Abstract
The many layers and structures of the skin serve as elaborate hosts to microbes, including a diversity of commensal and pathogenic bacteria that contribute to both human health and disease. To determine the complexity and identity of the microbes inhabiting the skin, we sequenced bacterial 16S small-subunit ribosomal RNA genes isolated from the inner elbow of five healthy human subjects. This analysis revealed 113 operational taxonomic units (OTUs; “phylotypes”) at the level of 97% similarity that belong to six bacterial divisions. To survey all depths of the skin, we sampled using three methods: swab, scrape, and punch biopsy. Proteobacteria dominated the skin microbiota at all depths of sampling. Interpersonal variation is approximately equal to intrapersonal variation when considering bacterial community membership and structure. Finally, we report strong similarities in the complexity and identity of mouse and human skin microbiota. This study of healthy human skin microbiota will serve to direct future research addressing the role of skin microbiota in health and disease, and metagenomic projects addressing the complex physiological interactions between the skin and the microbes that inhabit this environment.
The skin barrier is critical for survival, preventing the escape of moisture and invasion by infectious or toxic substances (Segre 2006). The skin is also an intricate habitat for a diverse population of microbiota. During the birthing process and subsequent exposure to the post-natal environment, the skin is colonized by a wide array of microbes, many of which are commensal or symbiotic. Proposed beneficial roles of resident microbiota include inhibition of pathogenic species and further processing of skin proteins, free fatty acids, and sebum (Roth and James 1988). The skin is composed of a variety of niches, including regions with a broad range of pH, temperature, moisture, and sebum content. Furthermore, skin structures such as hair follicles, sebaceous, eccrine, and apocrine glands comprise subhabitats that may be associated with their own unique microbiota (Marples 1965; Kearney et al. 1984).
Many lines of evidence suggest a role for microorganisms even in noninfectious skin diseases, such as atopic dermatitis (AD; eczema), rosacea, psoriasis, and acne (Holland et al. 1977; Thomsen et al. 1980; Till et al. 2000; Paulino et al. 2006). Resident microbiota may become pathogenic, sometimes in response to an impaired skin barrier (Roth and James 1988). This observation underscores the value of a comprehensive characterization of the healthy skin microbiota to understand its role in the pathogenesis of skin disorders and ultimately to facilitate novel translational therapies.
Until recently, knowledge of the skin microbiota was limited to culture-dependent assays, although it is estimated that less then 1% of bacterial species can be cultivated (Staley and Konopka 1985). The 16S small subunit ribosomal (rRNA) genes are universal among prokaryotes and contain species-specific variable regions that are useful for inferring phylogenetic relationships (Hugenholtz and Pace 1996; Pace 1997). Broad-range PCR primers that anneal to highly conserved regions flanking the variable regions of the gene allow amplification from the majority of known bacteria. A recent 16S rRNA gene survey from Gao et al. (2007) shed some light on the composition of the superficial skin microbiota; their findings suggest that the bacteria sampled by swabbing the volar forearm are highly diverse. Furthermore, their findings suggest a low level of interpersonal consensus and an extremely dynamic microbiota that fluctuates greatly in a short span of time.
The NIH Roadmap for Medical Research recently launched the Human Microbiome Project (HMP) with the mission to comprehensively characterize human microbiota and analyze its role in human health and disease state (http://nihroadmap.nih.gov/hmp/). Initial HMP efforts include sampling multiples sites of healthy volunteers to determine whether humans share core microbial diversity profiles. Obtaining accurate, representative samples is a major challenge when surveying microbiota. For large-scale skin sampling, the most practical method of collection would undoubtedly be swabbing because it is simple, quick, and noninvasive. However, swabbing may not accurately represent the resident microbiota of full thickness skin. Skin scraping offers intermediate benefits by allowing collection of superficial skin cells and associated microbes. Finally, punch biopsying, although invasive and poorly suited to highly visible areas of the body, such as the face, offers the best representation of skin microbiota. Resolving issues such as method of sampling at the outset is essential for the success of large-scale microbiome initiatives, such as the HMP.
We report here the analysis of a 16S rRNA gene survey of the resident skin microbiota of the antecubital fossae (inner elbow) region from healthy humans. This anatomical region is clinically significant as an area that is classically affected in patients with AD. We used three different sampling methods to survey the microbiota at different levels of the skin, and report that each method captures the same dominant phylotypes of microbiota. Moreover, we define a microbiome that is shared among the individuals sampled. Finally, mouse models have the potential to clarify the interactive roles that the environment and genetics play in skin disorders. We surveyed the microbiota of mouse ear skin and identified significant parallels with human antecubital fossa skin.
