Abstract
Using the massively parallel technique of sequencing by oligonucleotide ligation and detection (SOLiD; Applied Biosystems), we have assessed the in vivo positions of more than 44 million putative nucleosome cores in the multicellular genetic model organism Caenorhabditis elegans. These analyses provide a global view of the chromatin architecture of a multicellular animal at extremely high density and resolution. While we observe some degree of reproducible positioning throughout the genome in our mixed stage population of animals, we note that the major chromatin feature in the worm is a diversity of allowed nucleosome positions at the vast majority of individual loci. While absolute positioning of nucleosomes can vary substantially, relative positioning of nucleosomes (in a repeated array structure likely to be maintained at least in part by steric constraints) appears to be a significant property of chromatin structure. The high density of nucleosomal reads enabled a substantial extension of previous analysis describing the usage of individual oligonucleotide sequences along the span of the nucleosome core and linker. We release this data set, via the UCSC Genome Browser, as a resource for the high-resolution analysis of chromatin conformation and DNA accessibility at individual loci within the C. elegans genome.
The regulation of genetic information within eukaryotic cells involves a high degree of specificity both in the availability of individual DNA-binding factors in individual cells, and in availability in the genome of DNA sequences that are their potential targets. Modulating the accessibility of individual DNA sequences are many complex interactions, the most prevalent of which are the interactions between histone octamers and DNA in compacted chromosomes. Each histone core interacts with 147 bp of DNA, which coil 1.7 times around the histone octamer (Luger et al. 1997; Davey et al. 2002) to form the basic unit of chromatin structure, the nucleosome. Since the first description over three decades ago (Kornberg 1974), the nucleosome and its role in gene regulation has been the subject of intensive study and speculation.
As we have an increasingly detailed functional view of the genome, the tools of high-throughput molecular characterization have been used to begin obtaining a genome-wide description of nucleosome positions (Satchwell et al. 1986; Yuan et al. 2005; Johnson et al. 2006; Albert et al. 2007; Lee et al. 2007; Peckham et al. 2007; Schones et al. 2008; Shivaswamy et al. 2008). These data have in turn been used in attempts to reveal nucleosome positioning signals in DNA sequence (Satchwell et al. 1986; Ioshikhes et al. 1996; Segal et al. 2006; Yuan and Liu 2008). Although of great interest and value, sequence-based predictions of nucleosome position have been limited to date in their accuracy and resolution.
In the literature, nucleosome positioning has been defined as the ability of sequences in the genome to encode a nucleosome organization, with such features regulating the access of nonhistone proteins to DNA in vivo (e.g., Segal et al. 2006). A variety of global estimates for sequence-directed nucleosome positioning have been described for the single-cell eukaryote Saccharomyces cerevisiae (Segal et al. 2006; Lee et al. 2007; Peckham et al. 2007; Shivaswamy et al. 2008; Yuan and Liu 2008). More complex multicellular systems with multiple tissue and cell types provide a number of interesting challenges and opportunities for the global study of chromatin structure. In this study we use the new ultra-high-throughput Applied Biosystems SOLiD (sequencing by oligonucleotide ligation and detection) sequencing technology to characterize nucleosome positions in a mixed-stage mixed-tissue population of Caenorhabditis elegans cells. This work provides both a substantial in vivo data set of nucleosome positions for a metazoan organism and an experimentally derived high-resolution map of nucleosome constraints for the nematode C. elegans.
Modes of possible nucleosome positioning
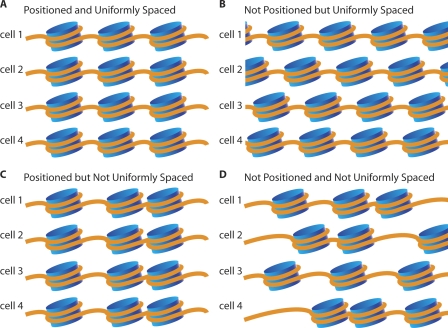
From the outset, one can conceive of four different combinatorial modes or patterns for the individual and relative positioning of nucleosomes (Fig. 1).
Figure 1.
Possible patterns of nucleosome positioning. Diagrams show four possible combinations for the local and relative position of nucleosomes. (A) Nucleosomes occupy reproducible positions in all cells and are regularly spaced relative to one another. (B) Nucleosomes are regularly spaced relative to one another, but have no strong tendency to occupy the same position in different cells. (C) Nucleosomes occupy the same position in all cells but are not regularly spaced relative to one another. (D) Nucleosomes neither occupy the same position in all cells nor have regular spacing relative to one another.
Pattern 1: Positioned and uniformly spaced
The first pattern consists of nucleosomes that are reproducibly spaced and positioned in the genome so that they occupy the same uniformly spaced set of positions regardless of cell type or developmental stage (Fig. 1A).
Pattern 2: Not positioned, but uniformly spaced
The second pattern consists of nucleosomes that are spaced at uniform intervals relative to each other within individual cells, but which lack any reproducibility in comparisons of absolute positioning between different cells (Fig. 1B). Differences in absolute positions could be specific to individual cell types or completely stochastic.
Pattern 3: Positioned, but not uniformly spaced
The third pattern consists of nucleosomes where the precise positioning is maintained as a function of the underlying DNA sequence, but for which there is no consistent pattern of spacing along the chromosome (Fig. 1C).
Pattern 4: Not positioned and not uniformly spaced
The fourth pattern invokes a scenario without any consistent positioning and spacing, so that nucleosomes are variably positioned on the genome with respect to both the underlying DNA sequence and each other (Fig. 1D).
It is important to note that the first and second patterns of nucleosome arrangement involve uniformly spaced nucleosomes and that either would be sufficient to explain the tight laddering bands of mono-, di-, tri-, and higher order nucleosome DNA fragments that are seen as a result of micrococcal nuclease (MNase) digestion (Noll 1974), whereas they would yield very different results in sequence analysis looking at reproducibility of individual positions. Additionally, across an individual genome, all four types of patterns could coexist at different loci, and similarly across cell types and states, all four patterns could exist and overlap at an individual locus. In order to dissect these possibilities, we took advantage of a new ultra-high-throughput sequencing technology, SOLiD.
