genomic Solutions). Search parameters used in this study were: 1) protein molecular mass search window of 10–300 kDa, 2) protein expectation p < 0.001, 3) minimum sequence coverage of 10%, 4) peptide mass tolerance limits of 30 ppm, and 5) complete cysteine modification by iodoacetamide (57 Da) and partial methionine oxidation (16 Da); two missed cleavage sites were also allowed. A positive identification was accepted when a minimum of six peptide monoisotopic masses matched a particular protein with a mass error tolerance of ≤30 ppm, sequence coverage ≥25%, and low expectation value (p < 0.001). In addition, the correlation of theoretical molecular weight and pI with the gel region was also considered (all the data are presented in supplemental Table 2 and supplemental Fig. 1, a–k).
In cases where the peptide mass fingerprint did not result in positive identification, we performed LC-MS/MS experiments. Thus, tandem mass spectra for 75 gel plugs were acquired using a Finnigan LTQ™ mass spectrometer (Thermo Electron Corp., Bremen, Germany) equipped with an Ettan multidimensional LC system (GE Healthcare). The protein digests were trapped onto a precolumn (C18, PepMap100, 300 μm × 5 mm, 5-μm particle size, 100 Å; Dionex) and separated with a reverse phase column (C18, 75 μm × 150 mm, 3 μm; Dionex) using mobile phases A (0.1% formic acid in a water) and B (84% acetonitrile, 0.1% formic acid in water) with a linear gradient of 2%/min, starting with 100% A. Subsequently the peptides were infused at a flow rate of 300 nl/min via a PicoTip emitter (New Objective Inc., Woburn, MA) and at a voltage of 1.8 kV. Full MS spectra (MS survey scan) were recorded in the ion trap cell, and dependent MS2 spectra for the three most intense ions were subsequently acquired by the LTQ under the collision energy of 35 eV and isolation width of 2.5 Da. The tandem mass spectra were annotated, and peak list files were generated, commonly referred to as .DTA files, by running SEQUEST extract_msn algorithm in Bioworks version 3.2 (Thermo Electron Corp.). The mass range m/z 400–3500 from full scan data with an absolute threshold of 100, a minimum ion count of 12, and a precursor ion tolerance of 1.4 Da was used to generate the .DTA file. Prior to searching, non-redundant databases of 3,893,302 sequences (release July 4, 2006) were downloaded in FASTA format via file transfer protocol from NCBInr. These databases were stored locally, and a subset database for rat species was created and indexed to produce faster search times. The resulting peak list (.DTA) files were then used to interrogate sequences present in an indexed rat subset database (36,274 sequences stored locally) by running SEQUEST SEARCH algorithm of Bioworks software version 3.2. SEQUEST searches were performed with maximum peptide and fragment ion mass tolerances of 2.5 and 1.0 Da, respectively, with partial methionine oxidation, complete carbamidomethylation of cysteine, and two missed cleavage sites allowed in the search parameters. The criteria for each protein identification was a minimum of two peptides with a significant peptide expectation (p < 0.001); peptide Xcorr of 1.9, 2.7, and 3.5 for the charge states +1, +2, and +3, respectively; and a minimum ΔCN (delta correlation) of 0.1. The correlation of theoretical molecular weight and pI with the gel region was also generally considered. In addition all the MS/MS spectra identified by SEQUEST were manually verified for spectral quality and matching y and b ion series (all the data are presented in supplemental Table 3).
Network Analysis—
The data set with a list of regulated proteins identified by 2D DIGE/MS was analyzed further by pathway analysis using the network building tool MetaCore (GeneGo, St. Joseph, MI). MetaCore consists of curated protein interaction networks on the basis of manually annotated and regularly updated databases. The databases consist of millions of relationships between proteins that are derived from literature publications on proteins and small molecules. The relationships include direct protein interaction, transcriptional regulation, binding, enzyme-substrate, and other structural or functional relationships. The networks can be visualized graphically as nodes (proteins) and edges (the relationship between proteins) alongside the empirical expression pattern. The data set from our study consisting of the proteins with statistically significant -fold changes, identified by 2D DIGE/MS, and gene name as a tab-delimited file was imported into the MetaCore. Hypothetical networks of proteins from our experiment and proteins from the MetaCore database were then built using the shortest paths algorithm, one of the several algorithms integrated within MetaCore. Furthermore once the list of altered proteins was uploaded, the size of the intersection between the subset of uploaded proteins and the proteins on all possible pathway maps in the MetaCore database was computed. The results (the maps with the list of proteins from the uploaded data set) were then compared with all the possible pathway maps for all the proteins in the database, and the p value (the probability of randomly obtaining an intersection of a certain size between the uploaded proteins and the proteins on the pathway maps follows a hypergeometric distribution) was calculated based on hypergeometric distribution probability test. The relevant pathway maps were then prioritized based on their statistical significance with respect to the uploaded data sets. Five pathway maps were considered to be statistically significant (p < 0.0001) for the uploaded data.
Immunoblotting Analysis—
The bladder tissue lysates (50 μg of protein/lane) were prepared from two biological replicates for STZ-induced diabetic and age-matched control and separated by electrophoresis under denaturing and reducing conditions on 4–20% polyacrylamide gels (Invitrogen). The proteins were then transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk for 1 h at room temperature with gentle shaking, washed, and incubated overnight with primary antibody (Santa Cruz Biotechnology, Inc.) at 0.2 μg/ml (at 1:1000 dilution). The membranes were washed, then incubated for 1 h at room temperature with 0.02 μg/ml (at 1:10,000 dilution) horseradish peroxidase-conjugated donkey anti-goat IgG or horseradish peroxidase-conjugated goat anti-mouse IgG, and then washed again. After three washes the blots were incubated for 5 min in the working solutions of the substrates that were prepared according to the manufacturer's instructions (Pierce). The membranes were removed from the substrates and placed in plastic sheet protectors, and the chemiluminescence was visualized by exposure of the blot on Eastman Kodak Co. X-Omat film. The blots were scanned, and the protein bands were quantified using ImageQuant-TL v2005 software (GE Healthcare). Equal loading of the protein samples was confirmed by reprobing the blots for β-actin.
RESULTS
To monitor hyperglycemia-associated protein expression changes in rat bladder at two specific time points compared with age-matched controls, total soluble proteins from smooth muscle strips at 1 week and 2 months after the onset of STZ-induced diabetes were analyzed using 2D DIGE/MS (27–29). A total of 16 samples, labeled with Cy3 and Cy5 (four biological replicates per group) were run in eight gels along with a pooled standard labeled with Cy2 giving a total of 24 images and 16 data points. As the pooled standard sample contained an equal aliquot from each of the 16 samples, it represents an average of all the samples being compared, and it is used as a normalization standard across all gels. For samples resolved in the same gel, quantification of the proteins were performed first by dividing the Cy3 or Cy5 signals by the Cy2 intensities, and then these intragel ratios were compared with the ratios from other gels by setting the standardized volume ratio for each standard image from the different gels to 1.0. Thus, for a given protein, the normalized abundance ratios from all 16 samples can be intercompared in an eight-gel experiment as the protein spot intensity in every gel is normalized to the same internal standard.
