Abstract
Cysteine mutagenesis and site-directed spin labeling in the C-terminal region of rhodopsin have been used to probe the local structure and proximity of that region to the cytoplasmic loops. Each of the native amino acids in the sequence T335–T340 was replaced with Cys, one at a time. The sulfhydryl groups of all mutants reacted rapidly with the sulfhydryl reagent 4,4′-dithiodipyridine, which indicated a high degree of solvent accessibility. Furthermore, to probe the proximity relationships, a series of double Cys mutants was constructed. One Cys in all sets was at position 338 and the other was at a position in the sequence S240–V250 in the EF interhelical loop, at position 65 in the AB interhelical loop, or at position 140 in the CD interhelical loop. In the dark state, no significant disulfide formation was observed between C338 and C65 or C140 under the conditions used, whereas a relatively rapid disulfide formation was observed between C338 and C242 or C245. Spin labels in the double Cys mutants showed the strongest magnetic interactions between the nitroxides attached to C338 and C245 or C246. Light activation of the double mutant T242C/S338C resulted in slower disulfide formation, whereas interactions between nitroxides at C338 and C245 or C246 decreased. These results suggest the proximity of the C-terminal residue C338 to residues located on the outer face of a cytoplasmic helical extension of the F helix with an apparent increase of distance upon photoactivation.
Keywords: G protein-coupled receptor, signal transduction, disulfide crosslinking, site-directed spin labeling, conformational change
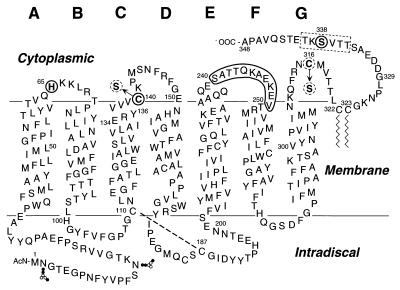
In common with the other G protein-coupled receptors, rhodopsin contains on its cytoplasmic face three interhelical loops and a peptide chain extending to the carboxyl terminus that includes multiple phosphorylation sites (Fig. 1). From several lines of evidence, we have previously concluded that light activation of rhodopsin results in a conformational change that is evidently coupled between the cytoplasmic transmembrane and intradiscal domains of the molecule (2–8). However, from the standpoint of understanding protein–protein interactions that ensue after light activation, the primary concern is to understand the nature of the conformational change in the cytoplasmic domain. A general approach that we have been using for this purpose involves systematic Cys substitutions in the cytoplasmic loops (Fig. 1). Single Cys substitutions enable a comparison of the chemical reactivity of the Cys residues, a feature that is determined in part by the accessibility of the residue and, hence, the local structure of the protein (6, 9, 10). Double Cys substitutions permit investigation of spatial proximity between two Cys in different regions of the molecule through a study of disulfide formation (7, 8, 11–13). In addition, Cys substitution enables the application of site-directed spin labeling, a technique that has been shown to provide direct information on secondary structure, tertiary structure, and inter-residue distances as well as information on the changes in these features that accompany structural changes (14). The application of these strategies has provided insights into the tertiary structure at the cytoplasmic face in both the dark- and light-activated states of rhodopsin (6–8, 10, 15–17).
Figure 1.
A secondary structure model of bovine rhodopsin shows the single and double Cys replacements in the cytoplasmic domain made in this work. The amino acids in the C-terminal peptide chain shown boxed (dotted rectangle) were replaced one at a time by Cys residues to prepare single Cys containing mutants. In every case, the double Cys mutants contained the first Cys at position 338 and the second Cys at position 65 in AB loop; position 140 in CD loop is shown by solid circles or one of the positions in EF loop is shown by the solid boundary. The native Cys at positions 140 and 316 were replaced by serines (dotted circles) in all the single and double Cys mutants except for C140/S338C, which contains the Cys at position 140.
The topology and structure of the C-terminal peptide chain continuing from Cys-322 and Cys-323, the palmitoylation sites (Fig. 1), remains unknown. One consequence of the structural change in the cytoplasmic loops upon light activation is the binding of rhodopsin kinase to the loops (18) with consequent activation of the enzyme and the phosphorylation(s) of one or more serine and threonine in the C-terminal sequence. Recently, from the NMR study of a peptide segment corresponding in sequence to the C-terminal tail, a three-dimensional structure for the region has been proposed (19). The main feature of the proposal is a short antiparallel β-sheet structure with a β-turn, which contains the rhodopsin kinase phosphorylation sites. Indeed, the NMR study has been extended by using soluble peptide segments corresponding to all the cytoplasmic loops, and based on those results (20, 21), specific structural proposals have been made for their tertiary structures in rhodopsin itself, as well as for an integrated structure in the dark state for the entire cytoplasmic domain (22). Clearly, additional work is required to determine whether the structures proposed from the NMR studies of the individual peptide segments alone can be extrapolated to the structures that are obtained in rhodopsin.