Results
Sequencing and coverage
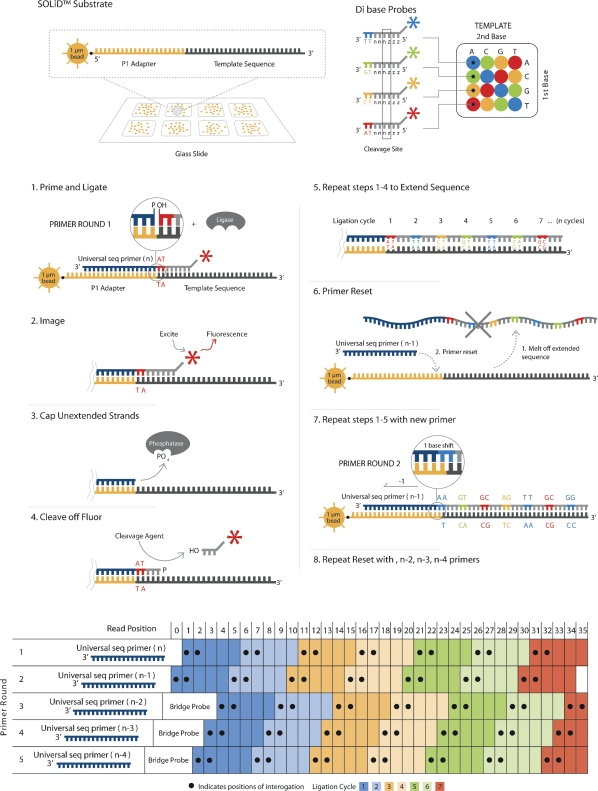
SOLiD sequencing (solid.appliedbiosystems.com) is a massively parallel, ligation-mediated sequencing method (Fig. 2) that extends a number of recent developments in nucleic acid chemistry (Kwoh et al. 1989; Kwoh and Kwoh 1990; Embleton et al. 1992; Macevicz 1998; Tawfik and Griffiths 1998; Ghadessy et al. 2001; Dressman et al. 2003; Shendure et al. 2005; McKernan et al. 2006). Tens of millions of 25–50 nt reads are generated by a single run of this sequencing technology.
Figure 2.
An outline of SOLiD sequencing technology.
To sample the global spectra of allowed positions for nucleosome occupancy within the C. elegans genome in vivo, we isolated DNA fragments associated with nucleosome cores from a mixed stage population of C. elegans (Johnson et al. 2006). Sanger sequencing of a limited number of these nucleosome core fragments revealed the average length to be 149.5 bp (Supplemental Fig. S1). C. elegans nucleosomal fragment libraries for SOLiD sequencing were constructed from this DNA population. A single experimental run on the SOLiD platform produced a total of 107 million 50 bp reads from the nucleosome core fragment experiment (Table 1). Allowing up to five mismatches (and no insertions or deletions), we were able to place 56.4 million reads using the standard SOLiD mapping pipeline onto the C. elegans (WS170 freeze) or the Escherichia coli (K-12) genomes (E. coli is the food source for C. elegans and, hence, a DNA contaminant). The 50 nt reads that contained a unique best match with least number of mismatches (using these criteria) were assigned to corresponding locations in the genome with 44.0 million reads being uniquely placed on the worm genome (core read data set). Of these 44.0 million, ∼10.7 million reads matched perfectly, ∼9.0 million reads had one mismatch, ∼7.5 million reads had two mismatches, ∼6.4 million reads had three mismatches, ∼5.5 million reads had four mismatches, and ∼4.8 million reads had five mismatches. As an experimental control and reference data set, we used the SOLiD technology to sequence C. elegans genomic DNA digested with MNase. The control library consisted of whole-genome DNA lightly digested with MNase and size selected in a range of from 400 to 900 bp. Fragments were circularized to produce a paired-end library and subjected to paired-end SOLiD sequencing, resulting in 25 bp reads from each end. Mate pairs were mapped onto the reference genome using standard SOLiD mapping pipeline, which produced 51.91 million mapped pairs in the correct size range matching either the worm or bacterial genomes. A total of 51.89 million reads were uniquely placed on the C. elegans genome: ∼27.9 million reads matching perfectly, ∼9.5 million with one mismatch, ∼5.7 million reads had two mismatches, ∼3.7 million reads had three mismatches, ∼2.5 million reads had four mismatches, ∼1.6 million reads had five mismatches, and ∼0.9 million containing six mismatches between the two 25-bp ends. By placing 50 nt reads uniquely on the genome, we exclude about 9 million base pairs of the C. elegans genome (i.e., 50-bp regions of the genome that are not unique and have an exact match elsewhere or have a second best match within one mismatch). Using this approach, our 44 million-placed putative nucleosome core reads correspond to, on average, one read every 2 bp of the genome. Since each placed read represents the position of a putative nucleosome covering 147 bp of DNA, our nucleosome coverage density of the genome is 71× or ∼484 nucleosomes per kilobase pair of unique genomic sequence.
Table 1.
Summary of SOLiD read placement
General caveats
With such a large data set sampling of nucleosome and control sequences across and throughout the genome, it is important to be aware of potential limitations of the tools used to generate the sampling. As part of the SOLiD sequencing sample prep, a library population or sample undergoes linker ligation, preliminary template amplification, and subsequent emulsion PCR to attach and extend clonal template species onto a single bead. In addition to the standard concerns of ligation and PCR bias, the emulsion PCR adds several possible caveats, including duplication of templates (multiple beads per reactor) and polyclonal beads (multiple templates per reactor). The latter could result in the obfuscation of individual sequence reads when multitemplated beads are removed from analysis due to multiple channels of fluorescence coincident on polyclonal beads, and the former could result when more than one bead extends the same molecule to result in over-representation of individual sequences. Additionally, in this data set there is an under-representation of coverage (both control and experimental) coincident with A/T-rich regions of the genome (mainly in introns and intergenic regions). This under-representation might reflect inefficient sequencing in these genomic loci due to the lower melting point of A/T-rich oligonucleotides in the ligation sequencing. In any case, for our analysis, sequencing biases such as that against A/T-rich DNA are greatly reduced by normalization with control experiment data.
The other tool used in this study with major caveats is micrococcal nuclease (MNase). While this enzyme is universally used to isolate nucleosome core DNA by preferentially digesting linker DNA to liberate the mononucleosome cores, its activity is not without sequence biases (Wingert and Von Hippel 1968; Horz and Altenburger 1981; McGhee and Felsenfeld 1983). These biases can result in the imprecise trimming of nucleosome core DNAs, cutting into the core DNA by a few nucleotides, or leaving behind a few linker DNA nucleotides. Such imprecise trimming, combined with some ability of MNase to cleave at preferred sequences will result in “blurring” of the resolution of the ends of nucleosome core DNAs by a few nucleotides. One manifestation of this is the strong bias for A or T at the first and G or C at the second nucleotide position in the beginning of nucleosome core DNAs (Johnson et al. 2006). MNase also has the potential to cleave nucleotides within the core of the nucleosome itself when highly accessible sites are on the outer surface of the core, thus de-enriching any mononucleosome core DNA library for cores that contain these preferred sites internally (McGhee and Felsenfeld 1983).