To compute the -fold changes in protein expression between the groups and their statistical significance, once the standardized abundance ratios for spots in every gel were computed, the corresponding Cy3 and Cy5 spot maps for 15 samples were grouped according to the group descriptions 1–4 as described in the experimental design (Table I). It should be noted that one of the gel images from the 2-month control group had very high background noise values and was excluded from the analysis. The pooled standard spot maps were given the group description of “standard” and were utilized as internal standards, and all the analysis gels were expressed as a function of the internal standard in the analysis. ANOVA (both one-way and two-way) was applied to matched spots, and data were filtered to retain spots that were observed in at least 80% of spot maps and that were confirmed with one-way ANOVA p value <0.05 to have -fold changes of 1.5 and above or −1.5 and below. Once the spots were detected to be significant, the spots were carefully checked for correct matching throughout all the gels. Of a total of 1515 spots that were matched in more than 80% of spot maps, the mean intensities of 100 spots were considered to be significantly different among the experimental groups and were further investigated as spots of interest.
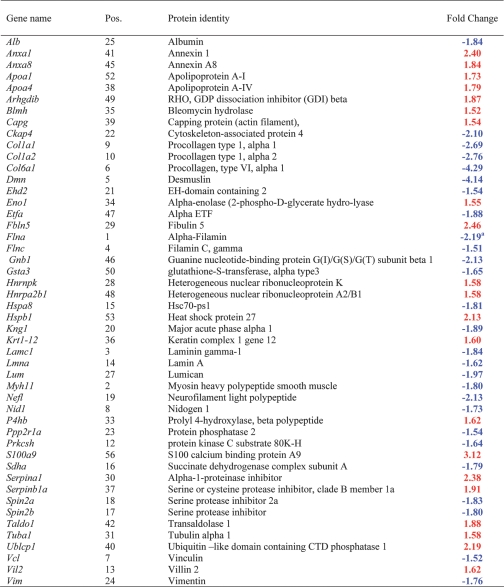
For these 100 spots, whose overall expression profile yielded a significant one-way ANOVA p value, a Tukey procedure was used to determine for which two groups the ratios of the expression changes were significant; these are indicated in boldface in supplemental Table 1. In addition, to evaluate the statistical significance of effects either of STZ-induced diabetes or time on protein expression and also to see whether there was an interaction effect between the STZ-induced diabetes and time, the data were further filtered by two-way ANOVA treatment, two-way ANOVA time, and/or two-way ANOVA interact (p ≤ 0.05). Of the total 100 spots that were differentially expressed among the four experimental groups, both STZ-induced diabetes and duration had a significant effect on the expression of 60 spots. For 21 spots STZ-induced diabetes resulted in significant changes in expression, whereas time had no effect. On the other hand, time had a significant effect on the expression of 19 spots, but STZ-induced diabetes had no effect on these spots. STZ-induced diabetes and time had a synergistic effect on the expression of 16 of 60 spots. In this way, we were able to distinguish the spots that are changing due to time but not treatment, due to treatment but not time, due to both but interfering, or due to both and in concert. We were also able to identify the proteins whose expression changes were due to STZ-induced diabetes alone. A total of 100 spots were excised from the preparative gel that was loaded with 500 μg of total protein, and these spots were in-gel digested for the subsequent MALDI-TOF and/or LTQ MS/MS analyses. Of this list, 93 spots were successfully sequenced, whereas the digest for the remaining seven spots did not reveal any peptides. A total of 39 spots were isoforms for 12 proteins due to either post-translational modification or proteolysis. As the overall expression patterns of the observed isoforms were similar, duplicates were omitted from the list in Table II and supplemental Table 1. Moreover for 10 spots, more than one protein per spot passed our stringent filtering criteria, and the results for these spots were excluded from the list (Table II and supplemental Table 1) because the average ratio observed corresponds to the combination of all the proteins present for a particular spot, and individual changes are not conclusive to date. Thus we report a total of 56 unique proteins along with the -fold changes that are summarized in Table II and detailed in supplemental Tables 1–3 and supplemental Fig. 1, a–k.
Table II.
Differentially expressed proteins 2 months (a) and 1 week (b) after the confirmation of STZ-induced diabetes
Position numbers (Pos.) correspond to the position of the proteins on the gel image in Fig. 1.
The -fold changes are displayed in the range of −∞ to −1 for decreases (blue font) in expression and +1 to +∞ for increases (red font) in expression.
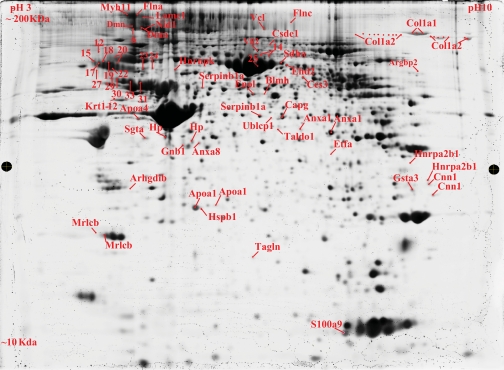
Fig. 1.
The 2D map of deep purple-labeled bladder smooth muscle proteins indicating pick location of a subset of proteins that changed in response to STZ-induced diabetes. Orientation of the pH gradients is indicated on the horizontal axes from 3 pH units (left) to 10 pH units (right), and approximate apparent molecular mass ranges are indicated along the vertical axes from 10 kDa (bottom) to 200 kDa (top).
A 2D map of a representative deep purple-stained gel is shown in Fig. 1. On this map the pick locations of proteins whose expression is significantly different based on the variance of the mean change among the group (p ≤ 0.05) are shown. In some cases, the same protein was identified in different spots across the 2D gel, suggesting the occurrence of post-translational modifications.
Principal Component Analysis—
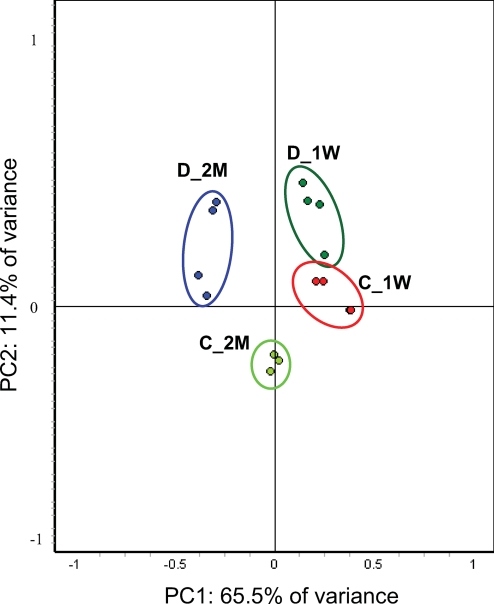
Global relationships among samples were visualized by performing a principal component analysis on the expression data (Fig. 2). Before dimensional reduction, each spot map existed in multidimensional space (one dimension for each of the expression values for a spot). The spot map comparisons were plotted in two-dimensional space, corresponding to the first and second principal components of variation. The first principal component for each spot map was the weighted linear combination of intensity values that shows maximum variation; the second principal component was a weighted linear combination orthogonal to the first component that has maximum variance. For the 100 spots, the first principal component distinguished 65.5% of the variance with 11.4% additional variation distinguished by the second principal component. The principal component analysis (PCA) indicated that the spot maps from each of the four experimental conditions resolved into four distinct groups and demonstrated high reproducibility between the replicates (Fig. 2). However, the 1-week STZ-treated and 1-week control groups were very close to each other, suggesting that the differences between the two experimental groups are not substantial. The PCA also demonstrated that the maximum amount of variation in the experiment is exhibited between the 2-month STZ-treated group and the rest of the experimental groups. One would not expect the individual samples to cluster in the way our experimental groups are clustered, shown in Fig. 3, if the -fold changes for the individual proteins reported in Table II arose by chance. In this way, the PCA further demonstrated the statistical significance of the -fold changes for the proteins reported in Table II.
Fig. 2.