Results
Features of the human skin 16S rRNA gene library
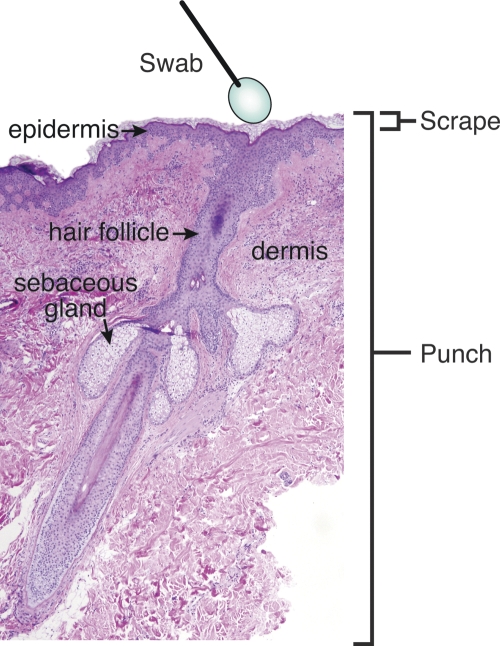
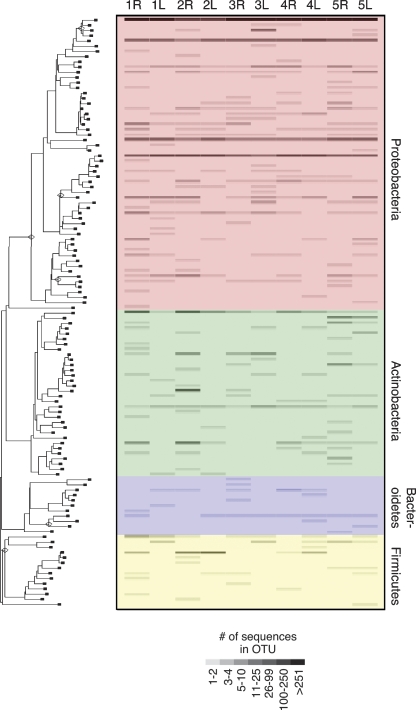
Skin samples were collected from left and right arms of five healthy human subjects with no history of dermatologic disease or other related disorders. To capture microbiota from all layers of the skin, we used three sampling techniques: swab, scrape, and punch biopsy (Fig. 1). Each sample was collected in a nonoverlapping area at the crease of the antecubital fossa (inner elbow). DNA was extracted from each sample and individually amplified with conserved 16S rRNA gene primers. The ∼1300-bp amplicons were cloned, and ∼200 clones were sequenced from each library. A total of 140 putative chimeric sequences, identified by Bellerophon, were removed from the data set (Supplemental Table S5). A total of 5373 quality near-full-length 16S rDNA sequences were subjected to phylogenetic analysis (Supplemental Table S1). These sequences were grouped into operational taxonomic units (OTUs; “phylotypes”) based on their genetic distance in a neighbor-joining tree with the DOTUR program (Schloss and Handelsman 2005). A species-level OTU has historically been defined as containing 16S rDNA sequences that are ≥97% identical (Gevers et al. 2005). Using the furthest-neighbor method of calculation and a similarity threshold of 97%, DOTUR assigned the 5373 sequences to 113 OTUs. Figure 2 illustrates the phylogenetic architecture of the 113 OTUs, and the abundance of those OTUs on the left and right arms of the five subjects we sampled (also see Supplemental Fig. S3). The majority of OTUs and sequences belonged to the division Proteobacteria (49% and 90%, respectively). The largest single OTU, a member of the division Proteobacteria and genus Pseudomonas, contained 3184 sequences (59%). The second largest OTU was also a member of the division Proteobacteria and of the genus Janthinobacterium. Other common Proteobacteria OTUs identified included Serratia, Halomonas, Stenotrophomonas, Delftia, and Comamonas genera. OTUs and sequences were also identified from the division Actinobacteria (28% and 5.6%, respectively), including species of the genera Corynebacterium, Kocuria, Propionibacterium, Microbacterium, and Micrococcus. OTUs and sequences identified in division Firmicutes (12% and 4.3%, respectively) were most commonly identified as Staphylococcus or Clostridium species. OTUs and sequences belonging to division Bacteroidetes (9.7% and <1%, respectively) were predominately Sphingobacterium or Chryseobacterium species. Two OTUs containing one sequence each were identified for divisions Acidobacteria and Cyanobacteria. Five hundred ninety-three sequences (11%) were 100% identical to at least one other sequence, and 4780 sequences (89%) were unique. We also identified one novel OTU that was <94% similar to any sequence in GenBank. This OTU was 93.9% similar to an unknown isolate from agricultural soil samples.
Figure 1.
A cross-section of the skin and the regions sampled by each collection method.
Figure 2.
Phylogenetic architecture and abundance of OTUs with ≥97% similarity in five human subjects. The left axis is a neighbor-joining tree composed of representative sequences from each of the 113 OTUs identified in the subjects. Abundance of OTUs in each subject is indicated by a grayscale value in the heat plot to the right. Each column of the heat plot corresponds to each arm of the five subjects (L is left arm; R is right arm). The heat plot is color-coded according to the major bacterial divisions observed: Proteobacteria (red), Bacteroidetes (blue), Firmicutes (yellow), and Actinobacteria (green).
To estimate the relative diversity captured by each sampling method for each subject, we calculated both Shannon and Simpson Diversity Indices, measures of ecosystem biodiversity (Supplemental Fig. S1). Each diversity index is associated with specific biases. The Shannon Index places a greater weight on consistency of species abundance in OTUs, while the Simpson Index gives more weight to abundant OTUs (Schloss and Handelsman 2005). Both indices suggest similar diversity profiles for all five subjects, with increased diversity in subject 2. These results suggest that the 16S rDNA sequencing equally represents the diversity found at all levels with the three sampling methods and the five subjects.
Using DOTUR, we also calculated observed and estimated (Chao1) total richness for each arm of each subject (Supplemental Fig. S3) and concluded that OTU richness will increase with additional sequencing of clones. Our estimates indicate that there is a minimum of 15–20 more OTUs per arm per subject yet to be uncovered. However, we recognize that no complex microbial community has ever been sampled to completion, and these are probably low estimates as the Chao1 curve rises as more clones are added.