In the present work, topology of the C-terminal tail and any interactions it may have with one or more of the cytoplasmic loops have been probed with Cys mutagenesis. A set of single Cys substitution mutants was prepared that replaced native amino acids 335–340, one by one. The chemical reactivity of each of these Cys was high, with no differences attributable to structural effects being observed. Furthermore, sets of dicysteine mutants were prepared with the first Cys at position 338 and the second Cys at different sites in the cytoplasmic loops. Both disulfide crosslinking and interaction between spin labels at the Cys in the sets provided evidence for spatial proximity of residue 338 in the C-terminal tail with amino acids 242, 245, and 246 in the E-F interhelical loop.
MATERIALS AND METHODS
Materials.
Dodecyl maltoside (DM) was obtained from Anatrace (Maumee, OH). Antirhodopsin mAb rho-1D4 was purified from myeloma cell lines provided by R. S. Molday (University of British Columbia) and was coupled to cyanogen bromide-activated Sepharose 4B (Sigma) as described (23). Cysteine-deficient DMEM and [35S]Cysteine (1,075 Ci/mmol; 1 Ci = 37 GBq) were from ICN. PDS was purchased from Sigma. Endoproteinase Asp-N was obtained from Boehringer Mannheim. Fluorescamine was obtained from Aldrich Chem, Metuchen, NJ. The sulfhydryl-specific spin label (1-oxyl-2, 2, 5, 5-tetramethylpyrrolin-3-methyl) methanethiosulfonate (MTSL) was a gift from Kalman Hideg (University of Pecs, Hungary). 11-cis-Retinal was a gift from Rosalie Crouch (Medical University of South Carolina and the National Eye Institute, National Institutes of Health).
Buffers used were: buffer A (NaCl 137 mM/KCl 2.7 mM/KH2PO4 1.8 mM/Na2HPO4 10 mM, pH 7.2), buffer B [buffer A plus 1% (wt/vol) DM/2 mM ATP/2 mM MgCl2/0.1 mM phenylmethylsulfonyl fluoride (PMSF)], buffer C (buffer A plus 0.05% DM), buffer D (5 mM Mes [2-(N-morpholino)ethanesulfonic acid] pH 6.0/0.05% DM), buffer E (2 mM sodium phosphate, pH 7.5/0.05% DM), buffer F (5 mM Mes, pH 6.0/0.1% DM), and buffer G (5 mM Mes, pH 6.0/0.02% DM).
Construction of the Cysteine Mutants of the Synthetic Opsin Gene.
The construction of single Cys mutants T335C, T336C, V337C, S338C, K339C, and T340C in the C-terminus. Fragment replacement mutagenesis was used to introduce the Cys mutations in the synthetic gene for the wild-type (wt) bovine opsin (24) as cloned in the expression vector PMT4 (25). Starting with a previously constructed opsin gene containing the C140S and C316S replacements (unpublished work), single Cys codons were introduced. For T335C, the restriction fragment BstEII/NarI (nucleotides 952-1038) was replaced by the corresponding synthetic oligonucleotide duplex containing the Cys codon TGC. For the mutants T336C–T340C, the restriction fragment SalI/NarI (nucleotides 1000–1038) was replaced by the synthetic oligonucleotide duplex containing the Cys codon TGC.
Construction of double Cys mutants.
The double Cys mutants were prepared so that C338 was constant whereas the second Cys varied from positions 240 to 250 in the EF loop. These are designated S240C/S338C to V250C/S338C and were derived from their corresponding single Cys mutants S240C–V250C in the EF loop (6). However, these single Cys mutants lacked Cys-322 and Cys-323, the palmitoylation sites, the Cys having been replaced by serines. To construct the above double Cys mutants with intact Cys-322 and Cys-323, restriction fragments MluI/NheI (1876 bp) in the EF loop single Cys mutants (S240C–V250C) were replaced by fragment MluI/NheI from the plasmid S338C (1,876 bp). This fragment contains the codon for Cys-338, as well as the codons for Cys-322 and Cys-323. Double Cys mutants H65C/S338C and C140/S338C also were prepared in which one Cys was kept constant at position 338, whereas the second Cys was at position 65 (AB loop) or at position 140 (CD loop). The PstI/NheI fragments (1925 bp) in the plasmids previously designated H65C (7) and C316S (26) were replaced by the PstI/NheI fragment from plasmid S338C.
All mutations were confirmed by the dideoxynucleotide sequencing method (27).
Expression of the Mutant Opsin Genes and Purification of the Expressed Opsins After Reconstitution with 11-cis-Retinal.
The procedures for the transient transfection of PMT4 vectors carrying opsin genes in COS-1 cells and reconstitution of the opsins with 11-cis-Retinal have been described (23). The mutant rhodopsins obtained from cells (five, 15-cm plates) were solubilized and the proteins were absorbed on 200 μl of antirhodopsin 1D4-Sepharose beads (≈binding capacity of 1 μg rhodopsin/μl) as described (23). The resin was washed first with 30 bed vol (6 ml) of buffer C, followed by another wash with 15 bed vol of buffer D (3 ml). The protein was eluted with buffer D containing 80 μM C′1–C′9 carboxyl terminal peptide and collected in 1.5-bed vol fractions.
UV/Vis Absorption Spectroscopy.
