Abstract
A key element for defining the centromere identity is the incorporation of a specific histone H3, CENPA, known as Cnp1p in Schizosaccharomyces pombe. Previous studies have suggested that functional S. pombe centromeres lack regularly positioned nucleosomes and may involve chromatin remodeling as a key step of kinetochore assembly. We used tiling microarrays to show that nucleosomes are, in fact, positioned in regular intervals in the core of centromere 2, providing the first high-resolution map of regional centromere chromatin. Nucleosome locations are not disrupted by mutations in kinetochore protein genes cnp1, mis18, mis12, nuf2, mal2; overexpression of cnp1; or the deletion of ams2, which encodes a GATA-like factor participating in CENPA incorporation. Bioinformatics analysis of the centromere sequence indicates certain enriched motifs in linker regions between nucleosomes and reveals a sequence bias in nucleosome positioning. In addition, sequence analysis of nucleosome-free regions identifies novel binding sites of Ams2p. We conclude that centromeric nucleosome positions are stable and may be derived from the underlying DNA sequence.
The eukaryotic centromere is a special genomic locus on which the kinetochore assembles and attaches to spindle microtubules for chromosome segregation during mitosis and meiosis. Two main questions are what defines centromeres and how the information is transmitted during cell division. The underlying centromeric DNA sequences display too much variation throughout evolution to serve as a conserved mechanism for defining centromeres, although some patterns of repeated sequences seem to participate in defining the identity of centromere in many organisms (Clarke et al. 1986; Mellone and Allshire 2003; Pidoux and Allshire 2004, 2005). The remarkably well-conserved entities across species are, instead, histone proteins—in particular, a centromere-specific H3 variant called CENPA (Cnp1p in the fission yeast Schizosaccharomyces pombe) (Earnshaw and Rothfield 1985; Palmer et al. 1991; Sullivan et al. 1994; Henikoff et al. 2001), which functions as an “adaptor” between kinetochore protein complexes and evolutionarily divergent centromeric DNA (Mellone and Allshire 2003). Since histones constitute nucleosomes—the fundamental structural units of chromatin—the high degree of conservation of canonical histones and centromere-specific CENPA indicates that the centromere identity may very well be established and maintained through conserved epigenetic mechanisms involving chromatin marks such as CENPA incorporation and nucleosome positioning.
In fact, CENPA is already known to be absolutely essential for functional centromeres (Palmer et al. 1991; Sullivan et al. 1994; Takahashi et al. 2000; Henikoff et al. 2001). Furthermore, nucleosomes containing CENPA were found to be associated with a set of kinetochore proteins in human cells, providing direct evidence that centromeric nucleosomes serve as the foundation for kinetochore assembly (Foltz et al. 2006). These results suggest that centromere-specific incorporation of CENPA may lead to the establishment of centromere identity and epigenetic regulation of kinetochore assembly.
Despite the recognized importance of CENPA, much remains unknown about the organization of centromeric chromatin. The first step toward unveiling the architecture of centromeric chromatin is to determine the positions of nucleosomes and whether potential changes of nucleosomes may influence kinetochore assembly. S. pombe is an excellent model organism for this purpose. Its three centromeres are relatively complex compared to Saccharomyces cerevisiae and representative of regional centromeres commonly seen in higher eukaryotes. Unlike higher eukaryotes with highly repetitive centromeric DNA, fission yeast also has the advantage of possessing almost 7 kb of unique DNA sequence in the central core of centromere 2 (cnt2), permitting the mapping of centromeric nucleosomes using tiling arrays (see Fig. 1A). In contrast, budding yeast centromeres are very short (125 base pairs [bp]), comparable to the size of DNA wrapping around one histone octamer (146 bp). Previous studies of centromeric chromatin structure in fission yeast, using micrococcal nuclease (MNase) digestion and Southern analysis, have revealed that the central centromeres, which include cnt and part of innermost repeats (imr), have a chromatin organization distinct from other parts of chromosomes (Polizzi and Clarke 1991; Takahashi et al. 1992; Marschall and Clarke 1995). Specifically, partial MNase digestion of cnt and imr produces a smeary electrophoresis pattern, whereas MNase digestion of outer repeats (otr) as well as the bulk of chromatin produces a ladder pattern (see Fig. 1B).
Figure 1.
Structure of S. pombe centromeres. (A) Schematic diagram of the centromeres. Each of the three centromeres contains central core cnt and flanking imr and otr repeat regions. imr and otr have repeat sequences so that a probe designed for these regions will hybridize to DNA fragments from multiple loci. Cross-hybridization thus renders nucleosome mapping unfeasible for imr and otr. Similarly, cnt1 and cnt3 on chromosomes 1 and 3 share >70% identical sequences, and thus, only the partial non-overlapping portions can be tiled unambiguously. On the other hand, cnt2 contains unique sequences and can be tiled densely for nucleosome mapping. (B) Gel electrophoresis and Southern. Chromatin or naked genomic DNA was treated with MNase digestion (see Methods for the details of digestion conditions), and the resulting DNA fragments were run on gel. Southern hybridization was performed using a cnt DNA fragment (pKT110) or an otr DNA fragment (pYC148) (Takahashi et al. 1992). Bands containing mono-nucleosomes or DNA of similar size from naked DNA digestion (white boxes) were extracted and hybridized onto the microarray. The otr region shows ladders for both the wild type and the mis12 mutant. In cnt, the mis12 mutant still exhibits distinct ladders, while the wild type shows a smear with only faint mono- and di-nucleosome bands.