PCA of the proteins mediated by STZ-induced diabetes. The protein expression profiles of experimental groups were visualized in two-dimensional Euclidian space by using the extended data analysis module of DeCyder software as described under “Materials and Methods.” The PCA distinctly clustered the 15 individual samples into four experimental groups (C_1W, 1-week control; D_1W, 1-week diabetes; C_2M, 2-month control; and D_2M, 2-month diabetes).
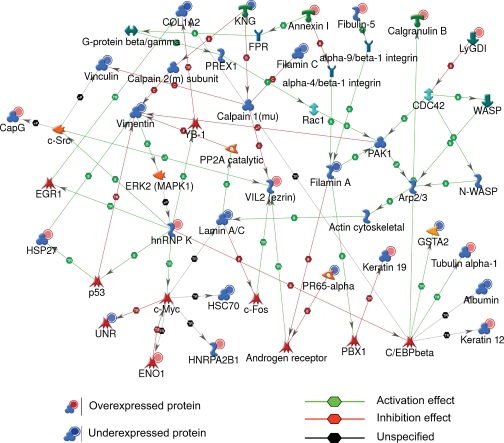
Fig. 3.
Protein networks associated with the proteins differentially expressed in response to STZ-induced diabetes. The network was generated by a shortest paths algorithm of MetaCore (GeneGo) software using the list of differentially expressed proteins identified by 2D DIGE/MS analysis. Individual proteins are represented as nodes, and the different shapes of the nodes represent the functional class of the proteins. The edges define the relationships of the nodes: the arrowheads indicate the direction of the interaction. P indicates phosphorylation, T indicates transformation, B indicates binding, C indicates cleavage, and TR indicates transcriptional regulation. The color of the hexagons on edges between nodes describe activation (green), inhibition (red), and unspecified (black) interactions. WASP, Wiskott-Aldrich syndrome protein; N-WASP, neural Wiskott-Aldrich syndrome protein; KNG, kininogen-1 precursor; CapG, capping protein; UNR, cold shock domain containing protein E1; FPR, fmet-leu-phe receptor.
Differentially Expressed Proteins—
The -fold changes we reported as significant (boldfaced and in red or blue font in Table II) for the proteins are ≥1.5 or ≤−1.5. A total of 56 unique proteins are in this category. Ten of these exhibited significant changes 1 week after STZ-induced diabetes prior to the development of any organ complications, whereas the changes for the remaining proteins occurred after 2 months of STZ treatment and the attendant hyperglycemia.
Differentially Expressed Proteins 1 Week after Confirmation of STZ-induced Diabetes—
Of a total of 10 proteins that exhibited significant changes 1 week after confirmation of the STZ-induced diabetes, five were down-regulated with STZ treatment, and the rest were up-regulated. From the down-regulated category, two proteins, Col1a1 and Col1a2, were decreased in the 1-week time period and remained decreased even after 2 months of treatment. The three other proteins, Cnn1, Hp, and Ces3, were down-regulated in STZ-induced diabetic rat models at 1 week, but no changes were seen at the 2-month time period. On the other hand, of the five up-regulated proteins, three proteins, Hspb1, Serpina1, and tubulin α1, showed increased expression levels that were STZ-induced and time-dependent. More specifically, tubulin α1, an abundant cytoplasmic protein in the microtubule family involved in the nucleation of the microtubule assembly during the cell cycle, showed a 43% STZ-induced increase at 1 week and was further increased at 2 months. Other proteins, including myosin regulatory light chain 2-B smooth muscle isoform (Mrlcb) and Argbp2 showed STZ-induced increases at 1 week but no significant change at 2 months. Consistent with our 2D DIGE results for Mrlcb and calponin (Cnn1), in a recent genome expression analysis Hipp et al. (20) found significant increases in Mrlcb and decreases in Cnn1 after 1 week of STZ-induced diabetes.
Differentially Expressed Proteins 2 Months after Confirmation of STZ-induced Diabetes—
Unlike the 1-week time point, 48 proteins showed STZ-induced differential expression 2 months after the confirmation of STZ-induced hyperglycemia. To understand the relationship of these proteins and their significance in the context of DM-associated organ dysfunction and their interactions with other proteins in known networks, the proteins were analyzed using MetaCore analysis tools (30, 31). Of the total 56 proteins imported into MetaCore, 50 were successfully mapped to the MetaCore database. A network of these proteins was generated using a “shortest paths algorithm.” Protein-protein interaction networks among the proteins identified by 2D DIGE/MS and proteins from the MetaCore database are shown in Fig. 3. The proteins that were unconnected in the network were omitted. On this network, nodes (proteins) with the additional overlaid red and blue circles are proteins identified with 2D DIGE/MS in this study. The red and the blue color of the circles represents the direction of the changes in the protein expression, up (red) and down (blue) with STZ-induced diabetes. The edges with arrowheads describe the nature and direction of the interaction. The colored hexagons on the edge describe the mechanism and the effect of interaction between proteins on the network, green for activation, red for inhibition, and black for unspecified.
For instance, according to this network (Fig. 3), the STZ-induced increase in hnRNP K is correlated with induction of cellular tumor antigen p53, nuclease-sensitive element-binding protein 1 (YB-1), proto-oncogene tyrosine-protein kinase Src (c-Src), and Myc proto-oncogene (c-Myc). Although none of these binding proteins and transcriptional factors was specifically detected with 2D DIGE/MS experiments, the nodes around these core proteins were identified by 2D DIGE/MS and showed a consistent up- and down-regulation in agreement with the predicted interactions on the network. Thus, the networks pointed toward several core transcriptional factors and binding proteins that hypothetically could be involved in diabetes-mediated bladder dysfunction but might have been unnoticed based solely on the list of proteins identified by 2D DIGE/MS (Table II).
To understand the biological relevance of the differentially expressed proteins and their -fold change implications in the context of diabetes-associated bladder dysfunction, the list of proteins with their expression data were visualized on MetaCore pre-existing pathway maps. The result of this mapping revealed the involvement of a set of structural and extracellular matrix proteins in statistically significant pathway maps including integrin-mediated cell adhesion, integrin outside-in signaling, cytoskeleton remodeling, neurofilaments, and extracellular matrix (ECM) remodeling pathway maps.
Validation by Immunoblotting—
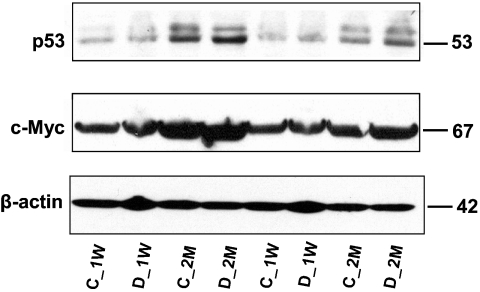
To further evaluate the nature and importance of the observed changes in protein expression listed in Table II, some of the critical transcriptional factors that were identified by network analysis were selected for semiquantitative immunoblotting. This list of targets includes p53, c-Myc, and c-Src. The relative -fold changes for these proteins were computed using ImageQuant-TL v2005 software and are summarized in Table III. At both the 1-week and 2-month time periods the relative abundance of p53 increased by about 1.5-fold in STZ-induced diabetic rats compared with age-matched controls (Fig. 4), whereas the relative abundance of c-Myc increased by only 1.2-fold in both early and late phases of the disease. The expression of c-Myc as predicted by the network analysis (Fig. 3) is regulated not only by hnRNP K but also by α-enolase in the opposite direction. Specifically the induction of α-enolase with STZ-induced diabetes may have inhibited the induction of c-Myc even though the induction of hnRNP K with STZ-induced diabetes activated c-Myc transcription. These immunoblot results for the two transcriptional factors are consistent with the network analysis prediction. These data are also consistent with the existing literature documenting that the induction of the tumor suppressor p53 is involved in negative regulation of c-Myc expression (37), although there are also reported cases where both p53 and c-Myc are co-expressed in colorectal and oral cancer (38, 39).