UV/Vis absorption spectra were recorded with a Perkin–Elmer λ-7 spectrophotometer (6). The molar extinction values (ɛ500) for mutant rhodopsins were determined as described (28); the value used for wt rhodopsin was 40,600 M−1CM−1. For photobleaching, the samples were illuminated for 30 sec with light at λ > 495 nm, and the spectra were recorded immediately. Metarhodopsin-II (Meta II) decay rates were measured by the fluorescence increase assay as described (29).
Quantitation of the Cysteine Groups in Rhodopsin Mutants by Reaction with PDS.
Quantitation of protein sulfhydryl groups with PDS has been described previously (30, 31). Appropriate mutant samples in buffer D were diluted to 200 μl with buffer E and then added to 50 μl of buffer E containing PDS, such that the solution contained 0.5 μM pigment and 25 μM PDS. The reaction was followed by the development of absorption at 323 nm by using the same concentration of PDS as a reference. The molar extinction coefficient of 4-thiopyridone at 323 nm was determined to be 19,000 M−1CM−1 by titration of l-cysteine with PDS, under the same conditions.
Determination of Rates of Disulfide Bond Formation.
The rhodopsin-double Cys mutants after elution from 1D4-Sepharose were all diluted with buffer D to give 2.5 μM concentration, and the pH of samples was then increased to 7.5 by adding 0.5 M sodium phosphate (pH 8.0). The time course of disulfide bond formation was monitored by taking aliquots (50 μl) at different time intervals and measuring the remaining free sulfhydryl content by reaction with PDS as described above. Thus, the extent of disulfide bond formation for the double Cys mutants was calculated from the decrease in the amount of the free sulfhydryl groups. For determining the rate of disulfide bond formation after light activation, samples were eluted in buffer F and maintained throughout at 4°C where the t1/2 of Meta II decay was 69 min.
Spin Labeling of the Cysteine Mutants and EPR Analysis.
Reaction of the rhodopsin mutants with the spin-labeling reagent MTSL while bound to the Ab 1D4-Sepharose and the elution of the derivatized protein from the Sepharose were carried out as previously described (15), except for the following modifications. The resin was washed first with 30 bed vol (6 ml) of buffer C, followed by a further wash with 15 bed vol of buffer G (3 ml), and then incubated with spin label (100 μM in 2 ml of buffer G) directly in the small spin column. After 3 h reaction at room temperature, the resin was washed with 30 bed vol (6 ml) of buffer G, and the proteins were eluted in buffer G as described above. The extent of spin labeling was assayed by titration of the remaining free sulfhydryl content with PDS. The first derivative x-band EPR spectra of labeled mutants were recorded at ambient temperature in a loop gap resonator as previously reported (17) and normalized to the same number of spins by using double integration. The EPR spectra for the singly labeled mutants in the sequence 240–250 were reported earlier (17).
Digestion of Rhodopsin Mutants with Protease Asp-N and Electrophoretic Analysis of the Digests.
The mutants (≈1.7 μg) in 20 μl of 50 mM sodium phosphate buffer (pH 8.0) were treated with a preparation of Asp-N in 10 mM Tris-HCl, pH 7.5 at a ratio of 20 (substrate) to 1 (enzyme) by weight. The mixture was incubated at room temperature for 16 h in the dark. The reaction was stopped by the addition of EDTA (final concentration, 10 mM). The digests were then analyzed by SDS-PAGE under both nonreducing and reducing (with 1 mM DTT) conditions as previously described (7). After electrophoresis, the gel was analyzed by immunoblotting with antirhodopsin Ab 1D4.
Preparation of 35S-labeled Mutant Opsin (K245C/S338C) in COS Cells.
The procedure used was as described previously (32) except for the following modifications. The DMEM lacking Cys was supplemented with heat-treated, dialyzed fetal bovine serum (10%) and [35S]Cys (50 μCi/ml) and was added to cells (10 ml). The cells were then incubated for another 24 h before harvesting. 35S-labeled rhodopsins were purified after addition of 11-cis-Retinal as described above.
Paper Chromatography.
Paper chromatography was performed by using the ascending technique with Whatman 3MM paper (33). The solvent system used was N-butanol:glacial acetic acid:H2O (4:1:5). After chromatography, the peptides were visualized by autoradiography or fluorescence with fluorescamine as described (34).
RESULTS
Characterization of the Rhodopsin Cysteine Mutants.
The single Cys mutants (T335C–T340C) and the double Cys mutants, each containing the S338C mutation together with a second Cys at S240C–S250C (EF loop), C140 (CD loop), or H65C (AB loop) (Fig. 1), were all expressed in COS cells and purified as described in Materials and Methods. Each mutant formed a wt-like chromophore with λmax at 498–500 nm, and spectral ratios (A280/A500) of ≈1.6–1.8 (Table 1). Upon illumination, all mutants formed the characteristic Meta II intermediate. The t1/2 for decay of the latter, as measured by retinal release (Materials and Methods), ranged from 8.2 min to 16.5 min, compared with 12.8 min for wt rhodopsin under the conditions used (Table 1).
Table 1.