Vague mono- and di-nucleosome bands can be observed underneath the smeared pattern associated with the centromere cores, leading to the speculation that either these regions were mostly devoid of nucleosomes (Polizzi and Clarke 1991; Takahashi et al. 1992; Marschall and Clarke 1995) or regular periodic arrays of nucleosomes were present but masked by kinetochore protein complexes (Marschall and Clarke 1995). Importantly, the smeared pattern of cnt and imr is converted to the ladder pattern by mutations in Cnp1p and additional proteins that are functionally related to the kinetochore (Goshima et al. 1999; Takahashi et al. 2000, 2005; Hayashi et al. 2004). These results indicate that in S. pombe a hitherto uncharacterized unique centromeric chromatin is intimately related to the functional kinetochore. To understand this relationship, it is necessary to obtain a detailed picture of the chromatin by mapping nucleosomes and investigate the dynamic role of chromatin in establishing the centromere identity.
In this vein, we have applied the recently developed high-resolution tiling microarray technology (Yuan et al. 2005; Lee et al. 2007; Ozsolak et al. 2007) to determine nucleosome positions within cnt2 in S. pombe. Our study shows that, despite the common belief, there are reproducible, periodic arrays of positioned nucleosomes in the core centromeres of wild-type cells and that those nucleosome locations are preserved in a number of mutant strains.
Results
Agreement with known nucleosome locations in ade6
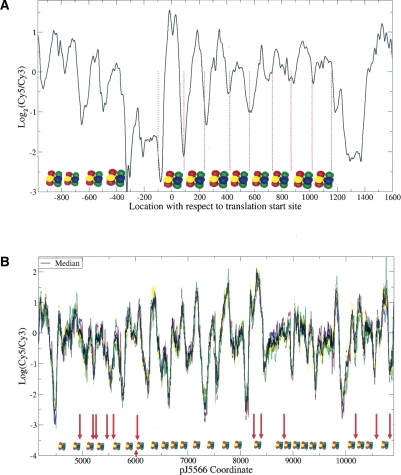
We tiled the central core centromere (cnt) regions, including the entire cnt2 and the parts of cnt1 and cnt3 that are of unique DNA sequence, at high density with a 2-nucleotide (nt) spacing between adjacent probes. A number of protein-coding genes, mainly histone-related loci, were also tiled. The fission yeast chromatin was digested with MNase, and DNA fragments were separated on agarose gel. Only the mono-nucleosome band was extracted, labeled, and hybridized. MNase-digested naked genomic DNA of similar size was used as the control (see Fig. 1B). To validate our experimental and computational procedures, we compared the nucleosome positions determined by our method with the known locations of eight positioned nucleosomes in the ade6 gene (Bernardi et al. 1991). As shown in Figure 2A, all eight nucleosomes were detected, confirming the accuracy and sensitivity of our approach. Furthermore, we also observed the characteristic nucleosome-free regions noted in Ozsolak et al. (2007) and Yuan et al. (2005) upstream of translation start sites.
Figure 2.
Chromatin structure of (A) ade6 and (B) cnt2. The mono-nucleosome band in Figure 1B was extracted, labeled with Cy5 dye, and hybridized onto the microarray, with MNase-digested naked DNA of similar size labeled with Cy3 as control. (A, black line) The mean of nucleosome data from four biological replicate experiments. (Red dotted lines) The MNase cleavage sites of eight previously known positioned nucleosomes (Bernardi et al. 1991). Positioned nucleosomes detected by our method are shown as histone octamers and agree very well with Bernardi et al. (1991). (B) The median nucleosome signal of three biological replicates, each with (bold black) two technical replicates; (colored lines) individual replicates. The technical replicates come from tiling both strands of cnt2. Positioned nucleosomes based on the median signal are shown as histone octomers. (Red arrows, including the one below pointing upward) The GATA motifs in troughs that are potential Ams2-binding sites.