Genome browser and individual loci
Our current core read data set is of sufficient density to allow analysis of the nucleosome footprint at any unique locus in the genome. To facilitate the global browsing of unique loci we have converted the data to a genome browser format, which can be viewed using the UCSC genome browser with the WS170 version of the C. elegans genome (http://genome.ucsc.edu/cgi-bin/hgGateway). There are nine custom tracks used to display our data and subsequent analysis. The first three tracks display the raw data from control and nucleosome core isolation experiments. Tracks two and three show the start of each placed nucleosome core read, with forward reads in blue and reverse reads in green. Each feature represents putative nucleosome cores extending 147 nt, with a thick bar of 50 nt in length demarking one end of and the portion of the nucleosome core actually sequenced, followed by a thin bar ending in a line that extends an additional 97 nt in the forward or reverse direction, representing the portion of the genome protected by the entire idealized putative nucleosome core and its respective extrapolated opposite end. Multiple reads starting at the same nucleotide were collapsed into the same feature and colored according to number of contributing reads (one read, two reads, three to five reads, six to 10 reads, and >10 reads). The control data (orange) was displayed in a similar fashion. The analysis tracks include coverage and nucleosome-positioning stringency plots. At every nucleotide, we provide coverage values from the control (orange) and nucleosome experiments (blue and green). They can be used to evaluate the frequency of nucleosome instances at every position. The nucleosome-positioning stringency track (purple) evaluates the degree of nucleosome positioning at every base pair (see below for formulaic description). Finally, the adjusted nucleosome coverage track (pink) provides the measure of abundance of nucleosome instances relative to control data to account for any local enzymatic and sequencing biases. When evaluating the nucleosome coverage of any particular locus, it is important to assess both the nucleosome and control read coverage. To aid in identifying regions of the genome with repetitive structure, we provided an additional browser track that shows masking of 25 base segments of genomic DNA that are not unique in C. elegans (Fire et al. 2006). A region devoid of nucleosome starts, but containing control starts, would indicate a bona fide nucleosome depleted region, or be devoid of both nucleosome and control starts, indicating a nonrepetitive region recalcitrant to sequencing by the SOLiD technology (discussed below).
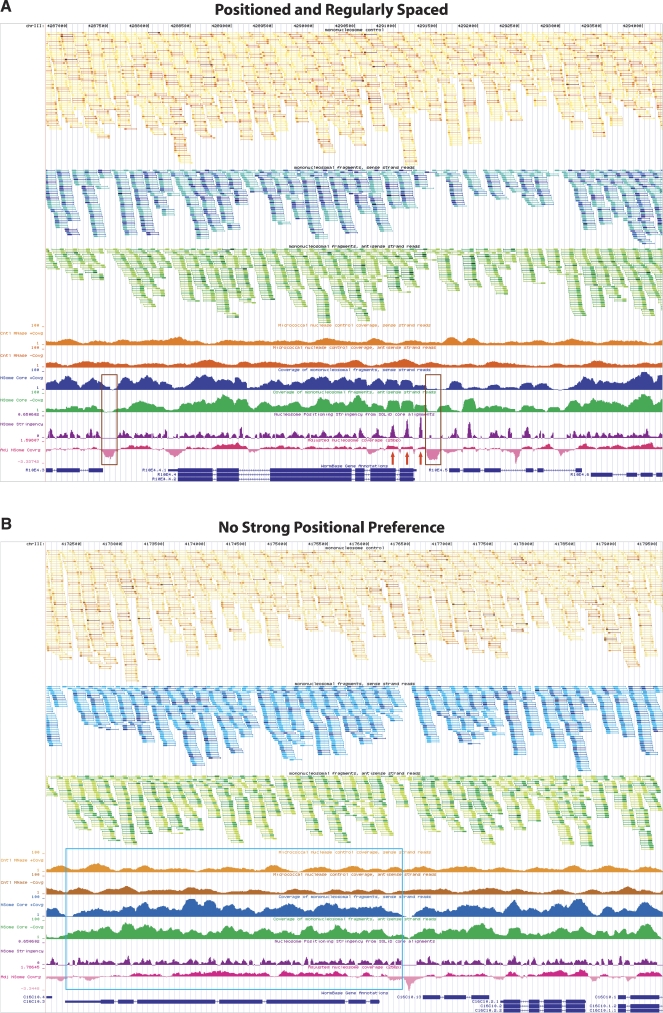
Using the nucleosome browser, we see multiple loci with reproducibly positioned and regularly spaced nucleosomes and areas of nucleosome eviction (Fig. 3A) and regions with no strong positional preference (Fig. 3B). The latter scenario is the predominant feature of C. elegans chromatin as assessed by our global analysis of neighboring nucleosomes (Start-to-Start and Start-to-End analysis, see below).
Figure 3.
UCSC Genome Browser displaying custom tracks of the Caenorhabditis elegans nucleosome position map. (A) Browser shot of the mcm-5 gene locus with several preferentially positioned nucleosomes in an organized array (red arrows). Nucleosome-depleted regions upstream of the ATG are highlighted by brown boxes, a common feature of many actively transcribed genes as noted by several groups (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Lin et al. 2007; Whitehouse et al. 2007; Schones et al. 2008; Shivaswamy et al. 2008; Yuan and Liu 2008). (B) A representative locus (displaying gene C16C10.3) in the genome demonstrating lack of nucleosome positioning particularly across a 3.5-kb stretch (highlighted by the blue box) possibly due to random nucleosome occupancy. In both A and B the top three tracks display raw data from control (orange) and nucleosome (forward blue and reverse green) experiments. Tracks two and three represent 147 nt stretches of putative nucleosome cores from forward (blue) and reverse (green) reads, with the first 50 sequenced nucleotides indicated by a thick portion of the track feature. Track one displays both forward and reverse control data (orange) in a similar fashion as the nucleosome data. The nucleosome core and genomic DNA control reads have been collapsed and colored such that multiple reads starting at the same nucleotide are represented by a single feature that varies in hue from lightest to darkest depending on the number of instances. Lightest to darkest hues correspond to the following categories: one read instance, two read instances, three to five instances, six to 10 instances, and >10 instances, respectively. Tracks four and five (coverage of nucleosome control, sense/antisense strand reads) display the coverage by 147 nt stretches from the control data. Tracks six and seven (coverage of mononucleosomal fragments, sense/antisense strand reads) show coverage by putative nucleosome cores inferred from reads that map to sense (blue) or antisense (green) strand of the reference genome. Track eight (purple) evaluates nucleosome positioning stringency at every base pair varying between 0 and 1, such that 1.0 corresponds to 100% positioning and 0.0 corresponds to no positioning or insufficient data. Track nine (adjusted nucleosome coverage, pink) displays nucleosome coverage (on a log2 scale) relative to control data to account for sequencing and enzymatic biases. Areas falling below 0.0 are on average more depleted for nucleosomes, while areas above 0.0 have increased frequency of nucleosome instances. ±1 indicates a twofold increase or depletion of putative nucleosome cores at that position.