Characterization of cysteine substitution mutants of rhodopsin
| Mutants | λmax (nm) | A280:A500 | t1/2 of retinal release (min) |
|---|---|---|---|
| A. Single cysteine mutants | |||
| wt | 500 | 1.6 | 12.8 |
| T335C | 498 | 1.8 | 12.4 |
| T336C | 498 | 1.7 | 11.6 |
| V337C | 498 | 1.7 | 11.7 |
| S338C | 498 | 1.7 | 12.0 |
| K339C | 498 | 1.8 | 11.7 |
| T340C | 498 | 1.7 | 11.4 |
| B. Double cysteine mutants | |||
| S240C/S338C | 499 | 1.6 | 13.9 |
| A241C/S338C | 499 | 1.6 | 8.9 |
| T242C/S338C | 499 | 1.6 | 10.0 |
| T243C/S338C | 499 | 1.6 | 11.4 |
| Q244C/S338C | 499 | 1.6 | 10.7 |
| K245C/S338C | 499 | 1.6 | 8.2 |
| A246C/S338C | 499 | 1.6 | 12.4 |
| E247C/S338C | 499 | 1.6 | 15.8 |
| K248C/S338C | 499 | 1.6 | 9.5 |
| E249C/S338C | 499 | 1.6 | 16.5 |
| V250C/S338C | 499 | 1.5 | 14.7 |
| H65C/S338C | 500 | 1.6 | 10.2 |
| C140/S338C | 499 | 1.7 | 10.7 |
Quantitative Derivatization of the Sulfhydryl Groups in Rhodopsin Mutants with PDS.
The reactions of wt rhodopsin and rhodopsin mutants were followed spectrophotometrically by monitoring the absorption at λ323 nm (30, 31). For quantitation of the absorption at this wavelength, the tailing absorption from PDS alone (around 0.007) and the absorption of rhodopsin alone were subtracted. The ratios of molar absorption at λ323 nm to that at λ500 nm corresponded in all cases to those expected for the mono- and dicysteine mutants. The reactions of all the monocysteine mutants (T335C–T340C) with PDS were completed by the time the first spectrophotometric measurement was performed (≈1 min), whereas the reaction of wt with PDS took >1 h to reach completion under the conditions used (unpublished data).
Disulfide Bond Formation in Double Cysteine Mutants in the Dark.
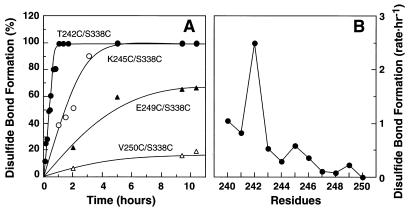
The mutants as purified by the immunoaffinity method at pH 6.0 were shifted to pH 7.5 (Materials and Methods) and kept in the dark at room temperature. Disulfide bond formation was followed by a decrease in the extent of reaction with PDS. Rapid disulfide bond formation occurred between Cys-338 and the Cys in the EF loop. As seen in Fig. 2A, the rates of disulfide bond formation varied with the location of the Cys in the EF loop. Thus, the mutant T242C/S338C showed a complete disulfide formation in ≈1 hr. The mutant K245C/S338C also showed a relatively rapid disulfide bond formation. However, the mutant E249C/S338C showed a much slower rate, and the mutant V250C/S338C showed almost no disulfide bond formation. In Fig. 2B, the disulfide bond formation rates for all the 11 mutants (S240C/S338C–V250C/S338C) are compared.
Figure 2.
Rates of disulfide bond formation in double Cys mutants S240C/S338C to V250C/S338C. All the mutant preparations after elution from the Ab 1D4-Sepharose column, were shifted to pH 7.5 as in “Materials and Methods.” The time course of disulfide bond formation was then monitored by measuring the decrease in sulfhydryl groups by reaction with PDS (Materials and Methods). (A) The extent of disulfide bond formation in mutants: T242C/S338C, K245C/S338C, E249C/S338C, and V250C/S338C as a function of time. (B) A plot of the rates of disulfide bond formation in the mutants S240C/S338C– V250C/S338C. The rate is represented by 1/t1/2 (h –1), where t1/2 is the time (h) required for reaching 50% disulfide bond formation. t1/2 for each mutant was derived from the data in A.
No disulfide bond formation was observed between Cys-140 in the CD loop and Cys-338. Mutant H65C/S338C showed a relatively slow rate, with <50% of disulfide bond formation after 8 h under the same conditions.
Characterization of the Disulfide Bond Between Cys-245 and Cys-338 in the Dicysteine Mutant K245C/S338C.