Nucleosomes are well positioned in cnt2
By imposing the minimum peak-to-trough ratio of 1.4 (Ozsolak et al. 2007), we identified 27 positioned nucleosomes within the 6.8-kb cnt2 region (Fig. 2B). The hybridization intensities of six replicates (three biological × two technical) were highly reproducible with a probe-level correlation coefficient of 0.95. Some of the nucleosome positions within cnt1 or cnt3 could not be assigned unequivocally because of high sequence similarity between the two (∼72% identity for cnt1 and 61% for cnt3); nonetheless, positioned nucleosomes were also reproducibly detected in unique regions within cnt1 and cnt3 (data not shown). The nucleosome positions at inner or outer repeat regions of the centromeres could not be determined due to the repetitive nature of the underlying DNA sequence (see Fig. 1A). The distribution of positioned nucleosomes in cnt2 (coding and promoter regions) is summarized in Table 1. The level of nucleosome occupancy in cnt2, which was comparable to that throughout the genome, is 11% less and 3% more than that in the coding and promoter regions, respectively. In summary, in wild-type cells, centromeric chromatin is occupied by orderly positioned nucleosomes.
Table 1.
Distribution of nucleosomes
For comparison, the average number of positioned nucleosomes is 3.3/kb in the S. cerevisiae genome (Lee et al. 2007) and ∼2/kb in human promoters (Ozsolak et al. 2007).
Altering Cnp1p loading does not affect the centromere nucleosome positioning
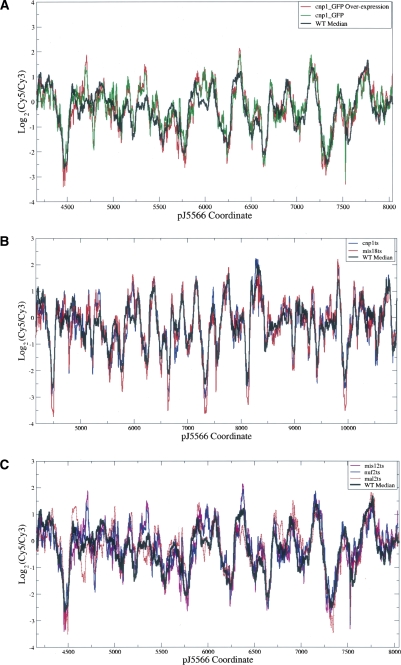
To study the possible effect of Cnp1p-containing nucleosomes on centromere chromatin, we first tested whether the increasing Cnp1p incorporation into centromeres alters the nucleosome positioning. A specific cnp1-GFP overexpression construct is known to increase the total Cnp1p concentration in the whole cell extract as well as the loading of Cnp1p into centromeres by fivefold to sixfold (Chen et al. 2003a; Joglekar et al. 2008). We compared the locations of nucleosomes among wild-type cells, cells that expressed cnp1-GFP at the endogenous level, and cells that overexpressed cnp1-GFP. As shown in Figure 3A, the nucleosome data from these cells exhibited a very high level of correlation above 0.9. We then tested mutations that cause Cnp1p mislocalization or reduction of Cnp1p loading, including temperature-sensitive mutations in Cnp1p (Takahashi et al. 2000) and Mis18p (Hayashi et al. 2004) and deletion of ams2 (Chen et al. 2003a). The Mis18 mutation also causes hyperacetylation of histones at the central centromeres (Hayashi et al. 2004). As shown in Figure 3B, the nucleosome data from wild-type and all kinetochore mutants showed a very high correlation. There was also no noticeable change in the locations of positioned nucleosomes and in the accessibility of the GATA motifs potentially recognized by Ams2p (see below), although Δams2 displayed more noise than other mutants (Supplemental Fig. S2A).
Figure 3.
Chromatin structure is preserved in mutants. (A) Wild-type versus Cnp1p overexpression. (B) Wild type versus mutants affecting Cnp1p localization. (C) Wild type versus outer kinetochore mutants. The array design in B covered a region longer than that in A and C.
Mutations in outer kinetochore proteins do not disrupt the nucleosome positioning in the centromere core
To assess directly the relationship between the outer kinetochore and the underlying centromere nucleosome positioning, we examined mutations within the two separable structural motifs of the kinetochore—mis12-537 and nuf2-1 in the NMS supercomplex and mal2-1 in the Sim4 supercomplex (Liu et al. 2005). It is known that the mis12-537 mutation severely disrupts the kinetochore architecture by mislocalizing the whole NMS complex (Goshima et al. 1999), whereas the mutant nuf2-1 disrupts microtubule binding activity of kinechores without detectable defects in the kinetochore structural integrity (Nabetani et al. 2001; Appelgren et al. 2003; X. Liu and X. He, unpubl.). The mal2-1 mutation causes a structural defect by mislocalizing Sim4 complex components and also leads to chromosome mis-segregation (Fleig et al. 1996; Jin et al. 2002). As shown in Figure 3C, we found no noticeable change in nucleosome positions in any of the tested mutants, compared to those in wild-type cells.
