Abstract
It has been speculated that before vertebrates evolved somatic diversity-based adaptive immunity, the germline-encoded diversity of innate immunity may have been more developed. Amphioxus occupies the basal position of the chordate phylum and hence is an important reference to the evolution of vertebrate immunity. Here we report the first comprehensive genomic survey of the immune gene repertoire of the amphioxus Branchiostoma floridae. It has been reported that the purple sea urchin has a vastly expanded innate receptor repertoire not previously seen in other species, which includes 222 toll-like receptors (TLRs), 203 NOD/NALP-like receptors (NLRs), and 218 scavenger receptors (SRs). We discovered that the amphioxus genome contains comparable expansion with 71 TLR gene models, 118 NLR models, and 270 SR models. Amphioxus also expands other receptor-like families, including 1215 C-type lectin models, 240 LRR and IGcam-containing models, 1363 other LRR-containing models, 75 C1q-like models, 98 ficolin-like models, and hundreds of models containing complement-related domains. The expansion is not restricted to receptors but is likely to extend to intermediate signal transducers because there are 58 TIR adapter-like models, 36 TRAF models, 44 initiator caspase models, and 541 death-fold domain-containing models in the genome. Amphioxus also has a sophisticated TNF system and a complicated complement system not previously seen in other invertebrates. Besides the increase of gene number, domain combinations of immune proteins are also increased. Altogether, this survey suggests that the amphioxus, a species without vertebrate-type adaptive immunity, holds extraordinary innate complexity and diversity.
Animal immunity comprises innate immunity and adaptive immunity. The diversity of non-self-recognition molecules in innate immunity is germline-encoded, while in adaptive immunity it is a product of somatic diversification and selective clonal expression (Rast et al. 2006). It was believed that only jawed vertebrates have a developed adaptive immune system, where the somatic rearrangement of IgV domains produces diversified B-cell and T-cell antigen receptors (BCRs and TCRs) and each B-cell or T-cell selectively expresses one type of receptor. (See Box 1 for a list of the most used abbreviations.) The BCR/TCR system also requires the MHC system to eliminate self-reactive BCR/TCR and to present intracellular antigens to extracellular TCR surveillance. However, recent reports have demonstrated that jawless vertebrates have variable lymphocyte receptors (VLRs) with somatically rearranged LRR ectodomains, suggesting that the BCR/TCR system is not the only form of adaptive immunity (Pancer et al. 2004; Nagawa et al. 2007). Moreover, comparative studies from invertebrates have revealed that: (1) Snail FREP genes are capable of somatic recombination (Zhang et al. 2004). (2) The Dscam gene of Drosophila melanogaster can produce 38,016 alternatively spliced mRNAs, and each cell randomly selects a set of variants to express (Neves et al. 2004). (3) The amphioxus VCBP proteins have highly variable IgV-like domains (Cannon et al. 2002). (4) The sea urchin has greatly expanded its immune receptor repertoire (Rast et al. 2006). These findings suggest that the great diversity and selective expression are not restricted to the vertebrate-type adaptive immune system (VAIS).
Box 1.
Most used abbreviations
The signaling network of immunity consists of two layers, namely, intercellular communication and intracellular signaling pathways. Intercellular communication is mediated by various cytokines, chemokines, growth factors, and their cognate receptors. Intracellular signaling pathways are downstream of immune receptors, which consist of adapters, GTPases, caspases, kinases, and transcription factors. Pathways of diversified immune receptors, including vertebrate BCR/TCR and perhaps the sea urchin TLRs, generally conform to the “funnel” model, in which molecular complexity is reduced rapidly from the cell-surface (receptor) level to the nucleus (transcription factor) level such that numerous initial signals will be sorted into a limited number of “simplified” pathways (Sansonetti 2006).