To calculate skin bacterial load and assess the technical aspects of sample collection and preparation, we utilized a quantitative PCR strategy based on a small amplicon of the 16S rRNA gene. First, we generated a standard curve with Escherichia coli DNA and demonstrated that admixture of human DNA in a range of 1:1 to 1:100,000 of bacterial to human DNA or ∼1000:1 to 1:100 of bacterial to human cells does not significantly alter the amplification (measured as Ct) for E. coli DNA (Supplemental Table S2A). Hence, the values calculated for punch biopsies, collected with significant human DNA, can be compared with swab, scrape, and purified E. coli DNA. All of the samples from all of the subjects demonstrated remarkable uniformity, suggesting that there is not significant heterogeneity in biological or technical aspects of this procedure in healthy subjects (Table S2B). Based on the molecular weight of an average bacterial genome and considering the surface area of sampling, we calculate that the swabs collect ∼10,000 bacteria/cm2, scrapes ∼50,000 bacteria/cm2, and punch biopsies ∼1,000,000 bacteria/cm2.
Similar microbial populations are captured by swab, scrape, or punch biopsy
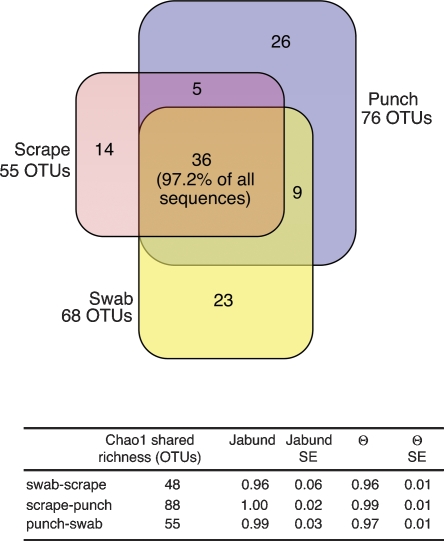
We collected bacteria from the skin using the three methods of swab, scrape, and punch biopsy (Fig. 1) to reach different dermatological levels and to evaluate the information captured by each collection method. Of the 113 total OTUs identified in the five subjects, 36 OTUs (31.9%) were identified by all three sampling methods (Fig. 3). However, these 36 OTUs represent 5225 of the 5373 sequences (97.2%). Although unique OTUs were identified by each sampling method, none of these OTUs contained more than four sequences. A priori it appears that the dominant OTUs are identified equally well by more and less invasive sampling methods. The rare OTUs may either be unique to the dermatologic level or rare enough to only be sampled with more exhaustive sequencing of the other levels. To further quantify these relationships, we utilized the SONS program, which applies OTU-based nonparametric richness estimators to calculate community structure and membership indices by taking into account species coverage and undetected species. The SONS program uses output from the DOTUR program to calculate these indices. Community membership only considers the list of OTUs in each sample, whereas community structure considers the list and abundance of OTUs. Jabund, an abundance-based Jaccard similarity index, is an estimator of community membership that reflects the probability that a randomly selected OTU in one community will be present in the other community (Schloss and Handelsman 2006). Values of Jabund can fall between 0 and 1: A value of 0 implies that the two communities do not share any OTUs, and a value of 1 implies that the two communities share all OTUs. The Jabund value between swab and scrape is 0.96 (standard error [SE] = 0.06), suggesting that if one was to sequence to exhaustion for both swab and scrape samples, the same OTUs would be identified (Fig. 3). The Jabund values for comparisons between swab and punch and between scrape and punch are 0.99 (SE = 0.03) and 1.0 (SE = 0.02), respectively, suggesting again that the same OTUs are present in the full thickness punch biopsy as in a swab or scrape of the same region (Fig. 3). Overall, these results indicate that any of the three sampling methods (swab, scrape, punch) are sufficient to obtain a representative profile of community membership.
Figure 3.
A Venn diagram that illustrates observed overlap of OTUs with ≥97% similarity according to sampling method. Among the five subjects, a total of 113 OTUs were detected. Thirty-six OTUs were detected by all three sampling methods; 5225 sequences of the total 5373 sequences fall within these 36 overlapping OTUs (97.2%). Jabund and θ estimates of OTU community structure and membership along with standard errors (SEs) are reported in the accompanying table.
Importantly, disease state may exhibit either the presence of a novel microbe or an alteration in the proportion of microbes. Thus, we also calculated a community structure estimator to compare sampling methods. The SONS program calculates values of θ, a community similarity index that takes abundance of OTUs into account (Yue and Clayton 2005; Schloss and Handelsman 2006). Values of θ can fall between 0 and 1: A value of 1 implies identical community structure, and a value of 0 implies dissimilar community structures. Again comparing the three sampling methods pairwise yielded θ values of 0.96 (SE = 0.01), 0.97 (SE = 0.01), and 0.99 (SE = 0.01) for swab versus scrape, swab versus punch, and scrape versus punch, respectively (Fig. 3). These estimations suggest that community structure captured by swab, punch, and scrape are virtually identical.
As an independent assessment, we employed phylogenetic tree-based comparisons, specifically the UniFrac metric and the phylogenetic (P) test. The UniFrac metric measures unique branch length present in each 16S rRNA library being compared as a proxy assessment of the evolution of divergent functions as a mechanism of adaptation (Lozupone and Knight 2005). The P test also assesses evolution and adaptation by comparing nonrandom distribution of lineages in each library (Martin 2002). Pooling all of the sequences from swabs, scrapes, and punches and then comparing each collection method pairwise by both the UniFrac metric and the P test, each sampling method was significantly different from each other (P < 0.03) after Bonferonni correction for multiple comparisons. To determine if patient samples clustered in two-dimensional space, we applied principle coordinates analysis (PCA) to the UniFrac metric. We could not detect biologically meaningful clustering based on PCA plots of the first three principle components (Supplemental Fig. S4). These tree-based tests differ from the above-described OTU-based tests in that they only consider observed species and do not take into account estimated species richness of the community. Therefore, it is not surprising that the UniFrac and P test reflect the Venn diagram depicted in Figure 3, as this is also a depiction of only those OTUs that we observed. The Jabund and θ estimators suggest that if we continue sequencing to the point of exhaustion we will find that there is virtually complete overlap of community membership and structure for all three sampling methods.