Table III.
Relative-fold changes for selected proteins that were identified either by 2D DIGE/MS and/or by network analysis as determined by conformational immunoblot analysis
| Proteins | STZ-induced diabetic/control a
|
|
|---|---|---|
| 1 week | 2 months | |
| α-Enolase | 1.11 ± 0.103 | 1.54 ± 0.049 |
| hnRNP K | 1.15 ± 0.025 | 1.30 ± 0.058 |
| Ly-GDI | 2.28 ± 0.050 | 1.62 ± 0.062 |
| p53b | 1.44 ± 0.067 | 1.49 ± 0.033 |
| c-Mycb | 1.17 ± 0.067 | 1.20 ± 0.049 |
| β-Actin | 1.03 ± 0.014 | 0.94 ± 0.020 |
Average of immunoblots from two independent biological replicates of the STZ-induced diabetic rat model or age-matched control.
Selected proteins that were identified by the network analysis.
Fig. 4.
Conformational immunoblots for selected proteins that were hypothetically identified by network analysis. Each lane is loaded with a sample from independent biological replicate (n = 2/experimental group). The blots were probed with antibodies to the proteins indicated at the left. C_1W, 1-week control; D_1W, 1-week diabetes; C_2M, 2-month control; and D_2M, 2-month diabetes.
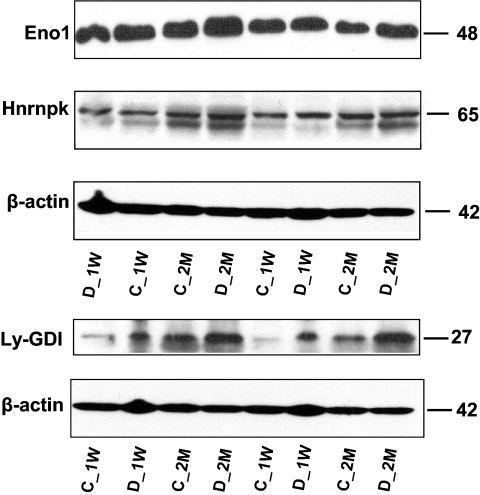
Three proteins that were identified by 2D DIGE were selected for semiquantitative immunoblotting on the basis of the availability of high quality antibodies. These include Rho-GDP dissociation inhibitor (GDI) β (Ly-GDI), enolase, and hnRNP K. Western blots, shown in Fig. 5, verified that both the direction and -fold changes for Eno1 and hnRNP K are consistent with the 2D DIGE results. However, for Ly-GDI, 2D DIGE showed no significant change at the 1-week time point, whereas the immunoblot showed about 2.28-fold up-regulation relative to the control. Despite this discrepancy, both 2D DIGE and immunoblot gave relatively similar results for Ly-GDI at the 2-month time period. In general immunoblotting confirmed the quantitative results detected by 2D DIGE as well as the theoretical predictions obtained by network analysis.
Fig. 5.
Conformational immunoblots for the selected proteins (Eno1, hnRNP K, and Ly-GDI) that were identified as differentially expressed by 2D DIGE. STZ-induced diabetic rat bladders at two time points was compared with age-matched control rats to identify changes in protein abundances. 50 μg of total protein was loaded in each lane, and the resulting blots were probed with anti-enolase, anti-hnRNP K, and anti-Ly-GDI. Relative -fold changes for these proteins in STZ-induced diabetic rats compared with the age-matched controls are given in Table III. C_1W, 1-week control; D_1W, 1-week diabetes; C_2M, 2-month control; and D_2M, 2-month diabetes.
DISCUSSION
Diabetes mellitus causes changes in both the function and morphology of the bladder. In exploring the possible causes of these changes at the proteome level during the initiation and progression of the disease, we identified multiple changes in protein expression that are consistent with the development of bladder dysfunction. The changes in expression for 85% of the proteins were identified at 2 months after the confirmation of STZ-induced hyperglycemia. Interestingly a recent report by Hipp et al. (20) using microarray analysis of gene expression after 1 week of diabetes in the same animal model demonstrated changes in expression of >300 genes. There are several possible explanations for the magnitude of the observed differences in protein and gene expression at the same time point in the same animal model. 1) Genes encoding membrane proteins detected by microarray would not be present in the soluble fraction of proteins used in the proteome analysis, 2) the resolution of genes is substantially greater on the microarray (genes are represented by a synthesized array of delineated nucleotides), than by 2D DIGE where overlapping spots could mask changes in protein expression, and 3) RNA turnover rates are faster than protein turnover. Although streptozotocin is a strong alkylating agent and can have nonspecific toxicity (40) besides an effect on insulin production causing hyperglycemia, only a very few proteins are changed in common at the two time points, suggesting that the effect of streptozotocin toxicity on protein expression is limited compared with the effects of an extended period of hyperglycemia.
Down-regulated Theme—
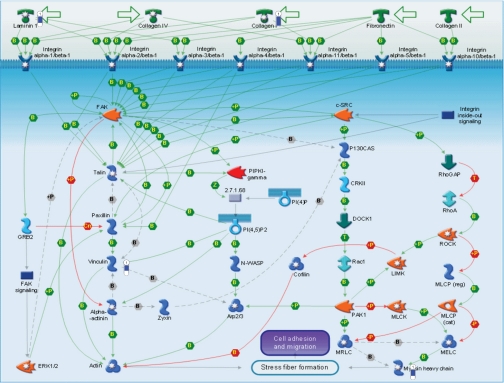
Down-regulation was observed for a group of structural and extracellular proteins with similar biological functions or involved in similar biochemical pathways. Examples of such STZ-induced alterations in protein expression include integrin-mediated cell adhesion, extracellular matrix remodeling, and cytoskeleton remodeling. Structural proteins such as vinculin, filamin a/c, and extracellular matrix proteins laminin γ1, nidogen, procollagens (in different isoforms; Table II), and protein phosphatase 2 (Ppp2r1a) are all down-regulated in STZ-induced diabetic rat models. Most of these proteins are known to regulate the very complex and dynamic processes of cell adhesion and migration (41–43). Four of these proteins are involved in the integrin-mediated cell adhesion pathway (Fig. 6), and essentially all of them were down-regulated. Within this group, vinculin is a cytoskeletal protein found in focal adhesion plaques. It is involved in linkage of integrin adhesion molecules to the actin cytoskeleton (44). In the past it has been shown that the loss of vinculin impacts a variety of cell functions, e.g. disruption of the formation of focal adhesions and prevention of cell adhesion and spreading (41, 45). Vinculin plays a key role in shape control based on its ability to modulate focal adhesion structure and function. Therefore, the decrease in adaptor proteins such as vinculin in diabetic bladder may modify the composition and behavior of focal adhesion complex and hence the mechanical link between the actin cytoskeleton and integrin. Potentially this could have a negative impact on integrin-mediated cell adhesion, disrupting signal transduction for migration and contraction of the cells, and therefore would be expected to be of great relevance to the development of bladder pathology/dysfunction.
Fig. 6.