Comparative immunological studies between human and D. melanogaster have led to several great discoveries, such as TLR and PGRP pathways. However, reliable inference can rarely be made across large evolutionary gaps because of the fast-evolving pace of the immune system. Therefore, better understanding of the evolution of vertebrate immunity requires collecting data from different intermediate species, such as echinoderms (the purple sea urchin), protochordates (amphioxus), urochordates (Ciona intestinalis), and agnathans (lamprey). Genome sequence is a good starting point to fill the gap (Hibino et al. 2006). Thus far, study of the C. intestinalis genome has provided some basic knowledge about the immunity of a chordate invertebrate (Azumi et al. 2003), while the sea urchin genome demonstrates great diversity of innate receptors in a basal deuterostome (Hibino et al. 2006).
Recently, amphioxus has been repositioned to the base of the chordate phylum (Delsuc et al. 2006), hence strengthening its role as a reference species in the understanding of chordate evolution (Schubert et al. 2006). Immunocytes (coelomocytes) of amphioxus were first discovered in the gut region 25 yr ago (Rhodes et al. 1982). Our studies have further confirmed that gill and gut represent the major immune region in amphioxus (Dong et al. 2005; Yu et al. 2005, 2007b; Huang et al. 2007a, b; Yuan et al. 2007). Unfortunately, since the year 1982, only a few studies address the amphioxus immunity. With newly established techniques such as embryonic microinjection, daily spawning, and breeding in captivity (Holland and Yu 2004; Fuentes et al. 2007), amphioxus has become a promising model for immunology. The recently released draft genome of the amphioxus Branchiostoma floridae (Putnam et al. 2008) has become an invaluable tool to the rapid identification of genes.
In this paper we report that more than 5000 out of 50,817 gene models in the draft genome may be relevant to immunity. Our comprehensive analysis of these models in the phylogenomic context provides in-depth insights into the evolution of chordate immunity and lays the framework for further comparative studies with amphioxus.
Results and Discussion
General considerations
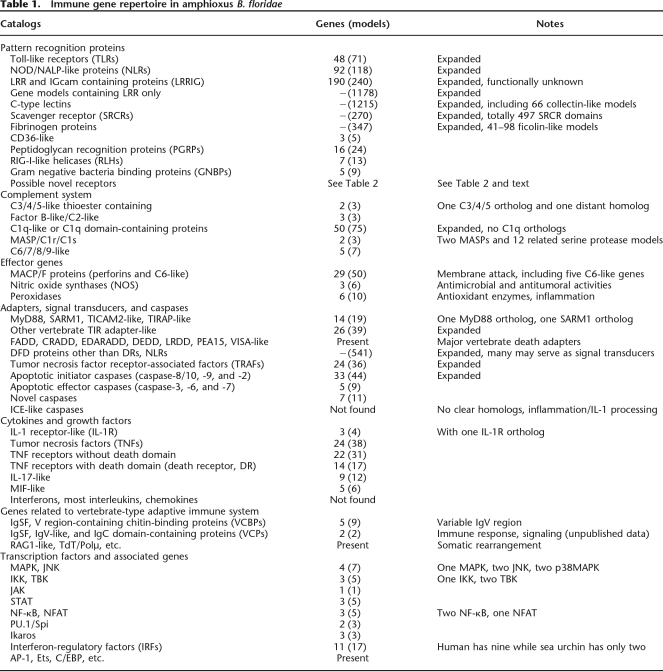
The current release v.1.0 of the B. floridae draft genome contains ∼50,817 gene models. Since ∼75% of genomic loci are represented by two haplotypes with 5%–10% allelic polymorphism (http://genome.jgi-psf.org/Brafl1/Brafl1.info.html), the actual gene number is one-half to two-thirds of the gene model number. This number would be further reduced if those pseudogenes and genes of split predictions, overlapping predictions, and false predictions were excluded. Therefore, the gene number mentioned in this study was estimated by phylogenetic analysis, and sometimes by syntenic analysis. Since for some greatly expanded families like CTL it is difficult to determine the allelic gene pairs, only the number of “gene models” was provided. Therefore, the terms “gene” and “model” (gene model) are not interchangeable in this paper. On average, the gene number is approximately two-thirds of the investigated model number. In the case of pseudogenes, they are obviously an important part of the evolutionary history of the expanded gene family, and we did not discriminate them. As for incorrect predictions, we also did not discriminate them unless they posed a problem for reliable analysis. Prior to detailed analysis, we presented a cross-species comparison of the immune-related protein domains (Supplemental Table S1), a summary of the immune gene repertoire (Table 1), and a list of all the investigated gene models (Supplemental Material B).