To explore the possibility that the tree-based analyses are significant because they are detecting differences at a phylogenetic level not revealed by SONS analyses, we increased the OTU threshold to ≥99% identity, which is stricter then the historical use of ≥97% identity to define a bacterial OTU (Gevers et al. 2005). At the 99% threshold, our data set sorts into a total of 261 OTUs. We obtained Jabund values 0.95 (SE = 0.03), 0.96 (SE = 0.04), and 0.99 (SE = 0.02) for swab versus scrape, swab versus punch, and scrape versus punch, respectively. The same 99% threshold revealed θ values of 0.94 (SE = 0.01), 0.91 (SE = 0.01), and 0.94 (SE = 0.01) for swab versus scrape, swab versus punch, and scrape versus punch, respectively. This would suggest that using a stricter OTU threshold reveals subtle differences that are not obvious when using a 97% threshold. This is not altogether surprising but may also explain why tree-based methods reveal significance where SONS does not.
Evidence for a common microbial diversity profile
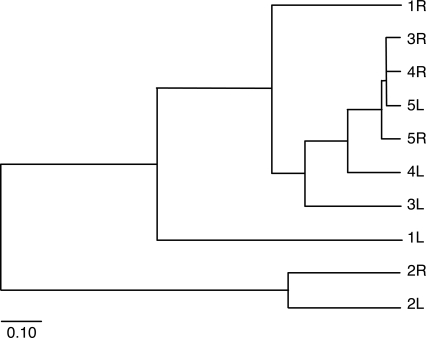
To determine whether individuals share a common skin microbial diversity profile, we utilized the four measurements previously described (Jabund, θ, UniFrac, and the P test) to assess intrapersonal and interpersonal variation. For each subject, the three sequence libraries (swab, scrape, and punch) for each arm were combined, and left and right arm libraries were compared to calculate intrapersonal variation. Next, the left and right sequence libraries from each subject were compared to the left and right sequence libraries of the other four subjects to calculate interpersonal variation. The Jabund values obtained for intrapersonal variation were not significantly different from those obtained for interpersonal variation. In all cases, the SE 95% confidence intervals overlap (data not shown). We then calculated θ values to examine community structure. Figure 4 illustrates how the samples cluster by UPGMA according to their θ indices. When taking into account SE, the calculated θ indices between subject 1 (right arm), subject 3 (right arm), subject 4 (left and right arm), and subject 5 (left and right arm) are not significantly different (for values and SE, see Supplemental Table S3). This analysis also revealed that subject 2 appears to be a major outlier in terms of community structure (Fig. 4). Upon further examination, the left and right swab libraries from subject 2 contained an increased proportion of Firmicutes sequences as compared to the other subjects (Supplemental Fig. S2). Because these sequences appear to be derived from Staphylococcus species, we presume that subject 2 was subclinically colonized with Staphylococcus, as is found by cultivation methods in ∼5%–10% of healthy adults (Marples 1965). Comparing subjects pairwise by the UniFrac metric and the P test did not reveal any significant differences. Taken together, these results point toward a common core skin microbiome among healthy human subjects.
Figure 4.
Interpersonal and intrapersonal variation. A UPGMA dendogram of pairwise θ values comparing community structure of OTUs from each arm of each subject. The length of the scale bar represents a distance of 0.10 (1 − θ). Values for θ were calculated for OTUs with ≥97% similarity (Supplemental Table 3). (L) Left arm; (R) right arm.
Significant parallels between mouse ear skin and human skin
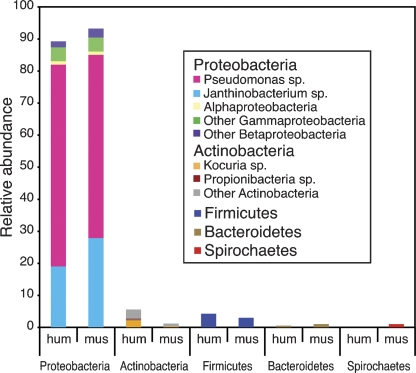
In order to take advantage of mouse models that could potentially clarify the interactive roles that the subject’s environment and underlying genotype play in skin disorders, we next sought to determine how closely mouse skin microbiota resembles human skin microbiota. We biopsied mouse ear skin since this body site, with substantially fewer hair follicles than the body, more closely mirrors the morphology of most regions of human skin. As well, the mouse ear is also commonly used in inflammation studies, underlying the biological relevance of studying this site. We biopsied the ears of four female adult C57BL/6J mice, a mother and three of her 2-mo-old offspring, housed independently for the last month in the same mouse facility. We sequenced 192 16S rRNA PCR products per mouse ear punch biopsy (total of 675 sequences; Supplemental Table S4) and compared this clone library to the human punch biopsy library (total of 1861 sequences; Supplemental Table S1). When comparing sequences at the division level, Proteobacteria is the dominant division, with predominance of the Pseudomonas and Janthinobacterium genera, in mouse ear skin, identical to the human antecubital fossae (Fig. 5). More detailed analysis of community structure using the Jabund and θ indices demonstrates that mouse ear skin microbiota is strikingly similar to human skin microbiota. At an OTU threshold of 97%, we calculated a Jabund index of 0.89 (SE = 0.14) and a θ index of 0.98 (SE = 0.01). At an OTU threshold of 99%, we calculated a Jabund index of 0.81 (SE = 0.08) and a θ index of 0.84 (SE = 0.03). Though the UniFrac metric and P test indicate significant differences in the tree branch length (P < 0.01 for both tests), these tests do not account for species coverage or unsampled species. Alternatively, these tests could be detecting differences at a phylogenetic level that are not detected by SONS. In summary, our murine ear skin studies have laid the groundwork for future studies aimed at dissecting the genetic and environmental components of human skin diseases.