DNA sequence may direct nucleosome positioning in the centromere core
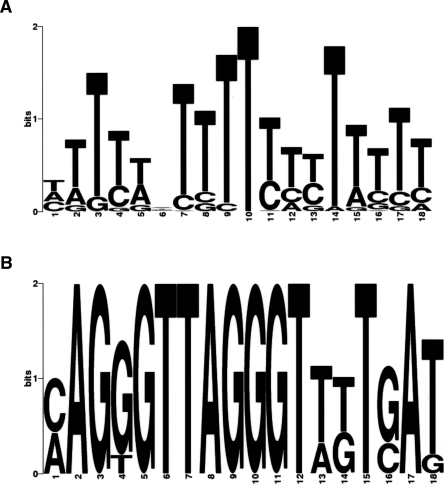
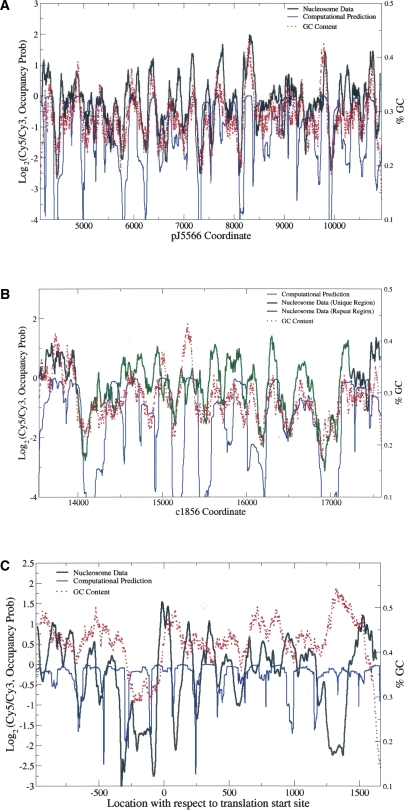
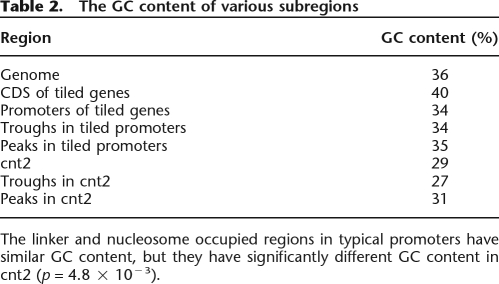
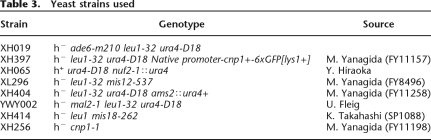
DNA sequence can provide a landscape of electrostatic potential energy and guide nucleosome positioning (Levitsky 2004; Segal et al. 2006). It was previously found that a stretch of T was enriched in linker DNA (Ozsolak et al. 2007). Similarly, using the de novo motif discovery program MEME (Bailey and Elkan 1994; Bailey et al. 2006), we found that a poly(T) motif (Fig. 5A, see below) was significantly enriched in our troughs (E = 8.0 × 10−6), agreeing with the previous finding. The enrichment of this poly(T) motif in troughs is consistent with the fact that it lacks the periodic non-T(A/T)G triplets that are known to favor nucleosomes (Stein and Bina 1999). The sequence-based predictions of nucleosome positioning described by Levitsky (2004) and Segal et al. (2006) utilize periodic dinucleotide probability distributions to calculate the DNA bending potential. We used the program from Segal et al. (2006) to compute the probability of nucleosome occupancy in cnt2. Figure 4A shows that ∼75% of nucleosomes detected by our method also have a high sequence probability of nucleosome occupancy. Another algorithm described in Yuan and Liu (2008) also yielded comparable results (data not shown). In addition, we computed the GC content of each peak and trough. Table 2 summarizes the average GC content in various regions. The GC contents of peaks and troughs in the 65 promoters investigated in this study were similar, while they differed significantly in cnt2 (p = 4.8 × 10−3, two-sample Welch t-test), suggesting that DNA sequence may guide nucleosome positioning in centromeres. To test this hypothesis, window averaged GC content was computed at each position in tiled regions and compared with nucleosome hybridization signals (see Fig. 4). The correlation between GC and nucleosome intensities was 0.66 for cnt2, well above the mean value of 0.15 for the tiled genes; it was also above 0.5 for cnt1 and cnt3. This high correlation of nucleosome signal with GC content was not an artifact of the probe behavior, as the GC content of cnt regions was actually lower than that of other parts of the genome, and the corresponding correlation was much lower outside the centromeres. This finding is also consistent with the recent work showing that the nucleosome formation potential may depend on the GC content, with GC-rich and AT-rich sequences favorable and unfavorable for nucleosome formation, respectively (Peckham et al. 2007).
Figure 5.
Enriched motifs in troughs. (A) Poly(T) and (B) histone motifs were found by scanning all trough regions with MEME.
Figure 4.
Nucleosome data correlate well with sequence-based predictions in (A) cnt2 and (B) cnt1 but not in (C) ade6. (Black line) The median nucleosome data for wild type; (blue line) the log nucleosome occupancy probability, computed as in Segal et al. (2006). (B, green line) The nucleosome data in the region repeated in cnt3; (red dotted line) the running window (size 120 bp) log GC content. In cnt1 and cnt2, the correlation between the nucleosome data and GC content is high, and the computational prediction of nucleosome locations works well. In ade6, however, the correlation is only 0.1, and the computational algorithm also fails to predict nucleosome-free regions.