Integrin-mediated cell adhesion pathway showing the response of some of the proteins on the map to STZ-induced diabetes. The various proteins on this map are represented by different symbols (representing the functional class of the protein). Thermometers with blue or red shading next to symbols depict proteins identified in the present study: blue color represents the proteins that were down in the STZ-induced diabetic relative to control group. P indicates phosphorylation, B indicates binding, Cn, competition; T, transformation; MLCK, myosin light chain kinase; ROCK, Rho-associated kinase; GAP, GTPase-activating protein; LIMK, LIM kinase; FAK, focal adhesion kinase; MLCP (cat), myosin light chain phosphatase, catalytic; MLCP (reg), myosin light chain phosphatase, regulatory; MRLC, myosin regulatory light chain; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(4)P, phosphatidylinositol 4-phosphate; PIPKI, phosphatidylinositol phosphate kinase type I; N-WASP, neural Wiskott-Aldrich syndrome protein; MELC, myosin essential light chain; CRKII, v-crk sarcoma virus CT10 oncogene homolog.
Although structural proteins such as vinculin function as physical links to the actin cytoskeleton, signaling molecules such as Ppp2r1a modulate integrin association to the cytoskeletal actin through tyrosine phosphorylation and inhibitory serine/threonine phosphorylation of β1 integrin for its cell adhesive function. As such, Ppp2r1a has an important role in the integrin-mediated cell adhesion pathways (46). In this scenario, a reduced expression of Ppp2r1a in diabetic rat bladder can lead to the serine/threonine phosphorylation of β1 integrin, which in turn may prevent integrin-actin association and lead to reduced cell adhesion.
In addition to the group of cytoskeletal and signaling proteins discussed above, several common ECM proteins that help the cell to endure stretching forces without being ripped out of the extracellular matrix, such as collagens (in different isoforms), nidogen, and laminin γ1 were down-regulated by up to 4-fold with STZ-induced diabetes. Our result is consistent with the previous studies that have shown decreases in collagen content in an enlarged rat urinary bladder due to diabetes complications (14, 47). The role of the cell adhesion site in maintaining the ECM components at the normal physiological level in tissues is as important as its role in modulating migration, contractility, gene expression, and cell fate (43, 48). Thus, the observed progressive decrease in the amount of the ECM in STZ-induced diabetic rat bladder can have negative consequences as it leads to loss of integrin binding sites or adhesion contacts. This may result in smooth muscle cells being unable to form adhesive contacts and stress fibers on the ECM.
Up-regulated Themes—
Proteins that were up-regulated in response to STZ-induced hyperglycemia include Mrlcb and Rho-GDIβ. Mrlcb is a major player of signal transduction in lower urinary tract smooth muscle (49). It is well established that an increase in the free intracellular calcium level is a prerequisite for the phosphorylation of Mrlcb and for the subsequent contraction of smooth muscle; however, the ability of the smooth muscle to sustain the contractile response is mediated by the Ca2+ sensitization pathway (50). In this pathway the phosphorylated Mrlcb is maintained at low intracellular Ca2+ concentration through regulation of myosin light chain phosphatase that involves small GTPase Rho (Ras-like GTP-binding proteins) and one of their downstream effectors, Rho-associated kinase (50–54). In this study the Mrlcb appeared at two positions on the gel, and both isoforms increased by about 80% in the early time point but not at the late time point after STZ-induced diabetes. The position of one of the two isoforms of the protein is actually shifted to the acidic side of the gel (Fig. 1) as compared with the protein theoretical pI, an indication of post-translational modification, most likely phosphorylation. Consistent with this result, Hipp et al. (20) recently reported an ≈3.4-fold increase of mRNA for this protein after 1 week of STZ-induced diabetes in the same rat model. The induction of myosin regulatory light chain in type 2 diabetes mice stomach tissue has also been reported (55). The increase in Mrlcb and its subsequent phosphorylation via either of the aforementioned pathways in 1-week STZ-induced diabetic rat compared with age-matched control reflects the presence of an imbalance between the myosin light chain kinase and myosin light chain phosphatase. This imbalance may lead to abnormal contraction of the detrusor and thereby play a role in, for example, diabetes-related detrusor overactivity. These results are consistent with epidemiological data that confirm the link between diabetes and bladder hyperactivity (3, 56) at the early times but failed to confirm it in subsequent disease progression. In addition, we found an STZ-induced diabetes-mediated induction of Rho-GDIβ but not Rho-GDIα. Rho-GDIβ has 67% sequence similarity to Rho-GDPα. Both isoforms of Rho-GDP dissociation inhibitors tightly regulate G-protein activities by inhibiting both the basal and guanine exchange protein; this protein stimulates dissociation of GDP from the GDP-bound form. This inhibition maintains the G-proteins in the GDP-bound form. It has been shown that Rho-GDIβ is expressed preferentially in lymphocytes and shows a preferential inhibition effect toward GTPases such as Cdc42 but is less potent than Rho-GDIα (57, 58). From our data we suggest that the increase in Rho-GDIβ is perhaps associated with the observed decrease of Mrlcb to the basal level (control level) at a later time point in diabetes. However, this is a hypothesis that requires further investigation.
Insulin regulates uptake of glucose from the blood primarily to the muscle and fat cells; deficiency of insulin plays a central role in many metabolic disorders. Although the actions of insulin are quite complex and involve glucose transporters, metabolic enzymes, and transcriptional regulation, our study detected only a few changes in proteins that are involved in glucose metabolism, including Ehd2, Sdha, Taldo1, and Enol. This could be due to the fact that diabetes-mediated changes in glucose metabolism could mainly be brought about via post-translational mechanisms (59), or it could also be associated with the partial proteome coverage of 2D DIGE. A recent report by Park et al. (60) demonstrated Ehd2 interactions with the insulin-responsive glucose transporter (Glut4) in rat adipocytes and suggested a key role in insulin-induced Glut4 recruitment to the plasma membrane. A decrease in the expression of Ehd2 identified in the present study may impair Ehd2-mediated insulin-dependent Glut4 transporter endocytosis and thus account for a decrease in the glucose uptake in the STZ-induced diabetic rat bladder smooth muscle cells. The reduced level of Sdha may also be associated with a decreased Ehd2 level. Because enolase catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate in the glycolysis pathway an increase in enolase after 2 months but not after 1 week of STZ-induced diabetes could be a cellular protective mechanism to counterbalance hypoxia (61, 62) in long standing diabetes mellitus.
In addition, several studies have demonstrated that enolase is a multifunctional protein, playing a role in various biological processes such as growth control (62). Up-regulation of Eno1 has been reported in highly tumorigenic and metastatic cell lines (63–65) and in lung cancer patients is tightly correlated with poor survival outcome (66). Its induction in tumor cells relative to normal cells has been associated with enhanced proliferative status of the cell rather than specifically relating to a neoplastic phenotype (62). It has also been demonstrated that the same gene encoding for Eno1 also encodes an alternative translated product, MBP-1, and both Eno1 and MBP-1 appear to function as negative regulators of c-myc proto-oncogene transcription (67). Thus the induction of Eno1 and p53, as identified by 2D DIGE/MS and network analysis, respectively, and further confirmed by Western blotting, could be protective responses designed to moderate the effects associated with the induction of c-Myc in STZ-induced diabetic rat bladder. Collectively the induction of Eno1, p53, and hnRNP, which has previously been associated with abnormal proliferation of the cell (68), with diabetes could have implications to the often observed enlargement of the bladder in diabetic patients/model animals.