Global analysis of nucleosome positioning and spacing in C. elegans
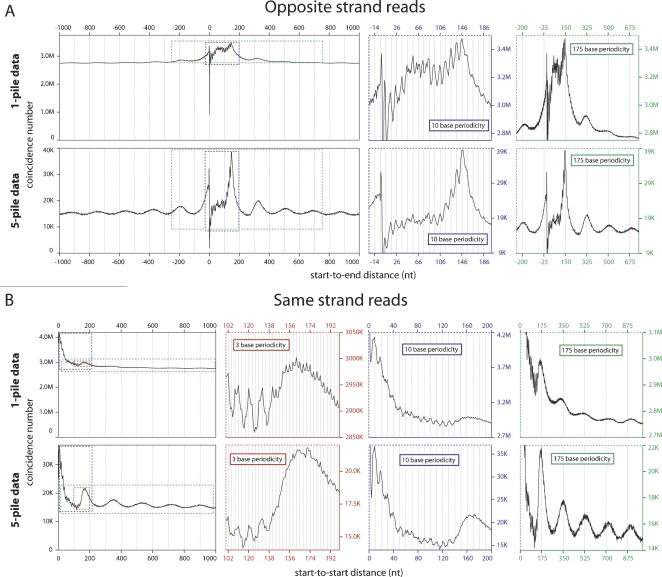
The dense nucleosome read coverage across the genome allowed us to perform a high-resolution global analysis of C. elegans chromatin, searching in particular for features that could not be seen by lower-density analysis. We derived a nucleosome read start set (1-pile data set) of 22,546,201 unique nucleosome core start-site positions in the C. elegans genome by taking all of the individual read start sites in the genome where one or more nucleosome core reads are detected. We observed some degree of nucleosome positioning constraint detected by using the 1-pile data set to compare distances between read starts on different strands (Start-to-End distances) (Fig. 4A, top). This positioning was evidenced by a major peak 147 nt downstream from the start site representing the opposite end of another nucleosome being sequenced from the opposite direction, indicating that there is at least a subset of genomic sites where nucleosomes exhibit a preferential localization. Additionally, more subtle peaks are detected downstream of this major peak at a periodicity of ∼175 nt, providing evidence for constrained positioning and spacing of neighboring nucleosomes in certain regions of the genome. Comparison of distances between starts of core reads on the same strand (Start-to-Start distances) revealed a consistent set of characteristics. (1) The major feature is the relative uniformity of the distribution, indicating that nucleosome positions for most of the C. elegans genome are flexible, (2) a series of peaks at ∼175 nt downstream and at further 175-nt intervals indicates some situations where positions are constrained and spacing is uniform (Fig. 4B, top).
Figure 4.
Global analysis of positional relationships between individual pan-cellular and neighboring nucleosomes. (A) Start-to-End distances for reads mapped to opposite strands. The graph shows total pairs of nucleosomes with a Start-to-End distance corresponding to the value on the X-axis. The dominant peak corresponding to position 146 (the 147th base from the start of the read) demonstrates reproducible positioning of nucleosomes at the same loci across cells. The colored graphs to the right zoom in on the same data (areas demarked by colored dotted lines), but highlight the 175-base periodicity reflecting the phasing on neighboring nucleosomes (green graph) and the 10-base periodicity indicative of rotational positioning of nucleosomes (blue graph). (B) Start-to-Start distances of reads mapped to the same strand. The graph shows total pairs of nucleosomes with Start-to-Start distance corresponding to the value on the X-axis. A subtle broad peak is located at approximately base 175, with echoes of this peak at a periodicity of 175 bases (green graph). A 10-base periodicity (blue graph) is also seen extending out from the start site, but is somewhat obscured by an underlying 3-base periodicity (red graph). In A and B the top set of graphs are generated form the total data (1-pile data) and the bottom graphs are generated from data enriched for positioned nucleosomes (5-pile data). This enrichment results in a greater relative amplitude of the major signal for reproducibility in absolute nucleosome positioning (A, bottom graph, major peak at 146) and in relative positioning (A,B, bottom, green graphs), but slightly decreases the resolution of the 10-base periodicity (A,B, bottom, blue graphs) and makes the 3-base periodicity (B, bottom, red graph) less prominent due to greater noise in the signal from the smaller data set (only 1/28th the size of nonenriched data set).
We can focus on regions of the genome where positioning of nucleosomes is partially or fully constrained by analyzing positions where at least five core reads define the same genome position. This “5-pile data set” contains 1,539,014 different start sites. On this data set, the “Start-to-End” analysis described above reveals an even stronger peak at ∼150 bp, independently reinforcing the preferred positioning of these nucleosomes. This plot also provides evidence of preferred spacing of neighboring nucleosomes, since we observe strong additional peaks at intervals of ∼175 bp (Fig. 4A, bottom). Start-to-Start analysis on the 5-pile data set reveals a strong peak at ∼175 bp (with echoes at 175-bp intervals), again supportive of preferred spacing of neighboring nucleosomes (Fig. 4B, bottom). Comparison of the analyses of the 5-pile versus the 1-pile data sets demonstrates an enrichment for positioned nucleosome signal (the amplitudes of the Start-to-End ∼150-bp peaks are 156% versus 26.4% of the baseline amplitude for the 5-pile versus the 1-pile data, respectively) and demonstrates that reproducible positioning tends to occur in neighboring clusters of nucleosomes (the amplitudes of the Start-to-End neighboring nucleosome peaks are 34.6% versus 6.58% and the amplitudes of the Start-to-Start neighboring nucleosome peaks are 45.4% for the first and 19.1% for the second versus 9.18% and 3.66% for the respective peaks in the 5-pile versus the 1-pile analyses).
In addition to the strong evidence for some degree of preferential positioning and uniform spacing of neighboring nucleosomes in the C. elegans genome, the Start-to-End and Start-to-Start analyses in both the 1-pile data and the 5-pile data sets reveal features that are consistent with rotational setting of nucleosomes. There is a striking 10-base periodicity of subpeaks that surround the major 147-base peak in the Start-to-End analysis (Fig. 4A). These peaks are very pronounced moving upstream and are much more subtle moving downstream from the major peak in the Start-to-End graph. The same evidence, consistent with rotational setting, exists in the Start-to-Start data, with subpeaks extending out from position zero and continuing at a period of 10 bases through position 130, thereafter being lost (Fig. 4B). These Start-to-Start subpeaks are somewhat blurred by a subtle, but persistent, three-base periodicity that is superimposed on the underlying data (presumably reflecting codon biases in protein-coding regions and their effects on true and/or measured nucleosome positions). While these data do argue in favor of rotational positioning/setting of nucleosomes, there are other mechanical explanations of these 10-base periodic subpeaks, which will be discussed below.