Table 2.
The GC content of various subregions
The linker and nucleosome occupied regions in typical promoters have similar GC content, but they have significantly different GC content in cnt2 (p = 4.8 × 10−3).
Locations of Ams2p-binding sites and enriched motifs
Since functional regulatory elements tend to lie in nucleosome-free linker regions (Yuan et al. 2005; Ozsolak et al. 2007), we examined whether Ams2p recognition sites are also free of nucleosomes. We scanned the cnt2 region with a GATA sequence motif matrix and found 18 occurrences of the motif. Of those 18, 13 sites were found in troughs (P-value = 0.02, binomial test with success probability normalized by the trough length), as indicated by the red arrows in Figure 2B.
To discover common motifs accessible to proteins, we used MEME to scan all troughs from our tiled regions (Bailey and Elkan 1994; Bailey et al. 2006). Interestingly, we found that the so-called histone motif (Fig. 5B), which is a conserved sequence of roughly 20 nucleotides identified in all histone gene promoters (Choe et al. 1985), was significantly enriched (E = 1.2 × 10−4), not only in the promoters of histone genes but also in the promoter of slp1 and SPAC22E12.19 (a putative histone deacetylase complex subunit). This motif is slightly less stringent than the previously characterized version (Choe et al. 1985). Using the resulting motif weight matrix, all 1-kb promoters of coding genes were scanned, and the histone motif was also discovered in SPAC631.02 (bromodomain protein) and spi1 (Ran GTPase). No other significant motifs were found.
Discussion
Cnp1 loading and outer kinetochore mutations do not affect the centromere nucleosome positioning
The unique chromatin structure of central centromeres as previously characterized by the smeared MNase digestion pattern has led investigators to hypothesize that the centromere-specific loading of Cnp1p may require disruption of the centromeric chromatin and that the smear is a manifestation of this disruption. In particular, it has been proposed that Ams2p may displace nucleosomes and guide the incorporation of Cnp1p (Chen et al. 2003a). If this model were true, the overloading of Cnp1p or ams2 deletion should affect the nucleosome positioning in the centromeres. However, our results in Figure 3, A and B, indicate that Cnp1p loading involves only the remodeling of individual nucleosomes and does not affect the positions of neighboring nucleosomes in the centromeres.
Mutations in multiple kinetochore proteins are also known to eliminate or reduce the smeared pattern of MNase digestion. These mutations are thus thought to introduce new nucleosome arrays and change the unique centromeric chromatin structure (Fleig et al. 1996; Goshima et al. 1999; Takahashi et al. 2000; Jin et al. 2002; Chen et al. 2003a; Hayashi et al. 2004), which was originally identified with nucleosome eviction (Polizzi and Clarke 1991; Takahashi et al. 1992; Marschall and Clarke 1995). As shown in Figure 3C, however, the mutants displayed no noticeable disruption in chromatin structure compared to the wild-type cells. We thus conclude that mutations in outer kinetochore proteins do not cause changes in nucleosome distribution and that centromere function and kinetochore assembly do not entail major chromatin remodeling by nucleosome relocation.
The smeary pattern observed from the Southern analysis of centromeric DNA after partial MNase digestion was probably caused by protection provided by the intact kinetochore. That is, the kinetochore may cover the centromeric DNA and allow only random cleavages while severely hindering cleavages at nucleosome boundaries, resulting in a continuous distribution of different sizes of DNA fragments. By specifically analyzing, using the tiling array, the DNA fragments extracted from a population of mono-nucleosomes after chromatins were treated with MNase, we were able to locate positioned nucleosomes and show that these nucleosomes are stable even when kinetochore components were mutated. The protection by the kinetochore must not be complete, as mono-nucleosomes derived from partial digestion hybridization yielded the same chromatin structure as the usual near-complete digestion (see Supplemental Fig. S2B). This finding may also explain the previous observation that in wild-type cells, mono- and di-nucleosome patterns were observed over a smear background in the centromere core regions by Southern analysis after chromatins were treated with MNase (Marschall and Clarke 1995; Goshima et al. 1999). As similar smeared patterns were observed in fission yeast centromeres during meiosis (Smirnova and McFarlane 2002), in Candida albicans centromeres (Baum et al. 2006), and in human telomeres (Tommerup et al. 1994), our model may be easily extended and shed light on the chromatin structure of those regions.