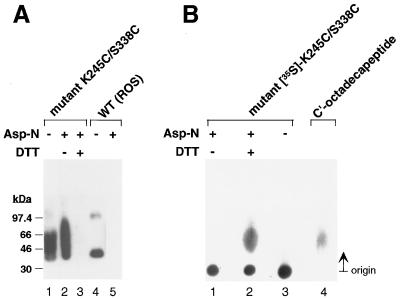
The enzyme Asp-N is known to cleave rhodopsin at position G329 in the carboxyl-terminal peptide chain of rhodopsin with the release of a l9-residue fragment (D330–A348) containing the Ab 1D4 epitope (35). Digestion of the disulfide-bonded mutant K245C/S338C was performed as in Materials and Methods. The digest was first subjected to SDS-PAGE, and the results of the examination by immunoblotting with Ab 1D4 are shown in Fig. 3A. The mutant without Asp-N treatment (lane 1) as well as after Asp-N treatment (lane 2) showed positive response to the Ab, indicating that the C-terminal epitope was linked to the protein. DTT treatment of the Asp-N–digested protein cleaved the disulfide bond, and the epitope was lost (lane 3). In a control experiment with wt rhodopsin (lanes 4 and 5), again after Asp-N treatment the epitope was lost. Because of the difficulty of identifying by immunoblotting the C-terminal nonadecapeptide released after DTT treatment of the Asp-N digestion product, 35S-labeled mutant K245C/S338C was prepared, and the digestion products were examined by paper chromatography (Fig. 3B). Lane 2, containing DTT-treated digestion product, now showed the 35S-labeled peptide. A synthetic sample of C′-octadecapeptide available in the laboratory from related work was used as a marker, and this peptide was visualized by reaction with fluorescamine (lane 4) (Material and Methods).
Figure 3.
Characterization of the disulfide bond in crosslinked K245C/S338C rhodopsin mutant. (A) SDS-PAGE and immunoblot analysis with the antirhodopsin Ab 1D4 after Asp-N cleavage. Lane 1, K245C/S338C before protease digestion; lane 2: Asp-N-digested mutant K245C/S338C applied in the absence of DTT; lane 3: Asp-N-digested mutant K245C/S338C applied after the addition of 1 mM DTT; lane 4: wt rhodopsin (ROS) before Asp-N digestion; lane 5: wt rhodopsin (ROS) after Asp-N digestion. (B) Identification of the C-terminal nonadecapeptide cleaved with Asp-N protease by paper chromatography. Lane1, [35S]K245C/S338C mutant after Asp-N digestion applied in the absence of DTT. Lane 2, [35S]K245C/S338C mutant after Asp-N digestion applied after DTT treatment. Lane 3, [35S]K245C/S338C mutant before Asp-N digestion. The ascending paper chromatogram was run for 3.5 h as in “Materials and Methods,” and the spots were visualized by autoradiography. Lane 4, visualization of the synthetic C-terminal octadecapeptide of rhodopsin by fluorescence after treating the chromatogram with fluorescamine (Materials and Methods).
Comparison of the Rates of Disulfide Bond Formation in the Mutant T242C/S338C in the Dark and After Illumination.
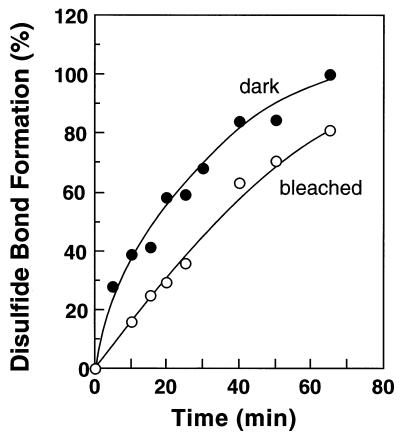
Mutant T242C/S338C showed the most rapid disulfide bond formation among all the double Cys mutants. The time course of disulfide bond formation between Cys-242 and Cys-338 in this mutant was followed both in the dark as well as after illumination as described in “Materials and Methods.” These rates were measured at 4°C, where the half-life of Meta II in the mutant was determined to be 69 min. The time points in the time courses were taken within the half-life of Meta II intermediate. As shown in Fig. 4, the formation of disulfide bond between Cys-242 and Cys-338 was slowed down after illumination of rhodopsin, with t1/2 of 16.1 min in the dark vs. 35.1 min after bleaching.
Figure 4.
Rates of disulfide bond formation in the mutant T242C/S338C in the dark and after light activation. The proteins were eluted from the Ab 1D4-Sepharose™ column in buffer F (Materials and Methods). The samples were maintained throughout at 4°C. After increasing the pH to 7.5, the decrease in sulfhydryl groups was monitored both in the dark and after illumination. The Meta II half-life of T242C/S338C at 4°C was ≈69 min.
Nitroxide–Nitroxide Interactions in Spin-labeled Double Cysteine Mutants.
The above data on disulfide formation suggest the proximity of residue 338 in the rhodopsin C-terminal domain and residues in the EF loop. To complement this data, we have evaluated the proximity of the Cys residues in the double mutants by spin–spin interaction between nitroxide side chains attached to the Cys residues. Spin–spin interactions are generally reflected in a broadening of the EPR spectral lineshape (7, 36–38). To reveal line width contributions caused by spin interactions, the spectra of the doubly labeled mutants were compared with the normalized algebraic sum of the spectra of the corresponding singly labeled mutants. In the absence of spin–spin interaction, the spectral sum should be the same as that of the doubly labeled mutant bearing nitroxides at the same sites. Any difference is attributed to magnetic interaction between the spins, which implies spatial proximity.