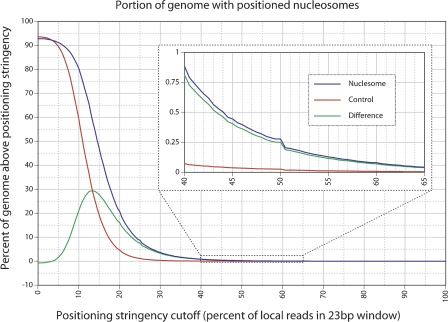
We used a separate analysis to quantify the degree of positioning as a fraction of total genomic sequence. This analysis is based on a compiled list of predicted dyad (center of nucleosome) sites (obtained by adding [for forward reads] or subtracting [for reverse reads] 73 bp from the coordinate representing each read). Positioning stringency for any site was calculated by counting putative dyads falling within a 23-bp window (the site in question plus 11 nt on each side) and dividing by the total number of infringing dyads within the flanking 150 bp (a 301-bp window). Sites were only considered as potentially positioned if both a forward-based and a reverse-based dyad were present within the 23-bp window. The analysis is illustrated for an exemplary locus in Supplemental Figure S2. Figure 5 shows the portion of the genome occupied by positioned nucleosomes as a function of the stringency (blue line). Control reads processed using an identical analysis are shown (red line), with the difference between the nucleosome and control data (green line), indicating the portion of the genome with putative positioned nucleosomes that is not due to random sampling. For low values of the cutoff score (e.g., <20%), the differences between the control and experimental curves may or may not be significant. The largest difference between the experimental and control curves (29.4%) is obtained only at a very low stringency (13.5%; so >85% of nucleosomes in such regions do not adopt the position of interest). At more significant stringency cutoffs of 20%, 25%, 33%, 50%, 66.5%, and 75%, the net portion of the genome with positioned nucleosomes (experimental vs. control) is calculated as 16.1% (20.7%–4.6%), 7.49% (8.64%–1.15%), 2.09% (2.30%–0.21%), 0.25% (0.27%–0.02%), 0.036% (0.040%–0.004%), and 0.014% (0.015%–0.001%), respectively, with the nucleosome to control differences in parentheses. Concordant results were obtained when the dyad position window was narrowed down from ±11 bp to ±5 bp (Supplemental Fig. S3).
Figure 5.
Portion of the C. elegans genome with positioned nucleosomes. The percent of the genome (vertical axis) that falls above as specified positioning stringency cutoff (horizontal axis). The blue line is obtained from the nucleosome data, the red line is obtained from the negative control non-nucleosomal data (background), and the green line indicates the difference between the two (net positioning). The inset graph is the same data expanded between the 40% and 65% stringency cutoff levels.
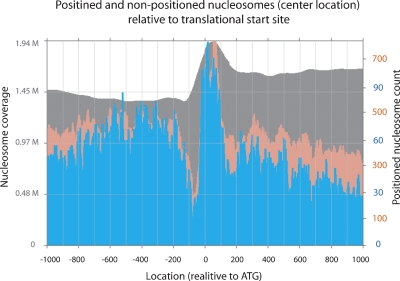
The C. elegans genome contains ∼20,000 genes. It was of considerable interest to address where positioned nucleosomes occur relative to transcriptional start sites. To assess the relationship between these sites and promoter sequences, we first needed at least an approximation to the list of C. elegans transcription start sites. Note that only a handful of transcriptional start sites are known for C. elegans, since trans-splicing tends to mask true initiation sites (Blumenthal 1995). As an alternative approximation, we have used defined and predicted translational start sites, which in many cases (due to the general brevity of C. elegans out-trons and 5′ UTRs) will provide a close mark for transcriptional initiation. We used moderate positioning stringency cutoffs of 20% and 33% to map the dyads of these putative positioned nucleosomes relative to the ATG of the translational start site for all RefSeq genes (UCSC genome browser) (Fig. 6). We observe a significant enrichment of positioned nucleosomes covering the translational start site, but centered slightly downstream (reaching maxima of 2.37- and 1.85-fold enrichment for 20% and 33% data relative to the mean across the respective plots). Notably, there is a pronounced depletion of positioned nucleosomes at ∼100 bp upstream of the start of the coding sequence (bottoming out with 6.13- and 2.40-fold depletion for 20% and 33% data relative to the mean across the respective plots). Similar results were observed genome-wide relative to transcriptional start sites in yeast (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Whitehouse et al. 2007; Shivaswamy et al. 2008; Yuan and Liu 2008) as well as in human promoters (Lin et al. 2007; Schones et al. 2008). The width of the enrichment peak and the lack of complete absence of positioned nucleosomes in the trough likely reflects a combination of variation in transcriptional activity between tissues, real variation in setting relative to the transcription start within individual tissues, and our use of translational instead of transcriptional start sites (since the former is not a fixed distance from the latter, there will be some “fuzzing” of the resulting positioning data). We also note a series of interesting enrichment peaks both upstream and especially downstream of the above-mentioned features.
Figure 6.
Positioned nucleosomes relative to the translation start site. Using the positioned nucleosome data from 20% (pink) and 33% (blue) positioning stringency cutoffs, the number of positioned nucleosome dyads (vertical axis) is plotted relative to the ATG of the translational start sites of all annotated RefSeq genes (UCSC genome browser) (horizontal axis). The gray plot in the background depicts the nucleosome coverage from the complete data relative to ATG over the entire genome.
Global sequence characteristics of nucleosome core segments
The 44.0 million sequence reads represent ∼22.5 million unique 147-bp putative nucleosome core sequences, which we designate the “22.5-M core sequence data set”. These provide a data-intensive framework to evaluate models for sequence-based nucleosome positioning. Many previous analyses of smaller nucleosome core data sets have reported a periodicity of k-mer usage within the DNA of the core of the nucleosome (e.g., Satchwell et al. 1986; Muyldermans and Travers 1994; Ioshikhes et al. 1996; Johnson et al. 2006; Albert et al. 2007), which is thought to be important in the positioning of nucleosomes due to the ability of certain regularly spaced dinucleotides to more easily facilitate the extreme curvature of the double-stranded DNA around the histone octamer (Luger et al. 1997). Consistent with these previous studies, a simple dinucleotide analysis of our core sequence data set reveals an oscillating ∼10-base periodicity of AA/TT with a counter phased ∼10-base periodicity for GC.