Possible structural differences between Cnp1-nucleosomes and the canonical nucleosomes may potentially complicate the interpretation of our results. For example, it has been observed that MNase digestion of reconstituted CENPA-containing nucleosomes yields 120–150-bp bands instead of the expected ∼146-bp length, indicating that CENPA nucleosomes are either more sensitive to MNase digestion or structurally distinct from canonical nucleosomes (Yoda et al. 2000). It was also shown that in the absence of CENPA nucleosome-associated complexes, CENPA nucleosomes are more sensitive to exonuclease III and disassemble more easily compared to canonical nucleosomes (Conde e Silva et al. 2007). Although these studies were done in vitro using reconstituted nucleosomes, they raise the question whether our method of mono-nucleosome extraction may have excluded the majority of CENPA nucleosomal DNA, thus leading to an alternative interpretation that none of the 27 identified peaks represent the CENPA nucleosomes. Our results, however, indicate that the alternative model is unlikely:
Our partial MNase digestion experiment (Supplemental Fig. S2B), which preferentially cleaves the linker DNA and likely preserves the CENPA nucleosomal DNA, shows that the distribution of positioned nucleosomes is virtually identical to that in the rest of the experiments.
In our protocol for gel extracting mono-nucleosomal DNA, we took a band corresponding to the DNA length from 110 to 160 bp, allowing for some ambiguity in MNase digestion. Thus, our sample would have included the CENPA nucleosomal DNA fragments even if they were overdigested to below 146 bp.
In a separate study, we have used quantitative fluorescence microscopy to show that each kinetochore contains on average six to seven copies of Cnp1p in wild-type cells (Joglekar et al. 2008), implying that on average three centromeric nucleosomes are Cnp1-nucleosomes. In contrast, when Cnp1 is overexpressed, the number of Cnp1-nucleosomes increases to 16–18 per kinetochore (Joglekar et al. 2008), which is more than half of the total nucleosomes that we have identified in cnt2. If none of the identified 27 peaks were Cnp1-containing nucleosomes, then the remaining centromeric DNA would not be long enough to accommodate the 16–18 additional Cnp1-nucleosomes in Cnp1-overexpressed cells.
We believe that these considerations together provide a strong support for our interpretation that CENPA nucleosomes were well represented in our study. The precise positions of Cnp1 nucleosomes are currently under investigation.
Ams2-binding sites
The GATA family of transcription factors associate with chromatin-remodeling complexes and share similar DNA-binding domains that recognize consensus sequences containing the core motif GATA, hence the name. Ams2p, a GATA-like transcription factor, has been shown to bind to the centromere core regions and plays an important role in localizing Cnp1p to centromeres during S phase (Chen et al. 2003a, b; Takahashi et al. 2005; Takayama and Takahashi 2007). The precise mechanism for how Ams2p facilitates Cnp1p loading is not known. Two proposed models are: (1) Ams2p may directly participate in a protein complex that remodels nucleosomes, and (2) Ams2p may initiate the transcription of non-coding centromeric RNA, which then interacts with Cnp1p (Chen et al. 2003a, b; Takahashi et al. 2005; Takayama and Takahashi 2007). In either case, our work shows that the activity of Ams2 may influence the composition but not the locations of centromeric nucleosomes and thus argues against the nucleosome eviction hypothesis for functional centromeres (see Supplemental Fig. S2A).
The preferential location of GATA motifs in troughs further supports the idea that Ams2p binds the centromeric core DNA. Ams2p also functions as an authentic transcription factor by binding to the promoters of histone genes and is required for their S-phase-specific transcription (Takayama and Takahashi 2007). Among the histone genes, only H2A.2, H3.3, and H4.3 had a GATA motif in the promoter (see Supplemental Fig. S1), indicating that the consensus recognition sequence of Ams2p may differ slightly from known GATA motifs. In fact, a recent study shows that the histone motif may also contain a GATA-like recognition motif for Ams2p (Takayama and Takahashi 2007).
An orderly nucleosome positioning pattern as an emerging common feature of centromere chromatin
A well-established common hallmark among centromeres from different species is the presence of CENPA-containing nucleosomes (Sullivan et al. 2001; Cleveland et al. 2003; Black et al. 2004). Here, we propose that the orderly nucleosome positioning may be another evolutionarily conserved feature. In the budding yeast S. cerevisiae, even though one nucleosome is sufficient to encompass the short centromeric DNA, the centromere-specific nucleosome is in phase with the flanking nucleosomes covering nonspecific sequences (Bloom and Carbon 1982; Yuan et al. 2005). Such a nucleosome arrangement is postulated to form the platform for kinetochore assembly (Espelin et al. 2003; Bloom et al. 2006; Westermann et al. 2007). In human centromeres, the nucleosomes between two adjacent CENPB boxes are in phase, implying that the CENPA/CENPB nucleosomes over the whole αI-type array are orderly positioned, reflecting the repetitive nature of the underlying DNA (Ando et al. 2002). Here we report that an array of nucleosomes is orderly positioned in the unique central core sequence of fission yeast centromere. Thus, orderly positioning of centromeric nucleosomes is not restricted to the repetitive sequences. Our results are consistent with the earlier finding that the fission yeast cnt DNA contained typical nucleosome arrays when carried on circular plasmids in S. pombe or artificially introduced into S. cerevisiae, indicating that there is no inherent sequence bias against nucleosome assembly in the fission yeast centromeres (Polizzi and Clarke 1991; Takahashi et al. 1992). Considering the rapid evolution of centromeric DNA in comparison to the bulk genome sequence (Meraldi et al. 2006), our finding that the underlying centromeric DNA sequence is likely to influence nucleosome positioning indicates that well-positioning of centromeric nucleosomes is evolutionarily selected for and has functional significance. The mechanism by which orderly positioned nucleosomes may influence kinetochore assembly awaits future investigation.
