Abstract
Chromatin structure is central for the regulation of gene expression, but its genome-wide organization is only beginning to be understood. Here, we examine the connection between patterns of nucleosome occupancy and the capacity to modulate gene expression upon changing conditions, i.e., transcriptional plasticity. By analyzing genome-wide data of nucleosome positioning in yeast, we find that the presence of nucleosomes close to the transcription start site is associated with high transcriptional plasticity, while nucleosomes at more distant upstream positions are negatively correlated with transcriptional plasticity. Based on this, we identify two typical promoter structures associated with low or high plasticity, respectively. The first class is characterized by a relatively large nucleosome-free region close to the start site coupled with well-positioned nucleosomes further upstream, whereas the second class displays a more evenly distributed and dynamic nucleosome positioning, with high occupancy close to the start site. The two classes are further distinguished by multiple promoter features, including histone turnover, binding site locations, H2A.Z occupancy, expression noise, and expression diversity. Analysis of nucleosome positioning in human promoters reproduces the main observations. Our results suggest two distinct strategies for gene regulation by chromatin, which are selectively employed by different genes.
Promoter chromatin structure has a key role in transcription regulation, but the exact mechanisms of chromatin regulation are still poorly understood (Kornberg and Lorch 1999; Li et al. 2007; Rando and Ahmad 2007). It is well accepted that nucleosomes decrease the accessibility of promoter DNA and thus hinder the binding of transcription factors to regulatory elements (Richmond and Davey 2003; Segal et al. 2006). Consistent with this, active promoters tend to be depleted of nucleosomes (Pokholok et al. 2005; Lee et al. 2007), and gene activation is often associated with nucleosome eviction (Lee et al. 2004; Hogan et al. 2006). Based on these results, the emerging picture portrays nucleosomes as an obstacle for transcription, and chromatin remodeling as the means to overcome repression by nucleosomes (Kornberg and Lorch 1999). However, this view is likely to be an oversimplification since many active regulatory elements are not depleted of nucleosomes, and since nucleosomes are regulated by numerous mechanisms whose exact functions are yet unclear, including nucleosome remodeling, histone modifications, and histone variants (Li et al. 2007).
Genes differ not only in their expression level but also in their capacity to alter expression levels in response to various signals. Some genes are constitutively expressed with little influence from the cellular state, while others respond to external and internal signals and dramatically modify their expression levels. Interestingly, expression of the latter genes appears to also have higher stochastic fluctuations (noise) and faster evolutionary divergence (Tirosh et al. 2006; Landry et al. 2007), perhaps indicating that their expression variability is hardwired in the corresponding promoters’ design.
What controls this property of expression variability? Several studies linked expression divergence to chromatin regulation (Lee et al. 2006; Choi and Kim 2008), while others linked it to the presence of a TATA box in promoters (Tirosh et al. 2006; Landry 2007). Interestingly, the TATA box has been associated with increased dependence on chromatin regulation (Basehoar et al. 2004) and with an atypical nucleosome occupancy (Ioshikhes et al. 2006; Albert et al. 2007; Tirosh et al. 2007). Taken together, these results suggest that expression variability could depend on promoter chromatin structure, although the nature of such association is not yet clear.
A recent study generated high-resolution data of nucleosome occupancy across the yeast genome (Lee et al. 2007). These valuable data now allow for a direct examination of the genome-wide patterns of nucleosome occupancy and their relationship with transcription regulation. Analyzing these data, we found that expression variability, as well as the dependency on chromatin regulation, are correlated with the patterns of promoter nucleosome occupancy. Furthermore, promoters can be classified by their patterns of nucleosome occupancy, defining two enriched promoter classes with high or low transcriptional plasticity, respectively. We analyze these classes of promoters and show how they differ in the positioning and dynamics of nucleosomes and in their regulation by transcription factors and chromatin regulators. Analysis of human promoters shows the same qualitative association between nucleosome positioning and gene transcriptional plasticity, suggesting a conserved function for the patterns of promoter nucleosomes.
Results
High nucleosome occupancy near the transcription start site is associated with transcriptional plasticity
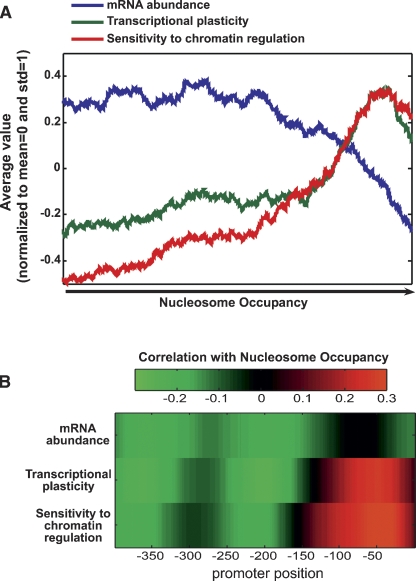
We collected a data set of more than 1500 transcription profiles (Ihmels et al. 2002) describing the expression changes of all yeast genes upon a variety of conditions (environmental stresses, mutations, or developmental transitions). Using these data, we calculated for each gene the average magnitude by which its expression was modulated across all conditions, or a subset of them (see Methods). Each gene was thus assigned a transcriptional plasticity measure, quantifying the dynamic range of its expression level. We then asked whether the transcriptional plasticity of a gene is correlated with nucleosome occupancy at the 150 bp upstream of the transcription start site (TSS) (Lee et al. 2007). As reported previously (Lee et al. 2004, 2007; Pokholok et al. 2005), average mRNA levels (mRNA abundance) tend to decrease with increasing nucleosome occupancy. In contrast, transcriptional plasticity increased with nucleosome occupancy (Fig. 1A), and this result also persisted when we excluded TATA-containing genes (data not shown).