Table 1.
Immune gene repertoire in amphioxus B. floridae
The TLR system
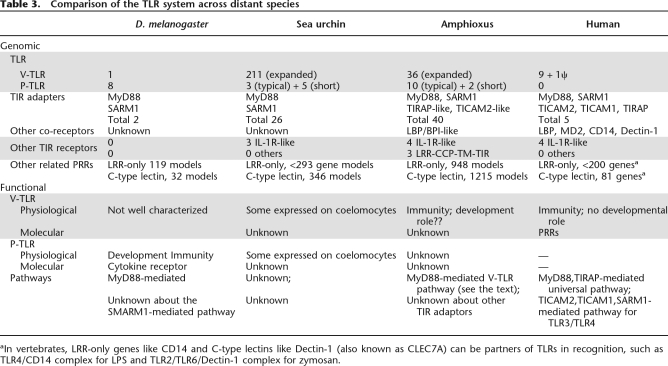
Toll-like receptors (TLRs) are present throughout virtually the entire animal kingdom and have important immune functions. A typical TLR is a type I transmembrane protein, consisting of a solenoid-like LRRNT-LRR-LRRCT ectodomain for ligand recognition and a cytoplasmic TIR domain for signal transduction. There are two structural types of TLRs (Supplemental Fig. S1): the vertebrate-like TLRs (V-TLR) and the protostome-like TLRs (P-TLR) (Hibino et al. 2006). P-TLRs have an extra LRRCT–LRRNT pair in the LRR ectodomains. In sea urchin and in amphioxus, some TLRs carrying short ectodomains without extra LRRCT–LRRNTs are also classified as P-TLRs because their TIRs are highly similar to typical P-TLRs. D. melanogaster has eight P-TLR genes and one V-TLR gene (Toll-9). The function of Toll-9 remains elusive, while the other eight P-TLRs encode dedicated cytokine receptors for insect cytokine Spatzles and have a role in development (Parker et al. 2001). The D. melanogaster P-TLR gene Tl also has a critical role in antifungal immunity (Lemaitre et al. 1996). Vertebrates have 10–20 TLRs. Unlike insect P-TLRs, vertebrate TLRs are all V-TLRs and are solely dedicated to immunity as PAMP recognition receptors (PRR). Recently, it has been reported that the sea urchin has a greatly expanded TLR repertoire with eight P-TLRs and 214 V-TLRs (Hibino et al. 2006).