Figure 5.
Comparison of relative abundance of bacteria in mouse ear punch and human punch biopsy libraries. 16S rDNA sequences are first grouped into division. Each division bar is then further broken down into class. When a particular genus dominated the class, the genus is noted. (hum) Human; (mus) mouse.
Discussion
We generated sequence from 5373 16S rRNA genes from bacteria, isolated by three skin collection methods of five healthy individuals, and found that the majority of these sequences derived from the Proteobacteria division and predominantly from the genera Pseudomonas and Janthinobacterium. With these sampling methods, we identified 113 OTUs within the divisions Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Cyanobacteria, and Acidobacteria. To our knowledge, representatives from all of these divisions have been isolated in culture, though from this analysis, we cannot determine whether these specific OTUs have been cultured or are even culturable. We are currently attempting to culture both aerobic and anaerobic bacteria from distinct skin subsites to create a large stock of bacteria from the skin of healthy subjects to contribute to the HMP reference genome sequencing project. Historically, dermatological microbiology has focused exclusively on rare pathogenic isolates that cause skin infections. Very few cultured commensal skin bacteria, such as Staphylococcus epidermidis and Propionibacterium acnes have been sequenced (Zhang et al. 2003; Bruggemann et al. 2004). We found 16S rRNA sequences that closely matched S. epidermidis and P. acnes, but they consisted of <5% of the microbiota captured in this study. This is contrary to the commonly held notion that S. epidermidis is the dominant aerobic bacteria residing on the skin (Marples 1965).
Just 10 OTUs account for >90% of the sequences, indicating that there are a few dominant OTUs that inhabit the antecubital fossa. Gao et al. (2007) has previously surveyed swabs of volar forearm skin microbiota and found much less representation from the Proteobacteria division with increased Actinobacteria and Firmicutes. When directly comparing the volar forearm data set with our data set, many of the OTUs do overlap. However, the most abundant OTUs (Proteobacteria division) in our data set are not represented in the volar forearm data set. We cannot conclude if this result is biological or technical, as there were significant differences in the methods in which the studies were conducted. Nonetheless, strong biological differences exist between the moist, sweaty hairless skin of the antecubital fossa and the dry, less sweaty, more haired skin of the volar forearm. One interesting proposition is that the unique population of microbiota of antecubital fossa skin is directly influenced by the retention of moisture and body secretions, and potentially increases the susceptibility of patients to episodic exacerbation of AD in this region.
The genera Pseudomonas and Janthinobacterium (both pseudomonads; gram-negative bacilli, aerobic, non-spore forming, motile by means of one or more flagella) are not typically thought of as skin microbes based on culture assays. Pseudomonads (and other Gram-negative bacilli) have historically been labeled as secondary invaders of wounds, most commonly referring to the colonization of burns by Pseudomonas aeruginosa. Overall, pseudomonads are extremely versatile organisms, primarily found in soil, water, or decomposing organic materials but also found in the intestinal flora (Marples 1965). The main requirement for pseudomonads to flourish is a moist environment. In fact, classical studies have shown that conditions on most areas of the skin surface are much too arid for multiplication of Gram-negative bacilli (Rebell et al. 1950; Marples 1965). The antecubital fossa, being moist and sometimes sweaty, might enable pseudomonads to flourish in this environment but be absent in the arid volar forearm.
The skin is a powerful system for microbial analyses with many characteristics that will enable us to answer fundamental questions of the role of microbial communities in both health and disease. This pilot study informs the diversity that exists between topographical regions of the skin surface and underscores the need to survey multiple subsites to explain why dermatologic disorders manifest at stereotyped body sites, e.g., psoriasis on the outside of and AD on the inside of elbows and knees. The noninvasive nature of sampling skin provides the unique opportunity to perform analyses on multiple subsites and address intrapersonal and interpersonal variation. We are currently undertaking a study to examine 20 skin subsites in healthy individuals, and foresee that this will further inform selection of skin subsites for the HMP Project.
The microbiota ascertained with a swab or scrape represents the history of skin differentiation, a process that should be considered when drawing biological significance from microbiota sampling of all epithelial surfaces. Skin cells turn over every 4 wk, differentiating from stem cells deep within the epidermis and hair follicles, and eventually slough off from the upper layer as cornified (enucleated, dead) cells (Blanpain et al. 2007). Although the 16S bacterial survey of the distinct layers of the skin (swab, scrape, punch) identified similar community membership and structure at all levels of the skin, a DNA-based analysis cannot distinguish between viable and nonviable bacteria. The bacteria may transit to the surface with differentiating skin cells and be surveyed from the top layers when they play a physiological role in a specific niche deeper within the skin. This linear program of terminal differentiation, resulting in enucleated skin cells at the surface, will also facilitate future metagenomic studies, another goal of the HMP project, since skin scrapes can produce high yields of microbiota DNA with little “contaminating” human DNA.
We demonstrate significant parallels between the human and mouse skin microbiota and are optimistic about the potential that mouse models offer for the elucidation of complex human skin disorders. Indeed, the St14hypo/− mouse model of filaggrin deficiency, which exhibits AD-like features, has demonstrated a selective microbial shift associated with the disease state (T.C. Scharschmidt, K. List, E.A. Grice, R. Szabo, NISC Comparative Sequencing Program, G. Renaud, C.C. Lee, T.G. Wolfsberg, T.H. Bugge, and J. Segre, in prep.). Filaggrin mutations have been demonstrated in ∼50% of patients with moderate to severe AD (Sandilands et al. 2007), and we are currently analyzing whether similar changes are found in human AD patients. Our findings provide the groundwork for future studies aimed at unraveling genetic and environmental contributions to skin disorders in both mice and humans.