Figure 1.
Correlation between nucleosome occupancy and transcription regulation. Nucleosome occupancy was compared with three aspects of transcription regulation: mRNA abundance (blue), transcriptional plasticity (average of the squared log2 expression ratio) to a range of perturbation (green), and sensitivity to chromatin regulation (red). These three measures were normalized by subtracting their means and dividing by their standard deviations. (A) Genes were ordered by their average occupancy at the 150 bp upstream of the TSS, and a sliding window (window size of 500 genes) is shown for the three measures of transcription regulation. (B) Correlations between nucleosome occupancy at each position (from −400 to 0, relative to the TSS) and the three measures of transcription regulation.
Notably, transcriptional plasticity was correlated with nucleosome occupancy also when restricting the analysis to genes with high expression levels in rich media (where nucleosome occupancy was measured), indicating that the high nucleosome occupancy of high-plasticity genes is not due to their specific repression in rich media (Supplemental Fig. 1). Furthermore, we measured transcriptional plasticity separately for activation or repression (Methods) and found that plasticity increases with nucleosome occupancy regardless of the direction of the expression change (Supplemental Fig. 2).
High nucleosome occupancy could make regulatory elements inaccessible, and thus the corresponding promoter would be less responsive to signaling pathways (Li et al. 2007). However, we observe the opposite, namely that promoters with high occupancy are in fact more responsive. An alternative explanation is that such promoters are subjected to more extensive regulation of chromatin structure and are thus more responsive to external and internal cues. To test this possibility, we utilized a compendium of 170 expression profiles of strains defective for various chromatin regulators (Steinfeld et al. 2007) and calculated the average expression change of each gene due to these defects, i.e., the sensitivity to disruption of chromatin regulation. As expected, sensitivity to chromatin regulation also increased with nucleosome occupancy (Fig. 1A).
Differential impact of TSS-proximal and TSS-distal nucleosome occupancy
In addition to the total number of promoter nucleosomes, mRNA abundance and transcriptional plasticity could also depend on the precise pattern by which nucleosomes are arranged across the promoter. To examine the possibility that different promoter regions carry distinct information about gene expression, we reanalyzed the correlation between nucleosome occupancy and mRNA abundance or transcriptional plasticity, considering this time the occupancy at each particular position among the 400 bp upstream of the TSS (Fig. 1B) (Lee et al. 2007). Consistent with the notion that nucleosome depletion facilitates transcription, mRNA abundance was negatively correlated with nucleosome occupancy at most promoter positions. Notably, the major influence of nucleosome occupancy appeared to be concentrated at distal upstream regions, whereas the occupancy at the ∼150 bp vicinity of the TSS had a relatively moderate correlation with mRNA abundance. In contrast, transcriptional plasticity was correlated with the occupancy across all promoter regions, but in a pronounced position-dependent manner: Transcriptional plasticity increased with the occupancy of nucleosomes at the ∼150 bp vicinity of the TSS, but in fact decreased with the nucleosome occupancy at more distal upstream regions. Notably, similar correlations were observed when gene activation and repression were considered separately (Supplemental Fig. 2). Furthermore, the correlation between nucleosome occupancy and the sensitivity of gene expression to disruption of chromatin regulation exhibited very similar position dependence (Fig. 1B).
Two patterns of promoter nucleosome occupancy
Our results suggest that the presence of a nucleosome at the ∼150 bp vicinity of the TSS has a moderate influence on transcript levels but largely facilitates the capacity to modulate these levels. Previous studies implicated this region as nucleosome-free (Yuan et al. 2005; Ioshikhes et al. 2006; Lee et al. 2007), following the observation that it is highly depleted of nucleosomes in many yeast genes. The observation that nucleosome occupancy in this region is still strongly correlated with transcriptional plasticity suggested to us that genes might differ in the organization of nucleosomes across their promoters, with different organizations conferring distinct logic of gene regulation and dependence on chromatin remodeling.
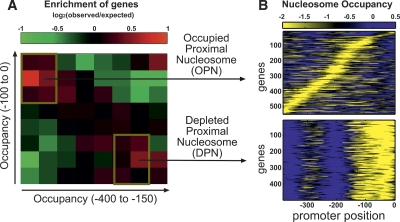
To examine the possibility of gene classes with different patterns of nucleosome occupancy, we summarized the patterns of nucleosome occupancy at each promoter by two numbers: the overall occupancy at the 100 bp proximal to the TSS (which is associated with high plasticity), and the overall nucleosome occupancy at the more distal region (−400 to −150), which is associated with low mRNA abundance and low plasticity. Reducing the overall nucleosome organization to just these two numbers is justified by our analysis in Figure 1, demonstrating a distinction between nucleosome occupancy at these promoter regions, and by the negative correlation between nucleosome occupancy at these different positions (Supplemental Fig. 3).