Methods
Yeast cell culture
The genotypes of the S. pombe strains used in this study are listed in Table 3. Cultures were grown to 1 × 107/mL in standard YE media at 25°C. The ts strains were grown to 5 × 106/mL at 25°C, then shifted to 36°C for 2–8 h as reported (Fleig et al. 1996; Goshima et al. 1999; Nabetani et al. 2001; Hayashi et al. 2004).
Table 3.
Yeast strains used
Micrococcal nuclease digestion and Southern analysis
A modified procedure based on Takahashi et al. (1992) was used. Exponentially growing yeast cells were harvested by centrifugation. The cell pellet was resuspended in two volumes of sorbitol solution (1 M sorbitol, 50 mM Tris-HCl at pH 7.5) and 10 mM β-mercaptoethanol; 2.5 mg/mL Zymolyase-20T (Seikagaku) was added to convert the yeast cells to spheroplasts for 1 h at 30°C. The spheroplasts were recovered by centrifugation, washed twice with sorbitol solution, and finally resuspended in Lysis buffer (500 mM spermidine, 1 mM β-mercaptoethanol, 0.075% NP-40, 50 mM NaCl, 10 mM Tris-HCl at pH 8.0, 5 mM MgCl2, 5 mM CaCl2) containing proteinase inhibitor cocktail (Roche).
To prepare nucleosomal DNA, the spheroplast suspension (8 × 108 cells/mL) was pre-warmed for 5 min at 36°C and then treated with micrococcal nuclease (MNase) (500 units/mL; Roche) for 15 sec–7 min at 36°C. The reaction was stopped by adding EDTA to a final concentration of 20 mM and SDS to a final concentration of 1%. The suspension was centrifuged at 14,000 rpm for 20 min. The supernatant containing the nuclease-digested nuclei was incubated with 0.1 mg/mL Proteinase K for 2 h at 50°C. DNA was extracted with phenol/chloroform, digested with RNase, re-extracted with phenol/chloroform, precipitated with ethanol, and resuspended in TE (10 mM Tris-HCl, 1 mM EDTA at pH 8.0).
To prepare naked DNA as control, the spheroplasts from wild-type yeast cells were incubated in Lysis buffer for 12 min at 36°C, then were stopped by adding EDTA to a final concentration of 20 mM and SDS to a final concentration of 1%. The total lyses were incubated with 0.1 mg/mL Proteinase K for 2 h at 50°C. DNA was extracted with phenol/chloroform and digested with RNase, phenol/chloroform re-extracted, precipitated with ethanol, and resuspended in Lysis buffer. The naked DNA was treated with 10∼500 units/mL MNase for 7 min at 36°C. The reaction was stopped by adding EDTA to a final concentration of 20 mM.
To purify DNA, electrophoresis was carried out in a 2.5% agarose gel. The 110∼160-bp band containing mono-nucleosome fragments and the corresponding 120–180-bp smear containing naked DNA fragments were cut and purified by the QIAEX II Gel Extraction Kit (QIAGEN).
After electrophoresis as above, except that a 1.6% agarose gel was used to ensure high efficiency in DNA transferring, Southern analysis was performed, using DNA fragments corresponding to cnt and otr as the probes for hybridization as previously described (Takahashi et al. 1992).
Microarray hybridization
The purified mono-nucleosomal DNA and the purified naked control DNA were labeled with Cy5-dUTP (PerkinElmer) and Cy3-dUTP (PerkinElmer), respectively, and purified using a BioPrime Array CGH Genomic Labeling Module (Invitrogen 45-0048) and Purification Module (Invitrogen 45-0049), respectively. One microgram of Cy5-labeled single nucleosome and 1 μg of Cy3-labeled naked control DNA were hybridized on Agilent’s 4 × 44k tiling array for 40 h at 65°C, and slides were washed following the standard Agilent Mammalian ChIP-on-ChIP protocol (Agilent version 9.0). The slides were scanned using a GenePix 4000B scanner and processed with GenePix 6.0 software.