Our results also demonstrate that the skin microbiome is vastly different from the gut microbiome, which consists primarily of members of Firmicutes and Bacteroidetes divisions (Eckburg et al. 2005). Furthermore, our results indicate a low level of interpersonal variation, which is not the case in gut studies. However, one shared feature of the gut and skin microbial communities is a low level of deep evolutionary lineage diversity. Of the more than 70 described bacterial divisions (DeSantis et al. 2006a), we found only six divisions associated with the skin, and only eight divisions have been associated with the gut (Eckburg et al. 2005; Ley et al. 2005). These two studies sampled approximately the same amount or twofold more clones then we sampled (5373 total human). The low level of deep diversity is in sharp contrast with environmental samples such as soil, which in one study was found to harbor 21 divisions of bacteria from sampling only 766 clones (Dunbar et al. 2002). Also, like microbial sequences derived from the human colon (Eckburg et al. 2005), the majority of the generated sequences (89%: 4780 of 5373) were represented only once in our data set. Skin sequences sort into a small number of OTUs (113), more like the human colon sequences than environmental samples (Eckburg et al. 2005). This type of phylogenetic architecture, thought to arise from adaptive radiation, has been suggested as a signature feature of the gut microbiome (Ley et al. 2006). In the case of the skin, it would appear that a small number of Proteobacteria were successful in adapting to the conditions of the antecubital fossa and thus gave rise to a wide variety of descendants.
These data represent a critical step for determining the diversity of the skin microbiota, and establish a platform for more large-scale studies. Future studies will address questions that cannot be answered by sequencing phylogenetic markers, such as the 16S gene. For example, what physiological functions are performed by skin microbiota? What are the complex ecological interactions that govern the physiology of the skin? Does the host genotype play a role in determining the skin microbiome? How do medications, clothing, sex, age, environment, and numerous other factors affect the skin microbiome? What is the role of the skin microbiome in disorders such as AD?
The essential role of microbiota in human health and disease is gaining recognition. The National Academies National Research Council Committee on Metagenomics recently issued a report with recommendations for tackling metagenomic studies, which aim to determine the DNA sequence of genomes from a mixed community of organisms (National Research Council 2007). The NIH Roadmap for Medical Research recently funded the HMP. The broad goals of the NIH Roadmap for Medical Research HMP include determining if there is a core microbiome, and whether changes in the core microbiome are associated with changes in human health. One funded HMP program initiative is to sequence 600 reference genomes of cultured and previously uncultured species in anticipation of the next step of deep metagenomic sequencing of specific body sites. Pilot studies, such as the one we have described here, are essential in determining which reference skin microbial genomes are most important to sequence to proceed with metagenomic analyses. Another funded HMP program initiative is to survey microbiota in healthy volunteers from five body sites (gut, oral, nasal, skin, and vaginal [for women]), possibly including multiple subsites of skin.
The concept that the human body is host to trillions of microbes and their genomes is revolutionizing the way we view complex medical challenges. A role for microbes in skin disease has long been hypothesized (Holland et al. 1977; Thomsen et al. 1980; Till et al. 2000; Paulino et al. 2006). The tools and knowledge are now in hand to examine this role, potentially opening the door to new strategies for the diagnosis, prevention, and treatment of disease. This report and others like it will contribute important information for implementation of ambitious large-scale microbiome projects such as the ones proposed by the NIH Roadmap for Medical Research.
Methods
Subject recruitment and sampling
Five healthy volunteers, male and female, of diverse race and ethnicities, ages 20–65, with no history of dermatologic disorders (mild acne or other common features acceptable) or other chronic medical disorders and with no current skin infections were recruited to the National Institutes of Health Clinical Center under an approved NIH/NCI IRB protocol. Subjects were instructed not to wash with anything, including water alone, for an 8-h interval prior to sampling, but otherwise were allowed typical daily hygienic practices. Informed consent was obtained from all participants. Study personnel wore sterile gloves for each sample collection. Biological samples were collected from nonoverlapping regions of the antecubital fossa (inner elbow) of both the left and right arm with no prior cleaning or preparation of the skin surface. Swabs were obtained using a sterile cotton pledget (Johnson and Johnson) soaked in sterile 0.15 M NaCl with 0.1% Tween 20 (Fisher Scientific) and wrung of excess solution. A 4-cm2 area of the antecubital fossa was gently rubbed using two steady strokes in a nonoverlapping manner. Superficial skin scrapings were obtained from a 4-cm2 area with two gentle steady strokes of a sterile disposable no. 15 blade in a nonoverlapping manner. The biopsy collection site was then anesthetized with injection of 0.1 mL of 2% lidocaine with epinephrine. The punch biopsies were taken as two separate samples. First, a 4-mm-diameter punch biopsy tool was used to mark a 4-mm diameter of skin with a depth of 1 mm, and then the epidermal layer was shaved at the 1-mm depth, parallel to the skin surface using a flat razor blade. After collecting the shave biopsy specimen, a 4-mm-diameter punch biopsy was used to mark the skin at the same site of the shave biopsy to a further depth of ∼3 mm. The dermal skin layer was removed using a sterile scissors and forceps. We sequenced clones (see below) from both the epidermal and dermal skin biopsies but ultimately combined the two libraries into one library for analysis to represent full thickness of the skin. All three samples (swab, scrape, punch/shave) were collected in nonoverlapping areas of the antecubital fossa. All samples were stored at −80°C until further processing.