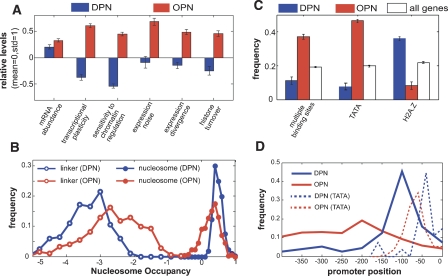
When positioned along these two axes, two prominent classes of genes emerged (Fig. 2A; Supplemental Tables 1, 2). Genes of the first class, depleted proximal-nucleosome (DPN), exhibited low occupancy close to the TSS and high occupancy at the more distal region (Fig. 2B). These genes were characterized by low transcriptional plasticity and low sensitivity to disruption of chromatin regulators. By contrast, genes of the second class, occupied proximal-nucleosome (OPN), exhibited relatively high nucleosome occupancy close to the TSS coupled with relatively low occupancy at a more distal region, whose exact positions are gene-specific (Fig. 2B). The expression of these genes is characterized by high transcriptional plasticity and sensitivity to chromatin regulation, as well as higher levels of stochastic fluctuations (Newman et al. 2006) and evolutionary divergence (Fig. 3A) (Tirosh et al. 2006). Notably, the difference between the expression variability of the two promoter classes is not confined to specific conditions but is rather prevalent: OPN genes were enriched with expression changes, compared with DPN genes, at 87% of all expression profiles and in particular at 89% of the defects in chromatin regulators, while the opposite (enrichment of DPN) was observed only in 2% and 5%, respectively (see Methods). In contrast to these widespread differences in expression variability, the absolute mRNA levels in rich media are comparable between the two classes (Fig. 3A).
Figure 2.
Two patterns of nucleosome occupancy. (A) The pattern of nucleosome occupancy at each promoter was summarized by two values: the average occupancy at −100 to 0 (proximal region), and that at −400 to −150 (distal region). For each of these values, the genes were sorted and divided into eight equal bins. We examined the number of genes in each combination of bins for the two values and compared it with the expected number if the two values were independent. Colors specify the log2 of the ratio between the observed and expected number of genes in each combination of bins: (red) positive values (enrichment), (green) negative values (depletion). (B) Two classes were defined based on their enrichment: low occupancy at the proximal region and high occupancy at the distal region (DPN), and high occupancy at the proximal region and low occupancy at the distal region (OPN).
Figure 3.
Differential properties of the two promoter classes. (A) Average values of properties that quantify the levels and variability of gene expression and the turnover of promoter H3 histones are shown for DPN genes (blue) and OPN genes (red). Values in each property were normalized to mean zero and standard deviation one. (B) Distribution of the maximal nucleosome occupancy within predicted nucleosomes (filled circles) and the distribution of the minimal nucleosome occupancy within predicted linker DNA (empty circles), for DPN (blue) and OPN (red) genes; distribution of the average occupancy within each element (rather than maximum or minimum) gives similar results (Supplemental Fig. 4). (C) Frequency of genes with multiple binding sites, TATA box, and histone variant H2A.Z is shown for DPN (blue), OPN (red), and all (white) genes. (D) Distribution of the promoter positions of transcription factor binding sites and TATA boxes (dashed lines) in the two classes. Error bars in A and C were calculated by bootstrapping.
In addition to the differential transcriptional plasticity, the two classes differed substantially in their overall promoter organization. DPN promoters displayed a characteristic pattern of nucleosome occupancy that includes well-positioned and high-occupancy nucleosomes at the distal promoter region, contrasted with a substantial nucleosome-free region (NFR) directly upstream of the TSS that is enriched with transcription factor binding sites. These promoters are enriched with the histone variant H2A.Z (Fig. 3B), which could help to establish the NFR (Raisner et al. 2005; Zhang et al. 2005). In contrast, OPN promoters had more uniform nucleosome occupancy, with a lower occupancy at the location of predicted nucleosomes and higher occupancy at predicted linker DNA, compared with DPN promoters (Fig. 3B; Supplemental Fig. 4). This pattern suggests that nucleosome positioning at these promoters is more dynamic, and thus the measured occupancies reflect averaging over multiple states. Indeed, promoters of this class were enriched with predicted “fuzzy” nucleosomes (Lee et al. 2007) (P < 10−4; see Methods). Furthermore, OPN promoters had significantly higher rates of histone H3 turnover (Dion et al. 2007) than those of DPN promoters (Fig. 3A; P = 3 × 10−12). Notably, the higher turnover rates at OPN promoters were found both for the proximal and distal regions (data not shown), and thus appear to be a general property of these promoters.