After 2 months of STZ-induced diabetes, a group of calcium-binding proteins that cause inflammation, such as S100A9, and proteins demonstrated to have anti-inflammatory responses, including two isoforms of Annexin (1 and 8, respectively), apoA-I, and apoA-IV (69–71), were up-regulated in the diabetic bladder. The expression of anti-inflammatory proteins could be secondary to the increased expression of MRP14 (S100A9), one of the two calcium-binding proteins that are specifically released by monocytes during the course of inflammatory reactions in various human diseases (72). It is most likely that the observed bladder urothelium layer erosion we observed2 could be associated with the induction of S100A9 following STZ-induced diabetes.
In summary, perhaps it is not surprising that diabetes differentially regulated far more proteins at the 2-month time period than at the 1-week time period because this is a time point when the animals are developing marked bladder pathophysiology (21, 22). Our data indicated that diabetes-induced bladder dysfunction is brought about by influencing the production of structural and extracellular proteins that are important for shape control, adhesion, and motility of the cell while inducing proteins that are involved in cell proliferation and inflammation. Our study also revealed additional proteins that, as far as we are aware, have not been reported previously to be altered in diabetes, including hnRNP and Ly-GDI. Although few proteins are altered by diabetes in the early stages (i.e. 1 week) of the disease process, proteins that showed temporal changes, such as Mrlcb, are interesting because they improve our understanding of hyperactive bladder during the initiation of the disease. Thus, this group of proteins can actually be used as markers to classify the bladder pathology. It is also important to mention that the 2D DIGE proteomics approach gave us statistically significant quantitative information from four biological replicate samples per experimental group on well over 55 proteins at the same time. However, proteins outside the molecular weight range and hydrophobic proteins difficult to resolve using 2D gel techniques could not be surveyed in this study. Nonetheless this work demonstrates the importance of using 2D DIGE techniques to analyze proteome expression in diabetic tissues. Potentially the technique, as described and applied herein, has some advantages over transcript analysis (i.e. microarray) especially because the technique is responsive to post-transcriptional regulation of protein expression, has the ability to detect changes in protein isoforms derived from splice variants, and can detect changes in proteins that result from post-translational modification.
Supplementary Material
Footnotes
Published, MCP Papers in Press, March 12, 2008, DOI 10.1074/mcp.M700563-MCP200
The abbreviations used are: DM, diabetes mellitus; STZ, streptozotocin; 2D, two-dimensional; DIA, differential in-gel analysis; BVA, biological variation analysis; ANOVA, analysis of variance; PCA, principal component analysis; ECM, extracellular matrix; hnRNP, heterogeneous nuclear ribonucleoprotein; BP, band pass; GDI, GDP dissociation inhibitor; Mrlcb, myosin regulatory light chain 2-B smooth muscle isoform; Ly-GDI, Rho, GDP dissociation inhibitor (GDI) β.
G. J. Christ, unpublished result.
This work was supported, in whole or in part, by National Institutes of Health Grants R21 DK-070229 and P01-DK060037 from the NIDDK and Grant P41-EB-1979 from the NIBIB. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Brown, J. S., Wessells, H., Chancellor, M. B., Howards, S. S., Stamm, W. E., Stapleton, A. E., Steers, W. D., Van Den Eeden, S. K., and McVary, K. T. ( 2005) Urologic complications of diabetes. Diabetes Care 28, 177–185 [DOI] [PubMed] [Google Scholar]
- 2.Faerman, I., Maler, M., Jadzinsky, M., Alvarez, E., Fox, D., Zilbervarg, J., Cibeira, J. B., and Colinas, R. ( 1971) Asymptomatic neurogenic bladder in juvenile diabetics. Diabetologia 7, 168–172 [DOI] [PubMed] [Google Scholar]
- 3.Salinas Casado, J., Virseda Chamorro, M., Adot Zurbano, J. M., Teba del Pino, F., Arredondo Martinez, F., Hernandez Lao, A., Fernandez Lucas, C., and Herrero Payo, A. ( 1999) Diabetic neurocystopathy: multivariate study. Arch. Esp. Urol. 52, 149–156 [PubMed] [Google Scholar]
- 4.Menendez, V., Cofan, F., Talbot-Wright, R., Ricart, M. J., Gutierrez, R., and Carretero, P. ( 1996) Urodynamic evaluation in simultaneous insulin-dependent diabetes mellitus and end stage renal disease. J. Urol. 155, 2001–2004 [PubMed] [Google Scholar]
- 5.Van Poppel, H., Stessens, R., Van Damme, B., Carton, H., and Baert, L. ( 1988) Diabetic cystopathy: neuropathological examination of urinary bladder biopsies. Eur. Urol. 15, 128–131 [DOI] [PubMed] [Google Scholar]
- 6.Liu, G., and Daneshgari, F. ( 2005) Alterations in neurogenically mediated contractile responses of urinary bladder in rats with diabetes. Am. J. Physiol. 288, F1220–F1226 [DOI] [PubMed] [Google Scholar]
- 7.Poladia, D. P., Schanbacher, B., Wallace, L. J., and Bauer, J. A. ( 2005) Innervation and connexin isoform expression during diabetes-related bladder dysfunction: early structural vs. neuronal remodelling. Acta Diabetol. 42, 147–152 [DOI] [PubMed] [Google Scholar]
- 8.Bianchi, R., Triban, C., Marini, P., Figliomeni, B., Paro, M., Italiano, G., Prosdocimi, M., and Fiori, M. G. ( 1991) Inner ester derivatives of gangliosides protect autonomic nerves of alloxan-diabetic rats against Na+, K+-ATPase activity defects. Diabetes Res. Clin. Pract. 12, 107–111 [DOI] [PubMed] [Google Scholar]
- 9.Hohman, T. C., Cotter, M. A., and Cameron, N. E. ( 2000) ATP-sensitive K+ channel effects on nerve function, Na+, K+ ATPase, and glutathione in diabetic rats. Eur. J. Pharmacol. 397, 335–341 [DOI] [PubMed] [Google Scholar]
- 10.Sasaki, K., Chancellor, M. B., Phelan, M. W., Yokoyama, T., Fraser, M. O., Seki, S., Kubo, K., Kumon, H., Groat, W. C., and Yoshimura, N. ( 2002) Diabetic cystopathy correlates with a long-term decrease in nerve growth factor levels in the bladder and lumbosacral dorsal root ganglia. J. Urol. 168, 1259–1264 [DOI] [PubMed] [Google Scholar]
- 11.Hellweg, R., and Hartung, H. D. ( 1990) Endogenous levels of nerve growth factor (NGF) are altered in experimental diabetes mellitus: a possible role for NGF in the pathogenesis of diabetic neuropathy. J. Neurosci. Res. 26, 258–267 [DOI] [PubMed] [Google Scholar]
- 12.Sasaki, K., Chancellor, M. B., Goins, W. F., Phelan, M. W., Glorioso, J. C., de Groat, W. C., and Yoshimura, N. ( 2004) Gene therapy using replication-defective herpes simplex virus vectors expressing nerve growth factor in a rat model of diabetic cystopathy. Diabetes 53, 2723–2730 [DOI] [PubMed] [Google Scholar]