Animals
Ear punch biopsies (2 mm in diameter on the outer margin) of the female C57BL6 mouse and her litter were taken when the pups reached 8 wk of age. Pups were weaned at 4 wk and subsequently housed independently. All samples were stored at −80°C until further processing. All experiments involving mice were performed under an NIH/NHGRI ACUC approved protocol.
DNA extraction and purification
All biological specimens were first incubated in a preparation of enzymatic lysis buffer (20 mM Tris at pH 8.0, 2 mM EDTA, 1.2% Triton X-100) and lysozyme (20 mg/mL) for 30 min at 37°C. Two 5-mm sterile stainless steel beads (Qiagen) were then added to the sample. Tubes were transferred to the TissueLyser (Qiagen) and processed for 20 sec at 15 Hz. Beads were removed, and samples were incubated overnight at 56°C in Buffer AL and Proteinase K from the DNeasy DNA Extraction Kit (Qiagen). The standard protocol for the kit was followed for all subsequent steps. The purified genomic DNA was resuspended in 100 μL of Buffer AE and stored at −20°C.
Quantification of total bacteria present in samples
A portion of the bacterial 16S gene was amplified using forward primer 63F (5′-GCAGGCCTAACACATGCAAGTC-3′) and reverse primer 355R (5′-CTGCTGCCTCCCGTAGGAGT-3′) (Castillo et al. 2006) to yield a 292-bp PCR product (for further details on primers, see Supplemental Table S6). A standard curve was constructed by amplifying serial dilutions of known quantities of E. coli cells. Real-time PCR was performed on an ABI 7300 system (Applied Biosystems Inc.) using optical grade 96-well plates. Each 25 μL reaction included 12.5 μL Platinum 2× Sybr Green Master Mix (Invitrogen), 0.25 μL MgCl2 (50 mM), 0.5 μL Rox Dye, 0.25 μL each primer (20 μM), 6.25 μL water, and 5 μL of DNA. Cycling conditions were as previously described (Castillo et al. 2006). The function used to calculate copy number is as follows: Ct = −3.42x + 34.06; R2 = 0.99; where Ct is the threshold cycle and x is the log copy number. Copy numbers calculated for the sample are adjusted to be relative to the surface area sampled.
PCR amplification of 16S rRNA genes
16S rRNA genes were amplified from purified genomic DNA using the primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1391R (5′-GACGGGCGGTGTGTRCA-3′) (Eckburg et al. 2005). For each 50 μL reaction, conditions were as follows: 5.0 μL of 10× buffer with MgCl2 (Roche Applied Science), 1 μL of dNTP mix (10 mM each; Invitrogen), 1 μL of each primer (20 μM; IDT), 3 μL of DMSO, 4 μL of bacterial genomic DNA, and 0.5 μL of FastStart High Fidelity Taq Polymerase (Roche). For each DNA sample, three replicates were performed. Thermocycling was as follows: Initial denaturation at 95°C for 5 min, followed by 25–30 cycles of a 30-sec 95°C denaturation, 30-sec annealing at 55°C, and 1.5-min elongation at 72°C, all followed by a final extension of 10 min at 72°C. Cycle number was determined on a case-by-case basis, such that amplification was still in the linear range of the reaction when stopped, but sufficient PCR product for cloning was produced (usually 25–30 cycles). PCR products were then separated on an agarose gel, and bands corresponding to the ∼1.3-kb product were extracted with a razor blade. Negative control PCR reactions were performed with each set of amplifications and in all cases did not produce an amplification product. PCR products were extracted using the Qiaquick Gel Extraction kit (Qiagen) per manufacturer’s suggested protocol. PCR products were resuspended in 30 μL of Buffer EB and stored at −20°C.
Cloning and sequencing of 16S rDNA PCR products
PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) per the manufacturer’s protocol. A total of 192 of the resulting bacterial colonies per ligation were picked, plasmid DNA was purified, and plasmid inserts were sequenced bidirectionally using the M13 primers on an ABI 3730xl sequencer (Applied Biosystems Inc.). Chromatogram data quality and quantity were evaluated using phred Q20 counts and non-vector sequence data remaining after cross_match screening. Bidirectional reads were assembled in a pipeline developed by NHGRI’s Bioinformatics and Scientific Programming Core. Sequences were extracted from chromatograms using phred, and bidirectional pairs were assembled using phrap. Vector sequence detected by cross_match was trimmed off. Only assembled sequences >1250 bp were studied further. Assemblies were screened for quality, and all sequences containing >20 consecutive bases of sequence <Q20 were discarded. Sequence traces and assemblies from OTUs containing less then four sequences were further examined in consed to verify quality of sequence and assembly.
Alignment and phylogenetic analysis of 16S rDNA sequences
Trimmed, assembled sequences were aligned to a core set of sequences using the NAST alignment tool (DeSantis et al. 2006b) on the Greengenes website (http://greengenes.lbl.gov) (DeSantis et al. 2006a). Alignments were manually curated. All sequences were examined for chimeras using the Bellerophon tool and standard settings (Huber et al. 2004), also available on the Greengenes website. One hundred forty putative chimeras were removed from further analyses (for further details on chimera removal, see Supplemental Table S5). A summary of quality sequences obtained is shown in Supplemental Tables S1 and S4. Sequences were inserted into a reference neighbor-joining tree in ARB using the parsimony insertion tool (Ludwig et al. 2004). Hypervariable regions were masked with lanemaskPH, and the Olsen correction was used when calculating pairwise distances.