Transcription factor binding sites at the two promoter classes
Transcription factor binding sites are enriched in OPN promoters compared with DPN promoters (Fig. 3C). Interestingly, the location of binding sites in the two classes is consistent with the patterns of nucleosome occupancy (Fig. 3D): Binding sites are highly localized to the NFR of DPN promoters, but are distributed more uniformly throughout OPN promoters with a weak enrichment around −200. The broad distribution of binding sites at OPN genes likely reflects the variability between different genes, whereby each promoter has a different requirement for the positions of binding sites according to its specific promoter structure. However, this distribution could also indicate that due to their dynamic nature, individual OPN promoters lack such requirements for the positioning of binding sites. To examine this possibility, we analyzed the differences in binding site positions among promoters of closely related yeast species (Doniger and Fay 2007). Changes in the positions of binding sites (i.e., binding site turnover) are fivefold more frequent at OPN promoters compared with DPN promoters (39 cases in OPN and seven cases in DPN; P < 10−5), suggesting that OPN promoters have fewer constraints on binding site position.
As expected, OPN promoters are also highly enriched with TATA boxes (Basehoar et al. 2004), whereas TATA boxes are underrepresented in DPN promoters (Fig. 3C). Previous studies have shown that TATA-containing genes are associated with increased expression variability and divergence (Tirosh et al. 2006; Landry et al. 2007). However, the enrichment of TATA-containing genes cannot account for the expression variability of OPN genes: Despite their enrichment, TATA-containing genes comprise approximately half of the OPN genes, and the other half of TATA-less OPN genes are also characterized by high expression variability. Two-way analysis of variance (ANOVA) indicates that both the DPN/OPN and the presence/absence of TATA have a significant effect on each of the expression variation attributes examined above, including transcriptional plasticity, sensitivity to chromatin regulation, expression noise, and expression divergence (Supplemental Figs. 5, 6). In contrast, only DPN/OPN has a significant effect on histone turnover, and only TATA has a significant effect on mRNA abundance. These results indicate that chromatin structure (DPN versus OPN) has an impact on expression variability irrespective of the presence of TATA, and vice versa that TATA has an impact on expression variability irrespective of chromatin structure, although the two effects may also act synergistically (Supplemental Figs. 5, 6).
Interestingly, the positions of TATA boxes also differ between the two classes (Fig. 3D). Unlike most eukaryotes, where the TATA box is located precisely 30 bp upstream of the TSS, yeast TATA boxes are more distant from the TSS, and their positions are variable among different genes (Giardina and Lis 1993). While the reason for this variability is yet unclear, previous studies suggested that Pol II binds the promoter near the TATA and scans the downstream DNA until reaching the TSS (Kuehner and Brow 2006). Our results suggest that nucleosome positions could partially account for the variability of yeast TATA positions, and may influence the promoter scanning of Pol II.
Additional patterns of nucleosome occupancy
Despite the enrichment of the two classes mentioned above, other patterns of nucleosome occupancy were also observed. For example, cytosolic ribosome genes tend to display the typical NFR directly upstream of their TSS but also have relatively low nucleosome occupancy at more distal promoter positions, consistent with the location of transcription factor binding sites and their high expression levels (Supplemental Fig. 7).
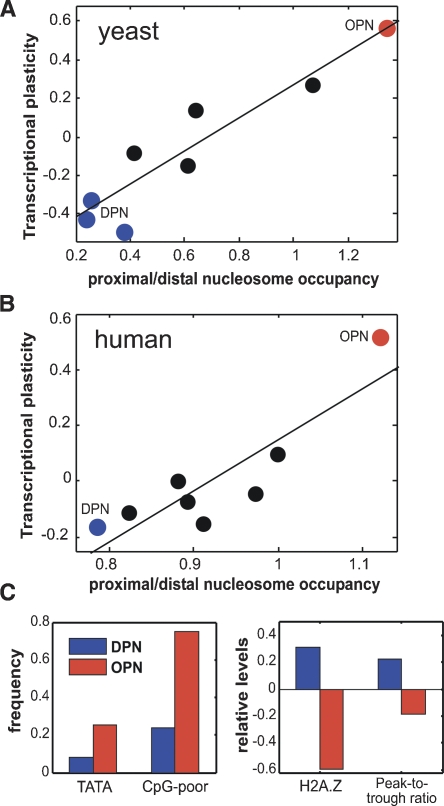
To characterize additional classes of promoter nucleosome organization in an unbiased manner, we clustered genes according to their patterns of nucleosome occupancy (Lee et al. 2007). Based on visual inspection we chose to focus on the division into eight clusters, although different divisions did not change our conclusions (data not shown). As expected, three clusters were associated with a distinct NFR directly upstream of the TSS and well-positioned nucleosomes at distal regions, thus corresponding to the DPN class. In contrast, two other clusters corresponded to the OPN class, with high occupancy directly upstream of the TSS and lower occupancy at more distal regions. The nucleosome patterns of each of the eight clusters, as well as their distribution of binding sites, expression properties, and functional enrichments are given in Supplemental Figure 8, reproducing our findings from the analysis of just the two classes (DPN and OPN). Importantly, the average transcriptional plasticity of each cluster is highly correlated with the ratio between nucleosome occupancy at the TSS-proximal and distal regions (Fig. 4A; r = 0.93; P < 0.01).