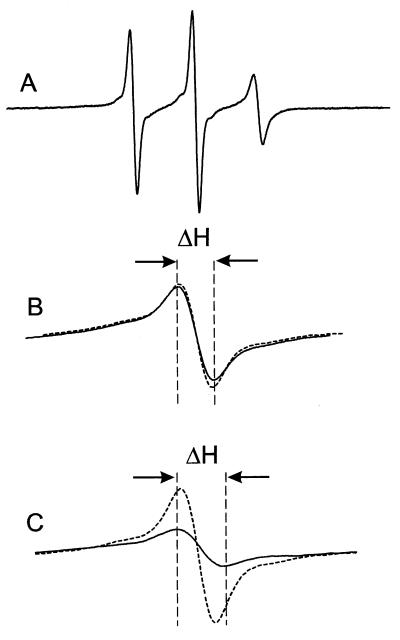
Fig. 5A shows the EPR spectrum of S338R1, the residue common to all of the spin pairs. The narrow linewidth indicates a major population of a highly mobile nitroxide with an effective correlation time of ≈1 ns. The R1 residues in the series 240–250 are all considerably less mobile with correspondingly broader EPR spectral lineshapes (17). As a result, the central line width (ΔH) in a composite spectrum arising from noninteracting nitroxides at 338 and a second site in the sequence 240–250 is dominated by the 338R1 spectrum. Thus, an increase in breadth of the central line in a spectrum of a double mutant relative to that of the sum of the single mutants (Δ(ΔH)) is a measure of the effect of the second spin on 338R1. Examples are shown in Fig. 5 B and C on an expanded scale for the center line. Fig. 5B shows the situation for 243R1/338R1, a spin pair that shows little evidence of interaction. Here the sum of 243R1 and 338R1 (dashed line) is essentially identical to the spectrum of the double mutant 243R1/338R1 (solid line), and the central linewidths are close to that of 338R1, alone. Fig. 5C shows the corresponding data for 245R1/338R1. Here the central linewidth of the double mutant is significantly broader and of correspondingly lower amplitude than that of the sum of the singles, indicating spin–spin interaction.
Figure 5.
The EPR spectra of spin labeled rhodopsin mutants. (A) the EPR spectrum of S338R1 in a solution of DM in the dark state. The scan width in the magnetic field is 100 G; (B) the center lines (m1 = 0) for the sum of the S338R1 and T243R1 single mutants (dashed line) and the T243R1/S338R1 double mutant (solid line); (C) the center lines for the sum of the S338R1 and K245R1 single mutants (dashed line) and the K245R1/S338R1 double mutant (solid line). In B and C, the magnetic field scan width is 17 G. ΔH is the peak-to-peak width of the center line of the corresponding double mutant.
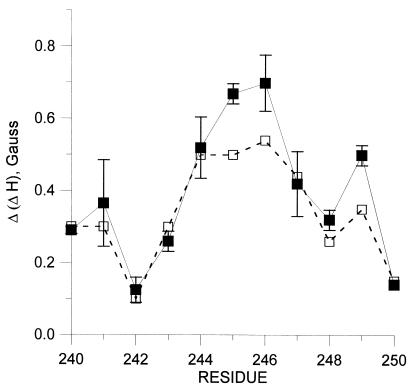
Fig. 6 (solid line) shows a plot of Δ(ΔH) derived from the spectra of spin pairs XR1/3381 in the dark state vs. sequence number X in the range of 240–250. As can be seen, the largest interactions of 338R1, as judged by the central linewidth measure, occur with residues 245 and 246. Residues 244 and 249 show the next largest effects, and a weaker but measurable interaction occurs with 247. Upon light activation, there is an apparent decrease in the interaction between 338R1 and 245, 246, and 249 (Fig. 6, dashed line). Essentially, no interaction is seen between 338R1 and 140R1 or 65R1 in the CD and AB interhelical loops, respectively (unpublished data).
Figure 6.
Resonance line broadening in spin labeled double mutants with one nitroxide fixed at 338 and the other in the sequence 240–250. The line broadening, Δ(ΔH) is the difference in the line width of the central (m1 = 0) resonance between the spectral sum of the single mutants and the spectrum of the corresponding double mutant. The error bars for the dark state were based on two independent measurements. Similar errors are expected for the data obtained after light activation.
DISCUSSION
Sulfhydryl Reactivity and Disulfide Formation.
Although evidence has been accumulating to support the plausible conclusion that the cytoplasmic loops in rhodopsin together are involved in a tertiary structure, the relationship of the C-terminal polypeptide to this structure is not understood. By introducing single Cys residues one at a time at positions 335–340 and studying their reactivity to the sulfhydryl reagent PDS, Cys at these positions showed the absence of any significant structural effects. These results should be contrasted with the frequent finding of a marked difference in reactivities found for Cys residues in the cytoplasmic loops. Thus, the two Cys of wt rhodopsin, Cys-140 and Cys-316, which are reactive in the dark state showed markedly different reactivity. Similarly, the Cys residues at positions 56–75 showed striking difference to reactivity with PDS in the dark (Judith Klein, unpublished work).