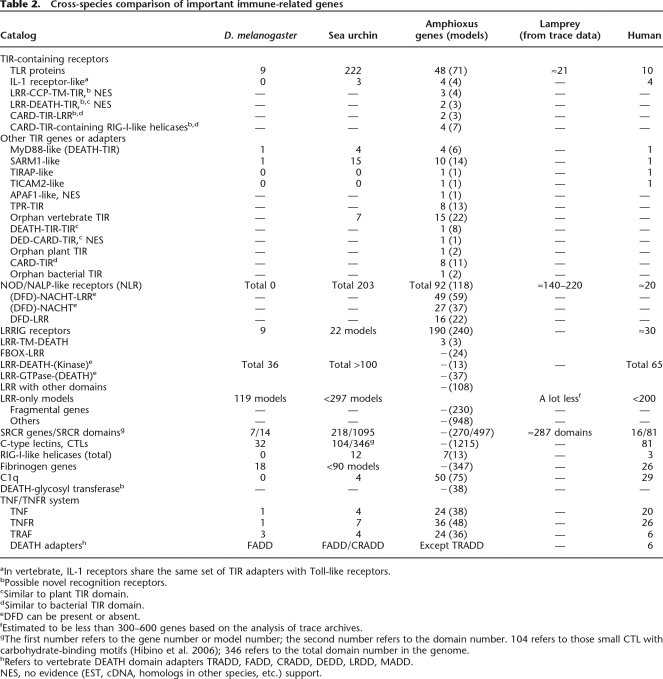
The draft genome of amphioxus contains at least 36 V-TLRs and 12 P-TLRs (Table 2), suggesting that the amphioxus V-TLR lineage has also greatly expanded and the P-TLR structure may only be lost in vertebrates. BLASTP analysis indicates that when compared with insects and sea urchins, amphioxus TLRs are more similar to vertebrate TLRs. Prompted by this, we performed a phylogenetic analysis of the amphioxus and vertebrate TLRs based on the TIR domain (Fig. 1). To our surprise, P-TLRs form a stable paraphyletic relationship with the vertebrate TLR4 lineage. Since P-TLR and V-TLR are supposed to have diverged before the protostome–deuterostome split, this phylogenetic pattern is highly dubious (for further discussion, see Supplemental Fig. S2). As for amphioxus V-TLRs, 33 form a paraphyletic cluster with the vertebrate TLR11 lineage (for further discussion, see Supplemental Material A). Furthermore, 19 of them comprise a distinct lineage, which is designated SC75 since 12 members are encoded on Scaffold_75. There are two pseudogenes and at least 12 intronless genes in the SC75 lineage. The SC75 lineage has conserved TIR domains (>85% amino acid identity) and highly variable LRR regions. These LRR regions are controlled by diversifying selection (Supplemental Material A). Taken together, the evolution of the amphioxus SC75 lineage is reminiscent of most of the sea urchin V-TLR lineages, which apparently have experienced a dynamic evolutionary history manifested by rapid tandem duplications, a high proportion of pseudogenes, and a rapid diversification of LRR regions relative to the conservation of TIR domains (Hibino et al. 2006). As for the V-TLRs outside the SC75 lineage, judging from the branching pattern and degree of TIR sequence divergence, the evolution is likely controlled by purifying selection, as is the case for most vertebrate TLRs (Roach et al. 2005). For example, the analysis of the dN/dS value of B. floridae TLR1 (bfTLR1) and its Branchiostoma japonicum ortholog (bjTLR1) indicates dominant purifying selection on both the LRR region and the TIR domain (S. Yuan, S. Huang, W. Zhang, M. Dong, H. Liu, M. Yang, K. Wu, T. Wu, T. Liu, L. Huang, et al., in prep.).
Table 2.
Cross-species comparison of important immune-related genes
aIn vertebrate, IL-1 receptors share the same set of TIR adapters with Toll-like receptors.
bPossible novel recognition receptors.
cSimilar to plant TIR domain.
dSimilar to bacterial TIR domain.
eDFD can be present or absent.
fEstimated to be less than 300–600 genes based on the analysis of trace archives.
gThe first number refers to the gene number or model number; the second number refers to the domain number. 104 refers to those small CTL with carbohydrate-binding motifs (Hibino et al. 2006); 346 refers to the total domain number in the genome.
hRefers to vertebrate DEATH domain adapters TRADD, FADD, CRADD, DEDD, LRDD, MADD.
NES, no evidence (EST, cDNA, homologs in other species, etc.) support.
Figure 1.