Figure 4.
Analyses of human promoters reproduce the distinction between promoter classes. The ratio between nucleosome occupancy at the TSS-proximal and -distal regions is correlated with transcriptional plasticity among the eight yeast (A) or human (B) promoter classes. Lines indicate the linear least-squares fit. Red and blue circles represent clusters corresponding to OPN and DPN, respectively. (C) Human OPN promoters (red bars) are enriched with CpG-poor promoters and TATA boxes, are depleted in H2A.Z, and have lower peak-to-trough ratios compared with human DPN promoters (blue bars). Similar results were obtained in analysis of human nucleosome positions from high-throughput sequencing (Supplemental Fig. 11; Schones et al. 2008). In addition, the ratio of human nucleosome occupancy was also significantly correlated with binding of CTCF and several histone H3 methylations (Supplemental Fig. 12; Barski et al. 2007).
Analyses of human promoters reproduce the distinction between promoter classes
To examine whether the connection we identified between nucleosome occupancy and gene regulation is conserved across evolution, we wished to extend the analysis to other organisms. Recently, the nucleosome occupancy of ∼3700 human promoters was characterized at high resolution in seven cell lines (Ozsolak et al. 2007). The patterns of promoter nucleosome occupancy were highly correlated among six cell lines, and, accordingly, we considered their average occupancy. Unlike the yeast data, the patterns of human nucleosome occupancy seemed to be more complex, and we could not divide the promoters reliably into two broad classes (see Methods). Instead, we grouped the promoters into eight clusters and examined the ratios between their occupancy at the TSS-proximal region (−150 to 0) and TSS-distal region (−400 to −200) (Supplemental Fig. 9). Out of the eight clusters, five had significantly lower occupancy in their proximal region (low proximal/distal ratios), consistent with a NFR, two clusters had approximately the same occupancy at both regions, and one cluster had significantly higher occupancy in the proximal region (high ratio). The two clusters with the lowest (425 genes) and highest (296 genes) ratios are reminiscent of the yeast DPN and OPN classes, and we thus refer to them as human DPN and OPN, respectively.
To examine the association of nucleosome patterns with transcriptional plasticity, we calculated the average expression changes of human genes in response to 164 distinct bioactive small molecules (Lamb et al. 2006). Strikingly, as in the yeast analysis, the average transcriptional plasticity of the eight promoter clusters was highly correlated with their ratios of occupancy between the proximal and distal regions (Fig. 4B; r = 0.88, P < 0.01). Furthermore, the human OPN cluster displayed particularly high transcriptional plasticity. This cluster was enriched with lowly expressed genes, but was nevertheless associated with plasticity for both activation and repression (data not shown). To further verify the correlation between transcriptional plasticity and the patterns of nucleosome occupancy, we separately calculated plasticity from another data set containing the expression patterns of 79 human tissues (Su et al. 2004) and found similar results (see Methods; Supplemental Fig. 10). Moreover, the OPN class is significantly enriched with CpG-poor promoters (Fig. 4C), which tend to have tissue-specific expression patterns (Saxonov et al. 2006). We thus conclude that the correlation between nucleosome patterns and transcriptional plasticity appears to be conserved from yeast to humans.
We next tested whether additional properties that distinguish the two promoter classes in yeast are conserved also in humans (Fig. 4C). First, we examined the presence of TATA boxes in the human DPN and OPN clusters (Tirosh et al. 2006). Consistently, we found that the percentage of TATA-containing genes in the OPN cluster is threefold higher than that in the DPN cluster. Second, we examined the localization of the histone variant H2A.Z (Barski et al. 2007) and found significant enrichment in the DPN cluster. Third, we examined the ratio between maximal occupancy within a nucleosome and minimal occupancy surrounding it (peak-to-trough ratio), as an estimate of dynamic nucleosome localization: High ratios indicate well-positioned nucleosomes, while low-ratios could indicate dynamic localization. OPN genes had significantly lower ratios, consistent with their dynamic nucleosome localization. Taken together, all three analyses reproduced the results found in the yeast data.
Discussion
We have shown that the pattern of promoter nucleosome occupancy is correlated with the capacity of genes to alter their expression levels. Promoters were found to be enriched with two patterns of nucleosome occupancy, corresponding to low or high plasticity. In particular, high-plasticity genes tend to have high nucleosome occupancy directly upstream of their TSS and dynamic nucleosome positioning with high turnover rate. Analyzing these promoter classes, we found that they also differ in other regulatory properties, including the frequency and distribution of transcription factor binding sites and the TATA box, the presence of histone variant H2A.Z, and their sensitivity to chromatin regulation. Notably, these features appear to be conserved from yeast to humans.