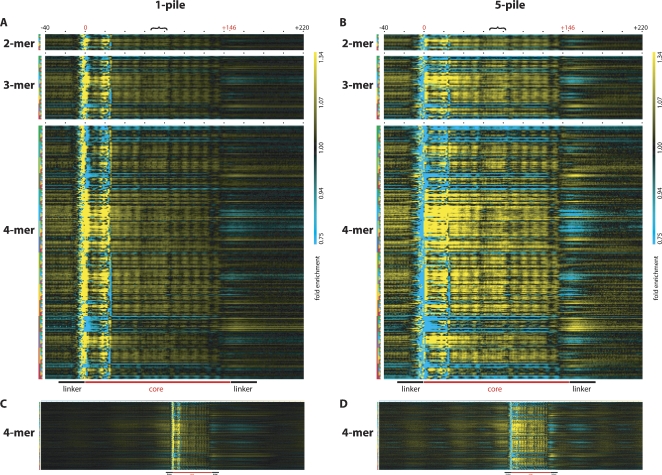
Our 22.5-M core sequence data set is sufficiently large to permit analysis of the tri-, tetra-, penta-, and even hexanucleotide (k-mer) prevalence as a function of position in nucleosome cores from C. elegans. To this end, we analyzed the over- or under-representation of each of the 64, 256, 1024, and 4096 tri-, tetra-, penta-, and hexanucleotide combinations (words) at each position in the nucleosome core relative to their prevalence in the control sequence data set inferred from the control read data set. To render the results in a visually accessible format, we generated intensity plots (“heat maps”) composed of grids, in which each column represents a position in the nucleosome core and each row represents an individual k-mer word. At any point on the plot, the over-representation or under-representation of a particular k-mer may be assessed by the intensity of the color, with the brightest yellow being a 1.3-fold or greater over-representation, black being no enrichment, and brightest cyan being a 1.3-fold or greater under-representation of the k-mer relative to the control set, with the intermediate colors representing a logarithmic scale within this range. Separate plots were generated for 2-mer, 3-mer, 4-mer (Fig. 7A,B), 5-mer (Supplemental Figs. S4, S5), and 6-mer (available on request) using both the 1-pile data (inclusive) and the 5-pile data (positioned-nucleosome-enriched set). An alternative means of representing the data that uses a wider color spectrum for enhanced visualization of subtle enrichment patterns is shown in Supplemental Figure S6. For each plot, any individual k-mer may be evaluated across the entire nucleosome core by reading the intensities across an entire row; conversely, an individual position within the core may be evaluated across all k-mers by reading down an entire column. Expanding the analysis to include the 500 nt flanking the start of the core sequences allowed us to look at the k-mer frequencies in the putative linker regions and also to detect neighboring nucleosomes, as evidenced by the fainter enrichment/de-enrichment patterns seen as echoes of the nucleosome core pattern both upstream and downstream of the core (Fig. 7C,D).
Figure 7.
Over- and under-representation of oligonucleotide words in and around nucleosome cores. The over- (yellow) and under-representation (cyan) of k-mer words in the nucleosome core is displayed for every position within the core. Each column represents the position in or around the nucleosome core starting with the position 40 nt upstream and ending with the position 220 nt downstream of the start position of the core as indicated by the black numbers −40 and +220. The putative start and end positions of the core are indicated with the red numbers 0 and +146, respectively, and black dots demark positions 20 nt apart, and the black bracket indicates the position of the putative nucleosome dyad. Each row represents one of the 16 possible 2-mers, 64 possible 3-mers, and 256 possible 4-mers, with the k-mer key to the left representing A in green, C in blue, G in yellow, and T in red. Thus, the key for the first row indicates AA as two green boxes, the second row indicates AC as a green, and then a blue box, the third row indicates AG as a green and then a yellow box, the fourth row indicates AT as a green and then a red box, etc. The over- or under-representation of each dinucleotide may be assessed at any position in or around the nucleosome core by looking at the intersection of any column (position relative to the start of the nucleosome core) and any row (individual dinucleotide word) with the color indicating over- (yellow) or under-representation (cyan) relative to the control value (black indicating no enrichment over sequenced control DNA). (A,B) The same depiction of data derived from the 1-pile or 5-pile data sets, respectively. (C,D) The same 4-mer analysis as in A and B expanded to show 500 positions both upstream and downstream of the nucleosome core start site. The magnification in C and D is reduced to allow visualization of all the positions. In all panels, the fold enrichment scale is the same ranging from 0.75- to 1.34-fold enrichment, and the color range is shown to the far right of A and B. Additionally, a cartoon depiction of the position of the nucleosome core along with the linker region relative to the graphical data is displayed at the bottom of A and B and at the bottom of C and D, scaled for panels. In all panels, sequencing and enzymatic biases affect the representation of k-mer words between positions −10 and +25, while the rest of the plot is expected to be free of these biases.
A visual inspection of these plots reveals a variety of distinctive features of nucleosome structure, including clear indications of the core termini, a periodic “shadow” of the individual helical turns of DNA that wrap around the core, and a uniquely structured dyad that sits in the middle of the core region. This detailed set of position-specific sequence biases should serve to refine current models for sequence-specific prediction of nucleosome seating preferences.
One striking feature of the incidence plots is a cluster of strong anomalies in virtually all k-mers at the beginning of the nucleosome core (around position 0). These anomalies can be explained by very localized sequence specificity in micrococcal nuclease trimming adjacent to the nucleosome core (e.g., an ability of MNase to trim terminal individual bases at nucleosomal termini much more efficiently if the following bases are not G/C base pairs). This bias is minimized due to use of the control data to normalize the heat maps. The asymmetric character of the anomalies at the cleavage site (i.e., lack of a similar set of anomaly at positions 146 ± 2 in the plot) argues against any definitive relationship between the cleavage-terminus sequence bias and nucleosome positioning. Conversely, the symmetric appearance of other features (such as the internal periodicity and dyad sites) is indicative of a fundamental relationship with nucleosome position.
The large data set allowed us to also address the extent to which k-mer strings of different lengths contributed to sequence preferences in nucleosomal positioning. In particular, we asked to what extent any preferences in incidence of any given (k + 1)-mer could be explained by preferences of the constituent (k)-mers. The resulting analysis, using Markov formalism, indicates a substantial contribution to nucleosomal positioning from 3-mer (as compared with 2-mer), with additional contributions of higher order (e.g., 4-mer) words (see Supplemental analysis and Supplemental Fig. S7).
Discussion
To sample the global spectra of allowed positions for nucleosome occupancy within the C. elegans genome in vivo, we have isolated (using MNase), sequenced (using SOLiD sequencing technology), and uniquely placed more than 44.0 million individual putative nucleosome core DNAs from a pan-cellular and mixed developmental stage sample. The density of this analysis provides a high-resolution map of nucleosome positioning in the entire unique portion of the genome, allowing scrutiny of any and all loci within this portion of the genome. We have made this map universally accessible through the UCSC genome browser.
We would envision the browser-based map being useful based on virtually any detailed analysis of association between DNA sequence and regulatory function in vivo. Circumstances in which a specific transcription factor has been shown to interact with a defined DNA sequence that is present in many places in the genome naturally lead to questions of the accessibility of the different putative sites in chromatin. Circumstances in which a region of the genome is a particularly active or inactive participant in modulating gene expression or in other activities of DNA (e.g., recombination, replication, repair, etc) similarly lead to questions of protected accessibility. While not all such circumstances would necessarily result from the simple nucleosomal coverage pattern, there is certainly strong precedent suggesting that this pattern will be of great use in evaluating and understanding the activities of individual loci.