The ME tree of the TIR domain of insect TLRs, 48 amphioxus TLRs and vertebrate TLRs. The gene ID in gray represents alleles. The vertebrate TLR3, TLR5, and TLR7 families are too diverged to include in this tree. (Dashed lines) The possible position of those divergent sequences not used in the tree construction; (dots) the truncated P-TLRs. Another tree of more reliable statistical significance is shown in Supplemental Figure S3. (m) Mouse; (f) Fugu; (tet) tetraodon; (ch) chicken; (xt) Xenopus; (hal) halibut; (md) Musca domestica; (ag) Anopheles gambiae; (d) Drosophila.
In addition to TLRs, two types of amphioxus TIR receptors are worth mentioning. One type comprises four IL-1 receptor-like, which in vertebrates share the same set of TIR adapters with TLRs. The other type comprises three novel genes with the architecture SP-LRR-CCP-TM-TIR, although EST evidence for this is lacking at present.
TLR proteins signal through the interaction between their TIR domains and the cytoplasmic TIR-containing adapters. Vertebrates have five TIR adapter genes, including MYD88, TIRAP, TICAM1, TICAM2, and SARM1. MYD88 carries DEATH and TIR domains and mediates a common pathway for all vertebrate TLRs but TLR3, while TIRAP sometimes functions as a partner for MYD88. TICAM1 and TICAM2 mediate a pathway specific to vertebrate TLR3 and TLR4. SARM1 contains TIR, SAM, and Armadillo domains; in vertebrates, it negatively regulates the TICAM1/TICAM2 pathway, whereas in Caenorhabditis elegans, it mediates a TLR-independent immune response (Couillault et al. 2004; Carty et al. 2006). It has been reported that there are no homologs for TIRAP, TICAM1, and TICAM2 in the sea urchin, but there are four MYD88-like, 15 SARM1-like, and seven orphan TIR genes (Hibino et al. 2006). The amphioxus genome encodes no TICAM1-like genes, but four MYD88-like (including one ortholog), 10 SARM1-like (including one ortholog), one TIRAP-like, and one TICAM2-like genes. Moreover, there are eight TPR-TIR (some encode both domains in the same exon), one APAF1-like (TIR-NBARC-WD40), and 15 orphan TIR genes. Taken together, amphioxus has 40 genes encoding adapter-like proteins, which is a great expansion whether compared to the TIR adapter repertoire of vertebrates and the sea urchin, or compared to the TLR repertoire of amphioxus.
Across-species comparison of TLR systems is presented in Table 3. As shown in the table, the amphioxus TLR system carries great genomic complexity. This may greatly affect the signaling pathway and its downstream cellular outcome, and may ultimately result in a functional mode considerably different from that in the sea urchin and in vertebrates. We have demonstrated that bjTLR1 (from B. japonicum), a typical amphioxus V-TLR expressed mainly in the gut and up-regulated by bacterial infection, can signal through bjMYD88 (from B. japonicum) in the HEK293 cell line. This suggests a conserved MYD88 pathway in amphioxus, hence laying the foundation for further exploration of the amphioxus TLR system (S. Yuan, S. Huang, W. Zhang, M. Dong, H. Liu, M. Yang, K. Wu, T. Wu, T. Liu, L. Huang, et al., in prep.).
Table 3.
Comparison of the TLR system across distant species
aIn vertebrates, LRR-only genes like CD14 and C-type lectins like Dectin-1 (also known as CLEC7A) can be partners of TLRs in recognition, such as TLR4/CD14 complex for LPS and TLR2/TLR6/Dectin-1 complex for zymosan.
Other TIR-containing genes
According to sequence homology, the TIR domain can be classified as bacterial-TIR, plant-TIR, and vertebrate-TIR. The TIR domain in plants also participates in host defense. Plant-TIR and bacterial-TIR have not been reported in vertebrates. In the amphioxus genome, we found TIR domains similar to plant-TIR and bacterial-TIR, although it is unclear whether they have common origins (Table 2). Plant-TIR-like genes of amphioxus include one LRR-DEATH-TIR, one DEATH-TIR-TIR, one DED-CARD-TIR, and one orphan TIR, whereas bacterial-TIR-like genes of amphioxus comprise four CARD-TIR-containing RIG-I-like helicases (RLH), two CARD-TIR-LRRs, eight CARD-TIRs, and one orphan TIR. The RLHs and the CARD-TIR-LRR appear to be receptors, while the CARD-TIR and the orphan TIR may serve as adapters. At present, none of these proteins are corroborated by cDNA evidence, but the DEATH-TIR-TIR has homologs in the sea urchin, and the CARD-TIR combination occurs multiple times, with two domains always adjacent to each other.