High-density tiling of core centromeres and histone genes
As a proof-of-concept experiment, arrays were designed to tile both strands of cnt2 at 1-bp spacing, both strands of cnt1 and cnt3 at 2 bp, and a 1-kb 5′ upstream region+ORF of 49 histone-related genes at 5 bp and six control genes (ade6, Ura4, Adh1, nmt1, mei4, rec8) at 2 bp. For each probe, the average number of occurrence in the S. pombe genome of all 15-mer subsequences was computed. Among the 44K probes, 2606 of them had an average 15-mer occurrence of greater than 2.0. From those 2606 probes, 1749 of them had a non-unique 40-mer subsequence and were classified as repeat probes, of which 1699 were interrogating the cnt1 and cnt3 regions. Importantly, all probes in cnt2 were unique. In the second array design, we increased the spacing to 2 bp in cnt2 and tiled only one strand from cnt1 and cnt3 regions; additional genes that are known to be periodic in cell cycle were added.
Nucleosome data analysis
Positioned mono-nucleosomes were detected as described in detail in Ozsolak et al. (2007) using wavelet denoising and the Laplacian of Gaussian edge detection algorithm. Less than 100 outlier probes arising from background subtraction were replaced with values linearly interpolated from neighboring probes.
Motif analysis
To search for Ams2 recognition sites, the cnt2 region was scanned with a position specific weight matrix for GATA (M00789) from TRANSFAC, with the threshold value set to 100% of its relative entropy. Sequences from troughs and peaks were analyzed using the de novo motif discovery program MEME (Bailey and Elkan 1994; Bailey et al. 2006). The resulting 18-mer histone motif shown in Figure 5B had a very strong consensus, so a lenient cutoff of 70% of its relative entropy was used for scanning the genome with its weight matrix; raising or lowering the cutoff did not affect the scanning result.
Acknowledgments
We thank J. Wang and Z. Zhou for helpful comments on the manuscript; G. Ira for help with Southern blotting analysis; and K. Takahashi, U. Fleig, Y. Hiraoka, M. Yanagida, and the Yeast Genetic Resource Center, Osaka City University for providing yeast strains. This work was supported by NIH RO1 GM068676 to X.H. and 1R01 HG004069-01 to X.S.L. Finally, we are grateful to X. Pan and R. Person for technical advice on array hybridization and data collection.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to Gene Expression Omnibus under accession no. GSE10742.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075374.107.
References
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelgren H., Kniola B., Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., Sanyal K., Mishra P.K., Thaler N., Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F., Koller T., Thoma F. The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast. 1991;7:547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Cleveland D.W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Bloom K.S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bloom K., Sharma S., Dokholyan N.V. The path of DNA in the kinetochore. Curr. Biol. 2006;16:R276–R278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Saitoh S., Yanagida M., Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003a;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Chen E.S., Yanagida M., Takahashi K. Does a GATA factor make the bed for centromeric nucleosomes? Cell Cycle. 2003b;2:277–278. [PubMed] [Google Scholar]
- Choe J., Schuster T., Grunstein M. Organization, primary structure, and evolution of histone H2A and H2B genes of the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1985;5:3261–3269. doi: 10.1128/mcb.5.11.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Amstutz H., Fishel B., Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N., Black B.E., Sivolob A., Filipski J., Cleveland D.W., Prunell A. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Espelin C.W., Simons K.T., Harrison S.C., Sorger P.K. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U., Sen-Gupta M., Hegemann J.H. Fission yeast mal2+ is required for chromosome segregation. Mol. Cell. Biol. 1996;16:6169–6177. doi: 10.1128/mcb.16.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Jin Q.W., Pidoux A.L., Decker C., Allshire R.C., Fleig U. The mal2p protein is an essential component of the fission yeast centromere. Mol. Cell. Biol. 2002;22:7168–7183. doi: 10.1128/MCB.22.20.7168-7183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Levitsky V.G. RECON: A program for prediction of nucleosome formation potential. Nucleic Acids Res. 2004;32:W346–W349. doi: 10.1093/nar/gkh482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J.R., He X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall L.G., Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J. Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B.G., Allshire R.C. Stretching it: Putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A.D., Rheinbay E., Sorger P.K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Koujin T., Tsutsumi C., Haraguchi T., Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Song J.S., Liu X.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Trong H.L., Charbonneau H., Margolis R.L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham H.E., Thurman R.E., Fu Y., Stamatoyannopoulos J.A., Noble W.S., Struhl K., Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Allshire R.C. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., Allshire R.C. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:569–579. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C., Clarke L. The chromatin structure of centromeres from fission yeast: Differentiation of the central core that correlates with function. J. Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova J.B., McFarlane R.J. The unique centromeric chromatin structure of Schizosaccharomyces pombe is maintained during meiosis. J. Biol. Chem. 2002;277:19817–19822. doi: 10.1074/jbc.M200765200. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina M. A signal encoded in vertebrate DNA that influences nucleosome positioning and alignment. Nucleic Acids Res. 1999;27:848–853. doi: 10.1093/nar/27.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Blower M.D., Karpen G.H. Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E.S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Takayama Y., Masuda F., Kobayashi Y., Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:595–606. doi: 10.1098/rstb.2004.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y., Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 2007;22:22. doi: 10.1093/nar/gkm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommerup H., Dousmanis A., de Lange T. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu J.S. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]