Next, we looked for possible proximity relationships in the dark state between amino acid residues in the C-terminal tail and the cytoplasmic loops. We introduced two Cys at selected positions and looked for disulfide formation and magnetic interactions between the spin labels attached to the sulfhydryl groups. No rapid spontaneous disulfide bond formation was observed, under the conditions used, between Cys-338 and Cys-65 (AB loop) or Cys-140 (CD loop). However, relatively rapid disulfide bond formation was observed between Cys-338 and Cys-242 (EF loop). Indeed the rates of disulfide bond formation varied when the position of the second Cys in the EF loop was varied between positions 240 and 250 (Fig. 2), and formed three groups, 240–243 (fast), 244–247 (intermediate), and 248–250 (slow). Also as seen in Fig. 4B, Cys-242, Cys-245, and Cys-249 showed higher rates of disulfide bond formation with Cys-338 than their respective neighboring Cys in each of the three groups. The results seem to be consistent with the previous findings (17) regarding the periodic variation in both accessibility and mobility of the spin-labeled side chains in the EF interhelical loop. Based on those results, it was concluded that the loop is largely α-helical, being formed by regular extensions of the E and F helices by ≈1.5–3 turns. The Cys at 240–250 now studied belong to the extension of helix F in aqueous phase. An extended helical structure in this region would place residues 242, 245, and 249 on the same outer face of the helix.
Finally, to probe structural change on light activation, we compared the rates of disulfide bond formation between Cys-338 and Cys-242 in the dark and after illumination. The experiment was carried out at 4°C, where the t1/2 for Meta II decay was ascertained to be 69 min. A decrease in the rate was found (Fig. 4), indicating an increase in distance between the two Cys. Because of a variety of interpretations that are possible for this result, further discussion is clearly premature.
As in previous studies (7, 8), we have complemented the disulfide-crosslinking approach for investigation of residue proximity with a second method based on site-directed spin labeling. The site-directed spin-labeling strategy is to introduce pairs of nitroxide side chains at the sites of substituted Cys residues and rank the proximities of the nitroxides based on the strength of their magnetic interactions. In favorable cases, quantitative values for interspin distances may be determined (7, 8, 36, 37, 39).
The data summarized in Fig. 6 reveal a periodic variation in apparent spin–spin interaction of 338R1 as a function of the position of the second R1 residue in the F helix, from 240 to 250. The maxima in interaction occur at positions 241, 245–246, and 249, although the maximum at 241 is uncertain because of experimental error. The interactions of 338R1 are clearly strongest with 245R1 and 246R1. The pattern and magnitude of spin interactions are consistent with the regular helical conformation of the sequence in the region 244–250 (17), if the mean position of 338 lies closest to 245 and 246.
The weak interaction of 338R1 with residues 241–243, which would lie one turn above 245 and 246 in a regular helical extension of helix F, requires comment. It is possible that the conformational flexibility of the R1 side chain may simply position these nitroxides beyond the limit of detectable interaction. All of the interactions are relatively weak, and a rather small displacement could move the nitroxides out of interaction range. This latter possibility is plausible considering that these residues are near the helix termination (17), and the side chain has a larger volume of conformational space in which to pack. On the other hand, it may be that residues 240–243 are in fact not in a regular helical conformation, perhaps forming part of the turn between helices E and F.
Unfortunately, it is not possible to make a quantitative estimate of the interspin distances because of the high degree of mobility of residue 338R1 (Fig. 5A). In this case, there are at least three mechanisms possible for the spin–spin interaction: modulation of the dipolar interaction because of reorientation of the interspin vector, modulation of the dipolar interaction because of time-dependent changes in interspin distance, and contributions from static dipolar interactions. In the absence of information on the relative contributions of these mechanisms and a suitable theory, the results may be used only to deduce relative proximities.
A comparison of Fig. 2B with Fig. 6 shows that in the sequence 244–250, the maxima (245, 249) and minima (248, 250) in disulfide crosslinking rates overlap with the maxima (245–246, 249) and minima (248, 250) of spin–spin interaction. Only in the region of 240–243 are there significant differences. The explanation for these differences likely lies in the very different requirements for observing interactions with the two methods. The degree of magnetic interaction is determined by an average interspin distance. In contrast, the disulfide crosslinking rate is not a measure of average distance between two Cys; the disulfide bond cannot be formed unless the sulfur atoms lie within a fixed distance with a particular geometry. Rather, the crosslinking rate is determined by the product of the probability that the sulfurs come within the required distance with a suitable geometry and the probability for reaction under those conditions. Thus in a system where one of the sulfhydryl groups has high flexibility, such as for 338, disulfide crosslinking may occur fairly rapidly because of the ability of the 338 residue to adopt a conformation suitable for disulfide formation, even though the time average distance may be too large to detect significant spin–spin interaction. This may be the case for crosslinking of 338 with residues in the 240–243 region, particularly because these residues themselves have an unusually high flexibility (17).
The above discussion emphasizes the difference in disulfide crosslinking and spin–spin interaction that must be considered in interpreting results. These differences render the methods complementary, each providing a different structural perspective. Disulfide crosslinking emphasizes accessibility and flexibility, whereas spin–spin interaction is a direct measure of an average distance.