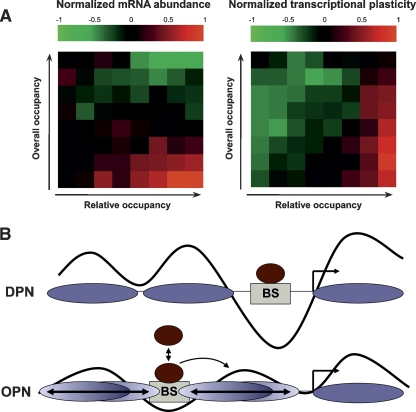
Importantly, the high occupancy near the TSS of high-plasticity genes does not imply their repression. First, the expression levels of these genes, at conditions similar to those where nucleosome occupancy was measured, are comparable to those of genes with a pronounced NFR (Fig. 3A; Supplemental Fig. 5). Second, expression levels are correlated with the overall nucleosome occupancy across the promoter but not with the relative pattern of nucleosome occupancy, whereas transcriptional plasticity is only correlated with the relative pattern of nucleosome occupancy (Fig. 5A; Supplemental Fig. 13). These results may suggest that overall nucleosome occupancy specifies the extent to which a promoter region is in “open” or “closed” chromatin, and is thus important to maintain high expression levels, while the relative pattern of nucleosome occupancy specifies the logic of gene regulation.
Figure 5.
A model for nucleosome organization at low-plasticity and high-plasticity promoters. (A) Average mRNA levels are correlated with overall nucleosome occupancy, whereas transcriptional plasticity is correlated with the relative pattern of nucleosome occupancy. Genes were sorted by overall nucleosome occupancy (Y-axis) or relative pattern of nucleosome occupancy (proximal/distal; X-axis), and divided to eight bins of equal sizes. The average normalized mRNA abundance (left) or transcriptional plasticity (right) is shown for each of the 64 bins. (B) The architecture of two promoter classes. Low-plasticity (DPN) promoters tend to have well-positioned nucleosomes and a strong NFR directly upstream of the TSS. High-plasticity (OPN) genes have “fuzzy” nucleosomes whose positions are heterogenic and dynamic, perhaps as a consequence of competition with transcription factors. Binding sites are distal to the TSS, and binding of regulators to these sites may influence nucleosome positions proximal to the TSS, thus affecting transcription. (Brown circles) Transcription factors in a binding equilibrium to their binding sites (BS), (ovals) nucleosomes, (curves) pattern of nucleosome occupancy.
Third, comparison of yeast nucleosome occupancy between normal and heat-shock conditions (Shivaswamy et al. 2008) indicates that >80% of the DPN and OPN genes retain their nucleosome patterns, even if their expression levels are altered upon heat shock (Supplemental Fig. 14). Furthermore, promoter changes in nucleosome patterns appear to be less frequent than changes in overall nucleosome occupancy both for yeast and among human cell lines (Supplemental Figs. 14, 15). Therefore, most genes appear to have an inherent pattern of nucleosome occupancy that may be important for their proper regulation and variability.
Our results suggest two general strategies for the regulation of gene expression by nucleosomes (Fig. 5B). Low-plasticity genes are associated with a static architecture of well-positioned nucleosomes and a substantial NFR directly upstream of the TSS. This pattern (DPN) may require deposition of the histone variant H2A.Z (Raisner et al. 2005; Zhang et al. 2005), which is enriched in DPN promoters (Fig. 3A). Regulatory elements are placed within the NFR and are thus accessible to transcription factors, whereas spurious elements are probably buried at well-positioned nucleosomes (Yuan et al. 2005; Liu et al. 2006; Narlikar et al. 2007). Therefore, binding of transcription factors to regulatory elements at the NFR might not require chromatin remodeling. This could explain the relatively low dependence of the corresponding gene expression on the disruption of chromatin regulators, and also the low level of expression noise (Fig. 3C), which is predicted to depend largely on chromatin remodeling (Raser and O’Shea 2004; Batada and Hurst 2007).
High-plasticity genes, on the other hand, are associated with more homogenous and dynamic nucleosome occupancy across promoters, and particularly high occupancy close to the TSS (Fig. 5B). In these promoters (OPN), transcription factor binding sites are more distant from the TSS and often are located within a region that is relatively occupied by nucleosomes, compared with the NFR of low-plasticity genes. It is possible that this organization reflects a dynamic competition between nucleosomes and transcription factors for binding the promoter (Miller and Widom 2003), consistent with the high rate of histone turnover in these promoters. Furthermore, the relative distance of the binding sites from the TSS may indicate that the binding of transcription regulators is only the first step in promoter activation, requiring the subsequent transient eviction of nucleosomes from the TSS-proximal regions to allow for the binding of additional regulators and/or general transcription machinery. This model is consistent with recent suggestions for the condition-dependent binding of Rap1 at certain promoters (Buck and Lieb 2006) and for the regulated accessibility of TATA boxes (Ioshikhes et al. 2006). As expected, such a design is more sensitive to the action of chromatin regulators that might be required for nucleosome eviction at the TSS and could also influence the competition between nucleosomes and transcription factors. The dependence on chromatin remodeling could be the underlying cause for the higher variability of the respective gene expression.