The NLR system
NACHT-LRR receptors (NLR) are cytosolic proteins that have important functions in apoptosis, inflammation, and intracellular innate immunity (Fritz et al. 2006). Vertebrate NLRs consist of a central NACHT domain for oligomerization, a C-terminal LRR region for ligand recognition, and N-terminal domains (often DFD domains) for signaling. There are 20–30 NLR genes in vertebrates, the majority of which encode NOD/NALP-like proteins that use CARD or PYD domains for signaling. Vertebrate NOD/NALP proteins are well-documented for their roles in intracellular PAMP recognition and intestinal inflammation (Inohara et al. 2005). Compared to vertebrates, the sea urchin genome encodes 203 NLRs, the majority of which consist of an N-terminal DEATH, a central NACHT, and a C-terminal LRR region (Hibino et al. 2006).
The amphioxus genome encodes at least 92 NLR genes. A tree of their NACHT domains indicates that these NLRs may have undergone a rapid expansion similar to the SC75 TLR lineage (Supplemental Fig. S6). Unlike NLRs of vertebrates and the sea urchin, many amphioxus NLRs lack one to two domains, including 14 NLR genes without detectable LRR regions, 30 without known N-terminal signaling domains, 22 without NACHT domains, and 21 without both LRR regions and known N-terminal domains (see EST evidence in Supplemental Material A). NLRs without detectable NACHTs are here termed DLRs (DFD-LRR). BLASTP analysis indicates that DFD domains and LRR regions of DLRs are similar to those of typical NLRs, suggesting a close link between DLRs and typical NLRs. The N-terminal region of vertebrate NLRs can be CARD, CARD-CARD, CARD-AD, PYD, or BIR-BIR-BIR, whereas that of sea urchin NLRs is mostly DEATH and sometimes CARD. In addition, TIR domain is also found in the N terminus of plant NLRs (Inohara and Nunez 2003). In amphioxus, typical NLRs use DEATH, CARD, and DED domains as N-terminal domains. DEATH-DEATH is also found in an NLR gene (Bf97362, with no EST evidence). NLRs without known signaling domains often have a long N-terminal sequence of unknown function instead. As for amphioxus DLRs, various N-terminal domains can be found, such as DED, CARD, CARD-TIR, and multiple DEATH and TIR-DEATH. However, since N-terminal regions of DLRs are usually encoded in complex exon structure and there is no EST evidence for most DLRs at present, these domain structures are dubious (more details in Supplemental Material A). Taken together, the amphioxus NLR repertoire contains more structural complexity than that of vertebrates and the sea urchin.