- 13.Goins, W. F., Yoshimura, N., Phelan, M. W., Yokoyama, T., Fraser, M. O., Ozawa, H., Bennett, N. J., de Groat, W. C., Glorioso, J. C., and Chancellor, M. B. ( 2001) Herpes simplex virus mediated nerve growth factor expression in bladder and afferent neurons: potential treatment for diabetic bladder dysfunction. J. Urol. 165, 1748–1754 [PubMed] [Google Scholar]
- 14.Pitre, D. A., Ma, T., Wallace, L. J., and Bauer, J. A. ( 2002) Time-dependent urinary bladder remodeling in the streptozotocin-induced diabetic rat model. Acta Diabetol. 39, 23–27 [DOI] [PubMed] [Google Scholar]
- 15.Kosan, M., Hafez, G., Ozturk, B., Ozgunes, O., Gur, S., and Cetinkaya, M. ( 2005) Effect of urothelium on bladder contractility in diabetic rats. Int. J. Urol. 12, 677–682 [DOI] [PubMed] [Google Scholar]
- 16.Poladia, D. P., and Bauer, J. A. ( 2004) Oxidant driven signaling pathways during diabetes: role of Rac1 and modulation of protein kinase activity in mouse urinary bladder. Biochimie (Paris) 86, 543–551 [DOI] [PubMed] [Google Scholar]
- 17.Poladia, D. P., and Bauer, J. A. ( 2003) Early cell-specific changes in nitric oxide synthases, reactive nitrogen species formation, and ubiquitinylation during diabetes-related bladder remodeling. Diabetes Metab. Res. Rev. 19, 313–319 [DOI] [PubMed] [Google Scholar]
- 18.Stothers, L., Laher, I., and Christ, G. T. ( 2003) A review of the l-arginine-nitric oxide-guanylate cyclase pathway as a mediator of lower urinary tract physiology and symptoms. Can. J. Urol. 10, 1971–1980 [PubMed] [Google Scholar]
- 19.Levin, R. M., Wein, A. J., Buttyan, R., Monson, F. C., and Longhurst, P. A. ( 1994) Update on bladder smooth-muscle physiology. World J. Urol. 12, 226–232 [DOI] [PubMed] [Google Scholar]
- 20.Hipp, J. D., Davies, K. P., Tar, M., Valcic, M., Knoll, A., Melman, A., and Christ, G. J. ( 2007) Using gene chips to identify organ-specific, smooth muscle responses to experimental diabetes: potential applications to urological diseases. BJU Int. 99, 418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daneshgari, F., Liu, G., and Imrey, P. B. ( 2006) Time dependent changes in diabetic cystopathy in rats include compensated and decompensated bladder function. J. Urol. 176, 380–386 [DOI] [PubMed] [Google Scholar]
- 22.Christ, G. J., Hsieh, Y., Zhao, W., Schenk, G., Venkateswarlu, K., Wang, H. Z., Tar, M. T., and Melman, A. ( 2006) Effects of streptozotocin-induced diabetes on bladder and erectile (dys)function in the same rat in vivo. BJU Int. 97, 1076–1082 [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura, N., Chancellor, M. B., Andersson, K. E., and Christ, G. J. ( 2005) Recent advances in understanding the biology of diabetes-associated bladder complications and novel therapy. BJU Int. 95, 733–738 [DOI] [PubMed] [Google Scholar]
- 24.Benko, R., Lazar, Z., Porszasz, R., Somogyi, G. T., and Bartho, L. ( 2003) Effect of experimental diabetes on cholinergic, purinergic and peptidergic motor responses of the isolated rat bladder to electrical field stimulation or capsaicin. Eur. J. Pharmacol. 478, 73–80 [DOI] [PubMed] [Google Scholar]
- 25.Bezuijen, M. W., Levendusky, M. C., and Longhurst, P. A. ( 2003) Functional response of bladder strips from streptozotocin diabetic rats depends on bladder mass. J. Urol. 169, 2397–2401 [DOI] [PubMed] [Google Scholar]
- 26.Zotova, E. G., Christ, G. J., Zhao, W., Tar, M., Kuppam, S. D., and Arezzo, J. C. ( 2007) Effects of fidarestat, an aldose reductase inhibitor, on nerve conduction velocity and bladder function in streptozotocin-treated female rats. J. Diabetes Complicat. 21, 187–195 [DOI] [PubMed] [Google Scholar]
- 27.Alban, A., David, S. O., Bjorkesten, L., Andersson, C., Sloge, E., Lewis, S., and Currie, I. ( 2003) A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3, 36–44 [DOI] [PubMed] [Google Scholar]
- 28.Friedman, D. B., Hill, S., Keller, J. W., Merchant, N. B., Levy, S. E., Coffey, R. J., and Caprioli, R. M. ( 2004) Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 4, 793–811 [DOI] [PubMed] [Google Scholar]
- 29.Lilley, K. S., and Friedman, D. B. ( 2004) All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteomics 1, 401–409 [DOI] [PubMed] [Google Scholar]
- 30.Nikolsky, Y., Ekins, S., Nikolskaya, T., and Bugrim, A. ( 2005) A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 158, 20–29 [DOI] [PubMed] [Google Scholar]
- 31.Mason, C. W., Swaan, P. W., and Weiner, C. P. ( 2006) Identification of interactive gene networks: a novel approach in gene array profiling of myometrial events during guinea pig pregnancy. Am. J. Obstet. Gynecol. 194, 1513–1523 [DOI] [PubMed] [Google Scholar]
- 32.Herbert, B., Galvani, M., Hamdan, M., Olivieri, E., MacCarthy, J., Pedersen, S., and Righetti, P. G. ( 2001) Reduction and alkylation of proteins in preparation of two-dimensional map analysis: why, when, and how? Electrophoresis 22, 2046–2057 [DOI] [PubMed] [Google Scholar]
- 33.Galvani, M., Hamdan, M., Herbert, B., and Righetti, P. G. ( 2001) Alkylation kinetics of proteins in preparation for two-dimensional maps: a matrix assisted laser desorption/ionization-mass spectrometry investigation. Electrophoresis 22, 2058–2065 [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 35.Yeung, K. Y., and Ruzzo, W. L. ( 2001) Principal component analysis for clustering gene expression data. Bioinformatics (Oxf.) 17, 763–774 [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 37.Ho, J. S., Ma, W., Mao, D. Y., and Benchimol, S. ( 2005) p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 25, 7423–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baral, R., Patnaik, S., and Das, B. R. ( 1998) Co-overexpression of p53 and c-myc proteins linked with advanced stages of betel- and tobacco-related oral squamous cell carcinomas from eastern India. Eur. J. Oral Sci. 106, 907–913 [DOI] [PubMed] [Google Scholar]
- 39.Rochlitz, C. F., Heide, I., Thiede, C., Herrmann, R., and de Kant, E. ( 1995) Evidence for a mutual regulation of p53 and c-myc expression in human colorectal cancer metastases. Ann. Oncol. 6, 981–986 [DOI] [PubMed] [Google Scholar]
- 40.Bennett, R. A., and Pegg, A. E. ( 1981) Alkylation of DNA in rat tissues following administration of streptozotocin. Cancer Res. 41, 2786–2790 [PubMed] [Google Scholar]
- 41.Ezzell, R. M., Goldmann, W. H., Wang, N., Parasharama, N., and Ingber, D. E. ( 1997) Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 231, 14–26 [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, K., and Takahashi, K. ( 2003) Reduced cell adhesion during mitosis by threonine phosphorylation of β1 integrin. J. Cell. Physiol. 197, 297–305 [DOI] [PubMed] [Google Scholar]
- 43.Geiger, B., Bershadsky, A., Pankov, R., and Yamada, K. M. ( 2001) Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat. Rev. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 44.Janssen, M. E., Kim, E., Liu, H., Fujimoto, L. M., Bobkov, A., Volkmann, N., and Hanein, D. ( 2006) Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell 21, 271–281 [DOI] [PubMed] [Google Scholar]