OTU determination and diversity estimation
Using the Olsen-corrected distance matrix exported from ARB, sequences were grouped into OTUs by the furthest-neighbor algorithm using DOTUR (Schloss and Handelsman 2005). We used a similarity cutoff of 97% or 99%, as indicated in the text, to define an OTU. This same cutoff was used for diversity indices and richness estimates that were calculated using DOTUR. We used both the Shannon and Simpson (1-D) diversity indices and Chao1 richness estimates calculated by DOTUR to estimate microbial diversity and richness.
Clustering and comparison of 16S libraries
All calculations using the SONS package were performed using an OTU cutoff of 97% or 99% similarity, as indicated in the text (Schloss and Handelsman 2006). All values are reported with their SE. The UniFrac package was used to compare unique branch length between communities (Lozupone and Knight 2005; Lozupone et al. 2006). The phylogenetic tree generated in ARB containing only the sequences generated in this study, along with environmental labels, was imported into UniFrac. The UniFrac metric was calculated along with its significance value, with and without regard to relative abundance of sequences. All significance values reported were corrected for multiple comparisons by Bonferonni correction. PCA analysis was performed within the UniFrac online suite of tools. The P test, also available on the online UniFrac suite of tools, assesses trees for nonrandom distributions of lineages according to environment (Martin 2002). All P test values reported were also corrected for multiple comparisons.
Acknowledgments
We thank all volunteers that participated in this study. We thank Susan Booher, RN (NIH/NCI) for assistance with sample collection. We thank all members of the Segre Lab for their underlying contributions to this work, including Clay Deming, Wei Liu, Tiffany Scharschmidt, and Cristina de Guzman Strong. We thank Eric Green (NIH/NHGRI), Andrew McCallion (McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University), and Mark Udey (NIH/NCI) for critical reading of the manuscript. We thank Jeffrey Gordon and Ruth Ley for initial advice. We thank Chenwei Wang for help with Figure 2 and Julia Fekecs for assistance preparing the figures. E.A.G. is supported by a Pharmacology Research Associate Training Fellowship, NIGMS. This work was supported by the NHGRI and the NCI Center for Cancer Research Intramural Research Programs.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. EU534411–EU540623.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075549.107.
References
- Blanpain C., Horsley V., Fuchs E. Epithelial stem cells: Turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H., Henne A., Hoster F., Liesegang H., Wiezer A., Strittmatter A., Hujer S., Durre P., Gottschalk G. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–673. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- Castillo M., Martin-Orue S.M., Manzanilla E.G., Badiola I., Martin M., Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 2006;114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006a;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Keller K., Brodie E.L., Larsen N., Piceno Y.M., Phan R., Andersen G.L. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006b;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J., Barns S.M., Ticknor L.O., Kuske C.R. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 2002;68:3035–3045. doi: 10.1128/AEM.68.6.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Tseng C.H., Pei Z., Blaser M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Cohan F.M., Lawrence J.G., Spratt B.G., Coenye T., Feil E.J., Stackebrandt E., de Van Peer Y., Vandamme P., Thompson F.L., et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Holland K.T., Cunliffe W.J., Roberts C.D. Acne vulgaris: An investigation into the number of anaerobic diphtheroids and members of the Micrococcaceae in normal and acne skin. Br. J. Dermatol. 1977;96:623–626. doi: 10.1111/j.1365-2133.1977.tb05206.x. [DOI] [PubMed] [Google Scholar]
- Huber T., Faulkner G., Hugenholtz P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Pace N.R. Identifying microbial diversity in the natural environment: A molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Kearney J.N., Harnby D., Gowland G., Holland K.T. The follicular distribution and abundance of resident bacteria on human skin. J. Gen. Microbiol. 1984;130:797–801. doi: 10.1099/00221287-130-4-797. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Hamady M., Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar A., Lai T., Steppi S., Jobb G., et al. 2004ARB: A software environment for sequence data Nucleic Acids Res. 321363–1371. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marples M. The ecology of the human skin. Charles C Thomas; Bannerstone House, Springfield, IL: 1965. [Google Scholar]
- Martin A.P. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council . The new science of metagenomics: Revealing the secrets of our microbial planet. National Academies Press; Washington, DC: 2007. [PubMed] [Google Scholar]
- Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Paulino L.C., Tseng C.H., Strober B.E., Blaser M.J. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebell G., Pillsbury D.M., De Saint Phalle M., Ginsburg D. Factors affecting the rapid disappearance of bacteria placed on the normal skin. J. Invest. Dermatol. 1950;14:247–264. doi: 10.1038/jid.1950.33. [DOI] [PubMed] [Google Scholar]
- Roth R.R., James W.D. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988;42:441–464. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- Sandilands A., Terron-Kwiatkowski A., Hull P.R., O'Regan G.M., Clayton T.H., Watson R.M., Carrick T., Evans A.T., Liao H., Zhao Y., et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.T., Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Thomsen R.J., Stranieri A., Knutson D., Strauss J.S. Topical clindamycin treatment of acne. Clinical, surface lipid composition, and quantitative surface microbiology response. Arch. Dermatol. 1980;116:1031–1034. doi: 10.1001/archderm.116.9.1031. [DOI] [PubMed] [Google Scholar]
- Till A.E., Goulden V., Cunliffe W.J., Holland K.T. The cutaneous microflora of adolescent, persistent and late-onset acne patients does not differ. Br. J. Dermatol. 2000;142:885–892. doi: 10.1046/j.1365-2133.2000.03467.x. [DOI] [PubMed] [Google Scholar]
- Yue J.C., Clayton M.K. A similarity measure based on species proportions. Commun. Stat. Theor. Methods. 2005;34:2123–2131. [Google Scholar]
- Zhang Y.Q., Ren S.X., Li H.L., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol. Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]