Beyond the examination of individual loci, these data allow assessment of the global state of nucleosome positions both relative to the underlying DNA sequence and in relation to other neighboring nucleosomes. The major feature we observed with C. elegans chromatin has been the lack of universal sequence-dictated nucleosome positioning for a substantial majority of the genome. We note a number of differences between this mapping project and the very interesting analysis that has been reported in yeast. First, C. elegans is a multicellular eukaryote with a diverse variety of cell identities and tissue types. A mixed stage population of C. elegans thus represents a wider diversity of physiological states than is observed in the relatively uniform yeast cell populations. This allows a critical test of sequence-dictated positioning as compared with uniformity in chromosome organization as a reflection of common metabolic states. Second, the sequencing-based analysis can provide discrete quantification of degrees of positioning in terms of fractions of nucleosomes that occupy preferred positions in a given genomic region. Critically, this analysis accurately reports situations in which a degree of constraint in a given region is superimposed on a background of relatively random positioning. Finally, the sequence-based analysis is of considerable value in interpreting genomic regions where several different positioning configurations coexist; such situations may produce relatively constant overall occupancy and thus be difficult to distinguish using standard microarray procedures from more constant “random positioning”. Certainly, sequencing and microarray approaches each offer unique advantages to chromatin analysis, and we anticipate that data from both approaches will be of considerable utility in generating comprehensive biological surveys of chromatin dynamics.
Although flexibility in positioning of nucleosomes is a major feature for large portions of the genome, we see clear examples of (and bulk evidence for) regions with strong preferences in precise positioning. On a bulk scale, the population of sites with preferred positioning is most evident from the ∼146-nt “Start-to-End” peak in the analysis in Figure 4. Both bulk and individual locus analysis also show some degree of uniform spacing of adjacent and preferentially positioned nucleosomes, with a spacer length of ∼28 nt. This spacing is somewhat larger than the 14–21-nt spacing reported from previous whole-genome biochemical analyses (Dixon et al. 1990; Johnson et al. 2006). One intriguing possibility is that the positionally constrained subset of the genome exhibits a longer spacer length than average for the larger portion of the genome in which relative spacing of nucleosomes is regular (e.g., Fig. 1B), but absolute positions show little or no constraint.
An additional salient feature of the bulk analysis (Fig. 4) is a modest 10-base periodicity in putative nucleosome residency along the genome. Although intriguing in that the 10-base periodic subpeaks can be interpreted as resulting from rotational positioning (Lu et al. 1994), it is also possible that these peaks could result from biases in the sequencing technique and/or MNase digestion (McGhee and Felsenfeld 1983). These nonphysiological explanations seem less likely (1) due to a lack of such periodicity in the control runs that were sequenced with the same technology, arguing against sequencing bias, and (2) due to the extent of periodicity across the entire 147 bases covering the putative nucleosome (and not just the region adjacent to [or 10n bases removed from] the MNase cleavage site).
Conclusion
Our data and analysis indicate that multiple scenarios of nucleosome positioning coexist for C. elegans. From the browser-based analysis of individual loci and from bulk informatic analysis, we know that there are sites in the genome where nucleosomes have strongly preferred positions and near-uniform spacing, and that there are sites where nucleosomes are preferentially positioned at one site but not organized into a reproducible local array. From the strong laddering pattern observed from partial micrococcal nuclease digestion, combined with the preponderance of genomic sites where multiple positions were observed in our sequence survey, we know that variable absolute positioning is a common feature even in regions where the spacing between nucleosomes is constrained. Finally, we have observed that the degree of local plasticity of nucleosome seating is a characteristic feature of individual localities in the C. elegans genome, and we look forward to further studies in which the details of chromatin architecture and nucleosome regularity can be associated with diverse nuclear functions on a genome-wide scale.
Methods
Isolation of mononucleosome core DNA fragments
Mixed stage, wild-type (N2) C. elegans were cultured on DH5α E. coli, flash frozen with liquid nitrogen in 0.34 M sucrose/Buffer A (15 mM Tris-HCl at pH 7.4, 15 mM NaCl, 1 mM DTT, 60 mM KCl, 0.5 mM spermidine, 0.15 mM spermine, 25 mM bisulfite), and ground to a fine powder in liquid nitrogen using a mortar and pestle. After thawing on ice, CaCl2 and micrococcal nuclease (Roche) resuspended at 300 U/μL were added for final concentrations of 1 mM and 25 U/μL, respectively, followed by incubation at 16°C for 12 min to liberate the mononucleosome cores. The reaction was stopped by the addition of an equal volume of worm lysis buffer (0.1 M Tris-HCl at pH 8.5, 0.1 M NaCl, 50 mM EDTA, 1% SDS), and proteins were removed by treating with one-tenth volume proteinase K (20 mg/mL in TE at pH 7.4) for 45 min at 65°C, followed by phenol, phenol/chloroform, and chloroform extractions and ethanol precipitation. After RNase treatment and phenol/chloroform, chloroform extraction, separation of the micrococcal nuclease-digested DNA into mono-, di-, tri-, and multinucleosome DNAs was done using a 2% UltraPure Agarose (Invitrogen) gel run at 100 V for 1.5 h, and DNA from the mononucleosome DNA band was extracted from the gel using the QIAquick Gel Extraction Kit (Qiagen) following the standard protocol, with the exception of allowing the isolated gel sample to incubate in Buffer QG at room temperature until dissolved.
End repair and linker ligation
The ends of isolated mononucleosome core DNAs were processed by treating 0.5 μg of the DNA sample with T4 DNA polymerase (New England Biolabs) at 12°C for 30 min, followed by purification (MiniElute reaction cleanup kit, Qiagen). T4 kinase treatment (New England Biolabs) for 30 min at 37°C was then used to add a 5′ phosphate and remove the 3′ phosphate, followed by purification as above. Linkering was accomplished by addition of 1 μL each of the appropriate SOLiD sequencing linkers (100 μM each), 3.6 μL 5× T4 ligase buffer (Invitrogen), and 2.4 μL HC T4 ligase (Invitrogen, 5 U/μL) with a 16°C overnight incubation. The ligation reaction was separated on a 1.8% agarose gel, and the relevant band isolated (MiniElute gel-extraction kit; Qiagen).
Library preparation
Sequencing runs were conducted using cycled ligation sequencing on a SOLiD Analyzer (Applied Biosystems) and were conducted using short fragment libraries. For the C. elegans micrococcal nuclease library, DNA fragmented into mononucleosomal units of ∼147 bp and ligated to unique forward (P1) and reverse (P2) adaptors (P1, 41 bp: 5′-CCACTACGCCTCCGCTTTCCTCTC TATGGGCAGTCGGTGAT-3′; P2, 23 bp: 5′-AGAGAATGAGGAA CCCGGGGCAG-3′) was used. For control libraries, genomic DNA (50 μg) from C. elegans (N2) in 0.34 M sucrose/Buffer A/1× BSA (New England Biolabs)/1 mM CaCl2 was digested with 200 units of micrococcal nuclease (Roche) (0.4 U/μL final concentration) in a total volume of 500 μL for 10 min at 23°C. The digestion was stopped by addition of 10 μL 0.5M EDTA, followed by ethanol precipitation. The digested DNA was run on a 2% agarose gel and the smear of DNA fragments from 400–850 bp was excised from the gel and purified using the QIAquick Gel Extraction Kit (Qiagen) as noted above. The size-selected fragment molecules were then subjected to SOLiD paired-end library preparation and sequencing (Applied Biosystems Inc).