Under any conditions, the differences in the results of crosslinking and spin–spin interactions assays do not detract from the clear general agreement regarding the topographical relationships of residue 338 to those in the 240–250 sequence.
Acknowledgments
We thank Dr. U. L. RajBhandary for the helpful discussions during the experiment. We also thank Ms. Judy Carlin for the assistance in the preparation of the manuscript. This work was supported by National Institutes of Health Grants EY05216 (W.L.H.) and GM 28289 (H.G.K.) and the Jules Stein Professorship Endowment (W.L.H.). R.L. is the recipient of NRSA from the National Eye Institute.
ABBREVIATIONS
- Opsin mutations are designated by using the single letter code for the original amino acid
the sequence number and the single letter code for the substituted amino acid, in that order. DM, Dodecyl maltoside, PDS, 4,4′-dithiodipyridine
- wt
wild-type
- Meta II
Metarhodopsin II, Ab, antibody
- R1
the designation for the spin-label side chain
Footnotes
This is paper No. 26 in the series Structure and Function in Rhodopsin. The preceding paper is ref. 1.
References
- 1.Cha K, Bruel C, Ingeles J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10577–10582. doi: 10.1073/pnas.94.20.10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resek J R, Farrens D L, Khorana H G. Proc Natl Acad Sci USA. 1994;91:7643–7647. doi: 10.1073/pnas.91.16.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson F F, Loewen P C, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farahbakhsh Z T, Ridge K, Khorana G H, Hubbell W L. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 6.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 7.Yang K, Farrens D L, Altenbach C, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]
- 8.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 9.Javitch J A, Li Z, Kaback J, Karlin A. Proc Natl Acad Sci USA. 1994;91:10355–10359. doi: 10.1073/pnas.91.22.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridge K D, Zhang C, Khorana H G. Biochemistry. 1995;34:8804–8811. doi: 10.1021/bi00027a032. [DOI] [PubMed] [Google Scholar]
- 11.Falke J J, Koshland D E., Jr Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 12.Falke J J, Dernburg A F, Sternberg D A, Zalkin N, Milligan D L, Koshland D E., Jr J Biol Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- 13.Milligan D L, Koshland D E., Jr J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 14.Hubbell W L, Altenbach C. Curr Opin Struct Biol. 1994;4:566–573. [Google Scholar]
- 15.Resek J F, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1993;32:12025–12031. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 16.Farahbakhsh Z T, Ridge K, Hubbell W L. Science. 1993;262:1416–1419. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- 17.Altenbach C A, Yang K, Farrens D F, Khorana H G, Hubbell W L. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 18.Thurmond R L, Creuzenet C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeagle P L, Alderfer J L, Albert A D. Nat Struct Biol. 1995;2:832–834. doi: 10.1038/nsb1095-832. [DOI] [PubMed] [Google Scholar]
- 20.Yeagle P L, Alderfer J L, Albert A D. Biochemistry. 1995;34:14621–14625. doi: 10.1021/bi00045a002. [DOI] [PubMed] [Google Scholar]
- 21.Yeagle P L, Alderfer J L, Salloum A C, Ali L, Albert A D. Biochemistry. 1997;36:3864–3869. doi: 10.1021/bi962403a. [DOI] [PubMed] [Google Scholar]
- 22.Yeagle P L, Alderfer J L, Albert A D. Biochemistry. 1997;36:9649–9654. doi: 10.1021/bi970908a. [DOI] [PubMed] [Google Scholar]
- 23.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferretti L, Karnik S S, Khorana H G, Nassal M, Oprian D D. Proc Natl Acad Sci USA. 1986;83:599–603. doi: 10.1073/pnas.83.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke R R, Sakmar T P, Oprian D D, Khorana H G. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 26.Karnik S S, Ridge K D, Bhattacharya S, Khorana H G. Proc Natl Acad Sci USA. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 30.Grassetti D R, Muttay J F JR. Arch Biochem. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y S, Hubbell W L. Membr Biochem. 1978;1:107–129. doi: 10.3109/09687687809064162. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Garriga P, khorana H G. Proc Natl Acad Sci USA. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett J C. Methods Enzymol. 1967;11:330–333. [Google Scholar]
- 34.Mendez E, Lai C Y. Anal Biochem. 1975;65:281–292. doi: 10.1016/0003-2697(75)90511-4. [DOI] [PubMed] [Google Scholar]
- 35.Palczewski K, Buczylko J, Kaplan W M W, Polans A S, Crabb J W. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 36.Mchaourab H S, Oh K J, Fang C J, Hubbell W L. Biochemistry. 1997;36:307–316. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 37.Rabenstein M R, Shin Y K. Proc Natl Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss J, Salwinski L, Kaback H R, Hubbell W L. Proc Natl Acad Sci USA. 1995;92:12295–12299. doi: 10.1073/pnas.92.26.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hustedt E J, Smirnov A I, Laub C F, Cobb C E, Beth A H. Biophys J. 1997;74:1861–1877. doi: 10.1016/S0006-3495(97)78832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]