Previous studies suggested that TATA-containing and TATA-less promoters have different chromatin structure (Ioshikhes et al. 2006; Albert et al. 2007; Tirosh et al. 2007), while other studies have shown that TATA-containing genes have higher expression variability compared with TATA-less genes (Landry et al. 2007; Tirosh et al. 2007). Our results suggest that the higher expression variability of TATA-containing genes may partially reflect the chromatin structure of these genes (OPN), although the TATA box appears to also have an independent effect on expression variability (Supplemental Figs. 5, 6), perhaps due to increased rates of re-initiation as previously suggested (Raser and O’Shea 2004; Blake et al. 2006; Tirosh et al. 2007). It would be interesting to examine if TATA boxes are mechanistically preferred at OPN promoters, or whether their enrichment at OPN promoters is an evolutionary consequence of the similar impact of TATA and OPN architecture on the dynamics of gene expression.
OPN genes display higher expression variation at multiple levels, including stochastic noise, response to perturbations, and evolutionary divergence, suggesting that the propensity for expression variation is hardwired in their promoters. This property could have been selected for in evolution if it is beneficial under certain circumstances. For example, upon changes in the environment, noise in gene expression could make some individuals in a population more fit due to unusually high or low expression of specific genes (Kaern et al. 2005). However, for these individuals to perpetuate and generate an adapted population, their unusual expression patterns should be inherited without sufficient time for fixation of genetic mutations. It would thus make sense, from an evolutionary perspective, to encode the property of expression variation in chromatin structure, thereby enabling its epigenetic inheritance.
Methods
Nucleosome occupancy
Nucleosome occupancy of ∼5000 yeast promoters (400 bp upstream of the TSS) was taken from Lee et al. (2007). The positions of TSSs were defined as in Lee et al. (2007). Restricting the data to unidirectional promoters did not change the results (data not shown). For each gene, we calculated the average occupancy at the TSS-proximal region (−100 to 0) and distal region (−400 to −150). The yeast regions (−100 to 0, −400 to −150) were determined from visual inspection of Fig. 1B and Supplemental Fig. 3: Nucleosome occupancy at −100 to 0 is positively correlated with transcriptional plasticity, and at −400 to −150 is negatively correlated with transcriptional plasticity (Fig. 1B); nucleosome occupancy is positively correlated among positions from −100 to 0 and is negatively correlated between these positions and −400 to −150 (Supplemental Fig. 3). Changing these definitions (e.g., −150 to 0 instead of −100 to 0) did not significantly alter any of the results (data not shown).
Genes were sorted and divided into eight bins with equal numbers (614 genes) for each region. We then examined the number of genes in each combination of bins (Supplemental Table 1) and compared it with the expected number if the two promoter regions were independent (i.e., 1/64 of all genes in each combination). The enrichment of each combination was quantified as log2(observed/expected). Combinations with either high occupancy at both regions or low occupancy at both regions were underrepresented, whereas combinations with low occupancy at one region and high occupancy in the other were typically enriched. We thus defined two broad classes of enriched combinations: low-occupancy at the proximal region but high occupancy at the distal region (DPN), and high-occupancy at the proximal but low occupancy at the distal region (OPN). To examine the different patterns of occupancy in an unbiased manner, we clustered the patterns of promoter nucleosome occupancy using deterministic annealing and defined eight distinct clusters.
Lee et al. (2007) predicted the location of nucleosomes from the raw data of nucleosome occupancy. Since many nucleosomes appeared to span a larger region than that of well-positioned nucleosomes, they divided the predicted nucleosomes into “well-positioned” and “fuzzy.” For each promoter (400 bp upstream of the TSS), we calculated the percentage of bases that are predicted to be occupied by a fuzzy nucleosome (out of all occupied bases). This percentage was significantly higher in OPN compared with DPN genes (P = 4 × 10−5, as determined by a Wilcoxon rank sum test).
Rates of histone H3 turnover were taken from Dion et al. (2007), normalized, and compared between OPN and DPN genes. We found that OPN promoters have significantly (P < 0.05) higher turnover rates than DPN promoters when considering the maximal rates across either all probes in each promoter (Fig. 3A), or across only probes in the proximal promoter region or only probes in the distal promoter region (data not shown).
Human nucleosome occupancy was taken from Ozsolak et al. (2007) and averaged over all experimental repeats and cell lines, except for A375, which had low correlations with all other cell lines. The data were mapped to human promoters (700 bp upstream of the TSS) using the TSS positions from the UCSC genome browser (Karolchik et al. 2004). We repeated the analysis in Figure 2A for the human data but did not find strong enrichment of specific patterns as in the yeast data. We therefore clustered the human nucleosome data using deterministic annealing, and examined the division into eight clusters. The two clusters with the most significant differences between the proximal and distal regions were denoted as human DPN and OPN. The human regions (−150 to 0 and −400 to 200) were defined based on visual inspection of the clusters in Supplemental Fig. 7. Also here, changing these definitions (e.g., −100 to 0 or −200 to 0 instead of −150 to 0) did not significantly alter any of the results (data not shown). Peak-to-trough ratios of predicted nucleosomes were averaged over the six cell lines and used to estimate dynamic nucleosome positioning.
Gene expression
Nucleosome occupancy was compared with three measures of gene expression:
mRNA abundance at rich media, which was averaged over multiple studies (Beyer et al. 2004) and log2-transformed.