Bead preparation
Double-stranded fragment libraries were diluted to 50 pg/μL into low 1× TE buffer (1 mM Tris-OAc, 1 mM EDTA) and supplemented into a 2800-μL PCR aqueous phase containing: 1× PCR Gold buffer (Applied Biosystems), 3.5 mM dNTP mix, 25 mM, 10 μM P1 primer (IDT, Inc.; 5′-CCTCTCTATGGGCAGTCGGTGAT 3′), 3 mM P2 primer (IDT, Inc., 5′-CTGCCCCGGGTTCCTCA TTCTCT-3′), 560–22400 pg library template, 150 U AmpliTaq Gold UP (Applied Biosystems), and 1.7 billion one-micron covalent P1 DNA-coated paramagnetic beads. Briefly, covalent P1 DNA beads were prepared by covalent attachment of 5′-aminated P1 oligonucleotides (P1, 41 bp: /5 AmMC6/CCACCACTAC GCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT) (IDT, Inc.) to one-micron carboxylic acid-coated paramagnetic particles (Dynal/Invitrogen). The PCR aqueous phase reaction mix containing covalent P1 beads was then added to a 50-mL vial containing 10 mL of oil phase (SOLiD reagent, Applied Biosystems). Emulsions were generated by vortexing at 2100 rpm for 9 min, 53 sec in an Emulsomatic Device (SOLiD System, Applied Biosystems). Emulsions were transferred to 96-well plates and cycled for 40 cycles in a Gene Amp PCR system 9700 (SOLiD System, Applied Biosystems). Post-amplification, emulsion beads (ePCR beads) were broken with butanol (Sigma) and washed with 1× TE buffer (10 mM Tris-OAc, 1 mM EDTA) supplemented with 0.01% Triton X-100 (Sigma). Washed ePCR beads were then enriched for template-positive beads by hybridization with P2-coated capture beads (SOLiD reagent, Applied Biosystems). Template-positive beads were collected and extended in the presence of terminal transferase and Bead Linker (SOLiD reagents, Applied Biosystems). Extended beads were then deposited and covalently attached onto 25 × 75 mm SOLiD slides at >20,000 beads per panel (SOLiD reagent, Applied Biosystems). Template bead slides were then loaded onto a SOLiD Analyzer, and cycled ligation sequencing using the SOLiD Sequencing System was performed.
Mapping of the SOLiD sequencing data
Sequence reads from both nucleosome fragment core isolation and control experiments were aligned to the reference genome using standard SOLiD mapping pipeline. Nucleosome experiment reads were aligned allowing up to six mismatches out of 50 bp. Reads that matched to a single position at the least number of mismatches and for which the second best match contained at least two more mismatches, contributed to the final nucleosome data. Control experiment sequences were comprised of two 25-bp reads derived from the fragment ends, which were matched to the reference genome using standard SOLiD paired-end rescue pipeline allowing up to six mismatches between both ends and falling in the correct size range (400–850 bp). All mapping was performed in the SOLiD color space corresponding to dinucleotide encoding of the sequenced DNA.
Acknowledgments
We thank Hiram Clawson and Jim Kent for their help with compiling and hosting the custom browser tracks, members of the Fire lab for their help and suggestions over the course of this work, and acknowledge the National Institutes of Health (Grant NIGMS RO1-GM37706 to A.F.) and American Cancer Society (postdoctoral fellowship PF-05-121-01-DDC to S.M.J.) for financial support.
Footnotes
[Supplemental material is available online at www.genome.org. SOLiD raw sequencing data from this study have been submitted to the Short Read Archive at NCBI under accession no. SRA001023.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076463.108.
References
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Trans-splicing and polycistronic transcription in Caenohabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Dixon D.K., Jones D., Candido E.P. The differentially expressed 16-kD heat shock genes of Caenorhabditis elegans exhibit differential changes in chromatin structure during heat shock. DNA Cell Biol. 1990;9:177–191. doi: 10.1089/dna.1990.9.177. [DOI] [PubMed] [Google Scholar]
- Dressman D., Yan H., Traverso G., Kinzler K.W., Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embleton M.J., Gorochov G., Jones P.T., Winter G. In-cell PCR from mRNA: Amplifying and linking the rearranged immunoglobulin heavy and light chain V-genes within single cells. Nucleic Acids Res. 1992;20:3831–3837. doi: 10.1093/nar/20.15.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Alcazar R., Tan F. Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics. 2006;173:1259–1273. doi: 10.1534/genetics.106.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy F.J., Ong J.L., Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad. Sci. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I., Bolshoy A., Derenshteyn K., Borodovsky M., Trifonov E.N. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J. Mol. Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Tan F.J., McCullough H.L., Riordan D.P., Fire A.Z. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kwoh D.Y., Kwoh T.J. Target amplification systems in nucleic acid-based diagnostic approaches. Am. Biotechnol. Lab. 1990;8:14–25. [PubMed] [Google Scholar]
- Kwoh D.Y., Davis G.R., Whitfield K.M., Chappelle H.L., DiMichele L.J., Gingeras T.R. Transcription-based amplification system and detection of amplified human immunodeficiency virus type 1 with a bead-based sandwich hybridization format. Proc. Natl. Acad. Sci. 1989;86:1173–1177. doi: 10.1073/pnas.86.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lin J.C., Jeong S., Liang G., Takai D., Fatemi M., Tsai Y.C., Egger G., Gal-Yam E.N., Jones P.A. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath L.L., Elgin S.C. Nucleosome positioning and gene regulation. J. Cell. Biochem. 1994;55:83–92. doi: 10.1002/jcb.240550110. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Macevicz S.C. DNA sequencing by parallel oligonucleotide extensions. Lynx Therapeutics, Inc.; Hayward, CA: 1998. [Google Scholar]
- McGhee J.D., Felsenfeld G. Another potential artifact in the study of nucleosome phasing by chromatin digestion with micrococcal nuclease. Cell. 1983;32:1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- McKernan K., Blanchard A., Kotler L., Costa G. In: Reagents, methods, and libraries for bead-based sequencing. McKernan K., et al., editors. Agencourt Bioscience Corp.; Beverly, MA: 2006. [Google Scholar]
- Muyldermans S., Travers A.A. DNA sequence organization in chromatosomes. J. Mol. Biol. 1994;235:855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974;251:249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Peckham H.E., Thurman R.E., Fu Y., Stamatoyannopoulos J.A., Noble W.S., Struhl K., Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S.C., Drew H.R., Travers A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik D.S., Griffiths A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wingert L., Von Hippel P.H. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim. Biophys. Acta. 1968;157:114–126. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu J.S. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]