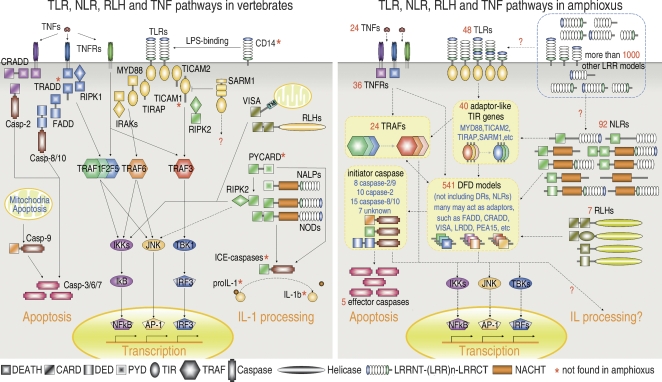
The signaling of vertebrate NOD/NALP proteins requires interactions of their CARD/PYD domains and the downstream adapters like CRADD, PYCARD, and RIPK2. These interactions lead to NF-κB activation and the processing of IL-1 proteins by ICE-like caspases (Fig. 2). Homologs of CRADD and RIPK2 are present in amphioxus, but PYCARD, IL-1 proteins, and ICE-like caspases are absent. PYCARD contains a PYD domain, yet no PYD has been detected in amphioxus. Since a conserved IL-1 receptor is present, IL-1 proteins may exist but may be too diverged to be detected. Although ICE-like caspases were reported in sea urchin (Robertson et al. 2006), no unambiguous ICE-like caspases have been identified in amphioxus. Despite the absence of some conserved downstream components, there are nevertheless hundreds of DFD gene models and 45 caspases (discussed below), some of which may serve for the amphioxus NLR system.
Figure 2.
A schematic comparison of TLR, NLR, RLH, and TNF pathways between vertebrates and amphioxus. Dashed lines indicate that the pathway has no functional evidence as yet. The colors used for different domains have no special meaning.
We have cloned three NLR cDNAs (GenBank accession nos. EU183366–EU183368), which encode CARD-NACHT-LRR, DEATH-NACHT-LRR, and DEATH-NACHT, respectively. Expression analysis shows that they all concentrate in the gut (data not shown), which is consistent with the case in vertebrates and the sea urchin, where NLRs are suggested to have a major role in monitoring the gut microflora and pathogens (Hibino et al. 2006; Pancer and Cooper 2006).
LRRIG proteins
A typical LRR and IGcam containing protein (LRRIG) consists of an N-terminal LRR region, one or more central IGcam domains, a transmembrane region, and a C-terminal cytoplasmic tail. There are approximately 30 vertebrate LRRIG proteins, which usually function in the nervous system (Chen et al. 2006). According to our analysis, the sea urchin genome encodes approximately 20 LRRIG models, whereas the amphioxus genome contains 240 LRRIG models (Table 2). The immunological relevance of amphioxus LRRIGs is not determined, but both LRR and IGcam are competent immune recognition modules. Please refer to additional data in Supplemental Material A.
Other LRR-containing genes
Besides the abovementioned TLRs, NLRs, and LRRIGs, there are also 1363 LRR-containing gene models in the amphioxus genome, of which 185 clearly contain LRR and other domains, whereas 948 contain LRR only (see the detailed description in Supplemental Material A). According to our analysis, most LRR-only models should represent distinct genes (Supplemental Material A), but because of the incorrect gene predictions and the limitations of homology searches, not all LRR-only models represent genes containing only LRR. In contrast, the human genome contains fewer than 265 LRR-containing proteins (various alternative spliced variants included) (Table 2). In vertebrates, CD14 and VLRs are typical LRR-only proteins with vital immune function. In amphioxus, there are no CD14 or VLR counterparts, but we have identified a full-length cDNA (GenBank accession no. DQ873591, from B. japonicum) from the gut of the immune-challenged amphioxus by suppression subtractive hybridization (Huang et al. 2007a). This cDNA encodes an LRR-only protein (amphiLRR1) with bacterial binding activity (G. Huang and A. Xu, unpubl.). We also identified an EST (GenBank accession no. EU183369, from Branchiostoma belcheri) for a gene from a family of eight LRR-only members encoded on Scaffold_88 (they may contain ankyrin and DEATH domains, but we haven’t cloned the full-length cDNA). This EST is mainly expressed in the gut and up-regulated by several hundred-folds in response to bacterial challenge (data not shown).