- 45.Goldmann, W. H., and Ingber, D. E. ( 2002) Intact vinculin protein is required for control of cell shape, cell mechanics, and rac-dependent lamellipodia formation. Biochem. Biophys. Res. Commun. 290, 749–755 [DOI] [PubMed] [Google Scholar]
- 46.Takahashi, K. ( 2001) The linkage between β1 integrin and the actin cytoskeleton is differentially regulated by tyrosine and serine/threonine phosphorylation of β1 integrin in normal and cancerous human breast cells. BMC Cell Biol. 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uvelius, B., and Mattiasson, A. ( 1984) Collagen content in the rat urinary bladder subjected to infravesical outflow obstruction. J. of Urol. 132, 587–590 [DOI] [PubMed] [Google Scholar]
- 48.Hynes, R. O. ( 2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 49.Christ, G. J., and Andersson, K. E. ( 2007) Rho-kinase and effects of Rho-kinase inhibition on the lower urinary tract. Neurourol. Urodyn. 26, 948–954 [DOI] [PubMed] [Google Scholar]
- 50.Somlyo, A. P., and Somlyo, A. V. ( 2003) Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol. Rev. 83, 1325–1358 [DOI] [PubMed] [Google Scholar]
- 51.Horowitz, A., Menice, C. B., Laporte, R., and Morgan, K. G. ( 1996) Mechanisms of smooth muscle contraction. Physiol. Rev. 76, 967–1003 [DOI] [PubMed] [Google Scholar]
- 52.Somlyo, A. P., and Somlyo, A. V. ( 2000) Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J. Physiol. 522, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballestrem, C., Hinz, B., Imhof, B. A., and Wehrle-Haller, B. ( 2001) Marching at the front and dragging behind: differential αVβ3-integrin turnover regulates focal adhesion behavior. Journal Cell Biol. 155, 1319–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Somlyo, A. P., and Somlyo, A. V. ( 1994) Signal transduction and regulation in smooth muscle. Nature 372, 231–236 [DOI] [PubMed] [Google Scholar]
- 55.List, E. O., Berryman, D. E., Palmer, A. J., Gosney, E., Okada, S., Kelder, B., Lichtenberg, J., Welch, L. R., and Kopchick, J. J. ( 2007) Application of bioinformatics and scalable computing to perform proteomic analysis of stomach tissue from diabetic mice. Scalable Comput. Pract. Exp. 8, 173–183 [Google Scholar]
- 56.Brown, J. S., Nyberg, L. M., Kusek, J. W., Burgio, K. L., Diokno, A. C., Foldspang, A., Fultz, N. H., Herzog, A. R., Hunskaar, S., Milsom, I., Nygaard, I., Subak, L. L., and Thom, D. H. ( 2003) Proceedings of the National Institute of Diabetes and Digestive and Kidney Diseases International Symposium on Epidemiologic Issues in Urinary Incontinence in Women. Am. J. Obstet. Gynecol. 188, S77–S88 [DOI] [PubMed] [Google Scholar]
- 57.Nomanbhoy, T. K., and Cerione, R. ( 1996) Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. Journal Biol. Chem. 271, 10004–10009 [DOI] [PubMed] [Google Scholar]
- 58.Scherle, P., Behrens, T., and Staudt, L. M. ( 1993) Ly-GDI, a GDP-dissociation inhibitor of the RhoA GTP-binding protein, is expressed preferentially in lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 90, 7568–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saltiel, A. R., and Kahn, C. R. ( 2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]
- 60.Park, S. Y., Ha, B. G., Choi, G. H., Ryu, J., Kim, B., Jung, C. Y., and Lee, W. ( 2004) EHD2 interacts with the insulin-responsive glucose transporter (GLUT4) in rat adipocytes and may participate in insulin-induced GLUT4 recruitment. Biochemistry 43, 7552–7562 [DOI] [PubMed] [Google Scholar]
- 61.Semenza, G. L., Jiang, B. H., Leung, S. W., Passantino, R., Concordet, J. P., Maire, P., and Giallongo, A. ( 1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 62.Wang, T., Marquardt, C., and Foker, J. ( 1976) Aerobic glycolysis during lymphocyte proliferation. Nature 261, 702–705 [DOI] [PubMed] [Google Scholar]
- 63.Peebles, K. A., Duncan, M. W., Ruch, R. J., and Malkinson, A. M. ( 2003) Proteomic analysis of a neoplastic mouse lung epithelial cell line whose tumorigenicity has been abrogated by transfection with the gap junction structural gene for connexin 43, Gja1. Carcinogenesis 24, 651–657 [DOI] [PubMed] [Google Scholar]
- 64.Zhang, L., Cilley, R. E., and Chinoy, M. R. ( 2000) Suppression subtractive hybridization to identify gene expressions in variant and classic small cell lung cancer cell lines. J. Surg. Res. 93, 108–119 [DOI] [PubMed] [Google Scholar]
- 65.Wu, W., Tang, X., Hu, W., Lotan, R., Hong, W. K., and Mao, L. ( 2002) Identification and validation of metastasis-associated proteins in head and neck cancer cell lines by two-dimensional electrophoresis and mass spectrometry. Clin. Exp. Metastasis 19, 319–326 [DOI] [PubMed] [Google Scholar]
- 66.Chang, G. C., Liu, K. J., Hsieh, C. L., Hu, T. S., Charoenfuprasert, S., Liu, H. K., Luh, K. T., Hsu, L. H., Wu, C. W., Ting, C. C., Chen, C. Y., Chen, K. C., Yang, T. Y., Chou, T. Y., Wang, W. H., Whang-Peng, J., and Shih, N. Y. ( 2006) Identification of α-enolase as an autoantigen in lung cancer: its overexpression is associated with clinical outcomes. Clin. Cancer Res. 12, 5746–5754 [DOI] [PubMed] [Google Scholar]
- 67.Subramanian, A., and Miller, D. M. ( 2000) Structural analysis of α-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J. Biol. Chem. 275, 5958–5965 [DOI] [PubMed] [Google Scholar]
- 68.Tockman, M. S., Mulshine, J. L., Piantadosi, S., Erozan, Y. S., Gupta, P. K., Ruckdeschel, J. C., Taylor, P. R., Zhukov, T., Zhou, W. H., Qiao, Y. L., and Yao, S. X. ( 1997) Prospective detection of preclinical lung cancer: results from two studies of heterogeneous nuclear ribonucleoprotein A2/B1 overexpression. Clin. Cancer Res. 3, 2237–2246 [PubMed] [Google Scholar]
- 69.Flower, R. J., and Rothwell, N. J. ( 1994) Lipocortin-1: cellular mechanisms and clinical relevance. Trends Pharmacol. Sci. 15, 71–76 [DOI] [PubMed] [Google Scholar]
- 70.Solito, E., Romero, I. A., Marullo, S., Russo-Marie, F., and Weksler, B. B. ( 2000) Annexin 1 binds to U937 monocytic cells and inhibits their adhesion to microvascular endothelium: involvement of the α4β1 integrin. J. Immunol. 165, 1573–1581 [DOI] [PubMed] [Google Scholar]
- 71.List, E. O., Berryman, D. E., Palmer, A. J., Qiu, L., Sankaran, S., Kohn, D. T., Kelder, B., Okada, S., and Kopchick, J. J. ( 2007) Analysis of mouse skin reveals proteins that are altered in a diet-induced diabetic state: a new method for detection of type 2 diabetes. Proteomics 7, 1140–1149 [DOI] [PubMed] [Google Scholar]
- 72.Seeliger, S., Vogl, T., Engels, I. H., Schroder, J. M., Sorg, C., Sunderkotter, C., and Roth, J. ( 2003) Expression of calcium-binding proteins MRP8 and MRP14 in inflammatory muscle diseases. Am. J. Pathol. 163, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.