Transcriptional plasticity, which was estimated as the average of the squared log2 expression ratio from a large number of microarray experiments (Ihmels et al. 2002). As controls, transcriptional plasticity was calculated separately for up-regulation (average square of positive log2 expression-ratios) or down-regulation (average square of positive log2 expression-ratios), or was calculated for a subset of the microarray experiments in which all reference samples corresponded to rich media. These alternative measures did not significantly change our results (data not shown).
Sensitivity to disruption of chromatin regulators, which was similarly defined based on a smaller data set compiled by Steinfeld et al. (2007); this measure quantifies the extent by which the expression of each gene depends on the activity of chromatin regulators.
These measures, as well as noise (Newman et al. 2006) and evolutionary divergence (Tirosh et al. 2006) of gene expression were centered and normalized, such that their means are zero and standard deviations are one.
In each of the expression profiles, we calculated the number of DPN/OPN genes whose expression changed by at least twofold, and we quantified the enrichment of DPN/OPN genes with a binomial test. This was done separately for down-regulated or up-regulated genes, and the most significant P-value of each expression profile was used to determine significance, with a P-value threshold of 0.025. The expected frequency of profiles with enrichment of OPN regulated genes is therefore 0.025 × 2 = 5%, while we observed 87% and 89% for all expression profiles and those of defects in chromatin regulators, respectively.
Human transcriptional plasticity was calculated as the sum of squares of the log2 expression ratios from Lamb et al. (2006), which measured the response of several cell lines to various small molecules. Restricting the data only to the MCF7 cell line did not change the results (data not shown). In addition, transcriptional plasticity was calculated from the expression patterns of genes among 79 human tissues (Su et al. 2004). In this data set, we divided each expression level by the average expression level of the corresponding gene among all tissues, thus generating expression ratios. Transcriptional plasticity was then defined as the sum of squares of the log2-transformed expression ratios.
Binding sites
Yeast transcription factor binding sites (MacIsaac et al. 2006) were mapped to promoters. The number of promoters with multiple binding sites was compared between the two classes, and the distribution of binding site locations (relative to the TSS) was examined in the two classes as well as in eight clusters of nucleosome occupancy. Similarly, the frequency and positions of yeast TATA boxes (Basehoar et al. 2004) was compared between the two classes. Human TATA boxes were defined as previously described (Tirosh et al. 2006). Binding site turnover was defined as in Doniger and Fay (2007), excluding cases where the change in position is >500 bp.
H2A.Z
Yeast genes with the 20% highest H2A.Z occupancy were defined as H2A.Z-containing, as in Zhang et al. (2005). Human H2A.Z data were taken from Barski et al. (2007), and the number of tags that were mapped to the 500 bp upstream of each TSS was calculated, log2-transformed, and normalized to a mean of 0 and standard deviation of 1.
Acknowledgments
This work was supported by the Helen and Martin Kimmel Award for Innovative Investigation and grants from the Kahn fund for Systems Biology at the Weizmann Institute of Science, the Israeli Ministry of Science, and the Bi-national Science Foundation (BSF).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076059.108.
References
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Batada N.N., Hurst L.D. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat. Genet. 2007;39:945–949. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- Beyer A., Hollunder J., Nasheuer H.P., Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- Blake W.J., Balazsi G., Kohanski M.A., Isaacs F.J., Murphy K.F., Kuang Y., Cantor C.R., Walt D.R., Collins J.J. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Buck M.J., Lieb J.D. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat. Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.K., Kim Y.J. Epigenetic regulation and the variability of gene expression. Nat. Genet. 2008;40:141–147. doi: 10.1038/ng.2007.58. [DOI] [PubMed] [Google Scholar]
- Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Doniger S.W., Fay J.C. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput. Biol. 2007;3:e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C., Lis J.T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- Hogan G.J., Lee C.K., Lieb J.D. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Friedlander G., Bergmann S., Sarig O., Ziv Y., Barkai N. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Ioshikhes I.P., Albert I., Zanton S.J., Pugh B.F. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Kaern M., Elston T.C., Blake W.J., Collins J.J. Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kuehner J.N., Brow D.A. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 2006;281:14119–14128. doi: 10.1074/jbc.M601937200. [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Landry C.R., Lemos B., Rifkin S.A., Dickinson W.J., Hartl D.L. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee S.I., Pe’er D., Dudley A.M., Church G.M., Koller D. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc. Natl. Acad. Sci. 2006;103:14062–14067. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu X., Lee C.K., Granek J.A., Clarke N.D., Lieb J.D. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.A., Widom J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar L., Gordan R., Hartemink A.J. A nucleosome-guided map of transcription factor binding sites in yeast. PLoS Comput. Biol. 2007;3:e215. doi: 10.1371/journal.pcbi.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.R., Ghaemmaghami S., Ihmels J., Breslow D.K., Noble M., DeRisi J.L., Weissman J.S. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Song J.S., Liu X.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O.J., Ahmad K. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Raser J.M., O'Shea E.K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld I., Shamir R., Kupiec M. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat. Genet. 2007;39:303–309. doi: 10.1038/ng1965. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Weinberger A., Carmi M., Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- Tirosh I., Berman J., Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007;23:318–321. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang H., Roberts D.N., Cairns B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]