C-type lectins
C-type lectin proteins (CTL) contain one or more C-type lectin domains (CTLDs) that are capable of binding a variety of ligands, including pathogenic carbohydrates, lipids, proteins, CaCO3, and ice. Vertebrate CTLs have seven “large CTL” families with multiple CTLDs or complex domain combinations, and 10 “small CTL” families with single CTLDs and few non-CTLD domains (Zelensky and Gready 2005). According to our analysis, the human genome has 57 small CTLs, 47 of which are immune-related, such as NK-cell receptors, collectins, and macrophage/dendritic-cell receptors, and the like. NK-cell receptors recognize endogenous ligands and regulate immune homostatsis, while collectins and macrophage/dendritic-cell receptors recognize pathogenic carbohydrates. CTLDs capable of carbohydrate binding usually have sugar-binding motifs (mostly EPN/QPD + WND). Half of the human CTL genes encode CTLDs with sugar-binding motifs, in constrast to 20% of D. melanogaster CTLDs and 10% of C. elegans CTLDs, respectively (Dodd and Drickamer 2001).
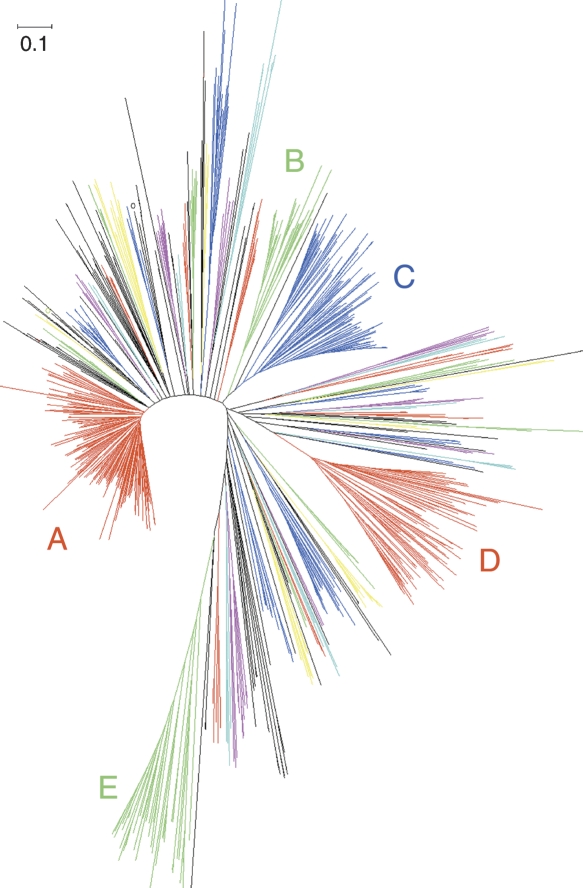
The amphioxus genome contains a vast CTL repertoire of 1215 models (Table 2). It has been reported that CTLs from C. elegans, D. melanogaster, and vertebrates are highly diverged from each other (Dodd and Drickamer 2001). This is also the case for amphioxus CTLs, because only certain human CTL families have weak homologs in amphioxus, such as SEEC, DGCR2, polycystins, collectins, and KLRD1-like proteins. Additionally, dual-CTLD genes are also present in amphioxus, which play a role in the immunity of D. melanogaster and bonyfishes. There are 113 models with multiple CTLDs and complex (or novel) domain combinations (Supplemental Material A). However, here we focus on the 1102 models encoding single CTLDs because the majority of them are small CTLs and may have potential immune function. Of these, 927 contain well-predicted CTLDs, the phylogenetic analysis of which identifies 43 CTL subfamilies. The five largest subfamilies are designated A, B, C, D, and E and account for 189, 36, 108, 100, and 40 CTLs, respectively (Fig. 3). There are 32 subfamilies including the aforementioned A–E subfamilies, with a total of 692 models encoding small CTLs with simple protein structures (Supplemental Table S2). There are 483 out of 927 CTLDs containing canonical EPN or QPD sugar-binding motifs, but there are also derivative motifs such as EPS, EPK, EPE, EPD, QPS, and QPN, which add up to more than 650 CTLDs (Supplemental Table S2). Such a variety of derived motifs may suggest diversified sugar-binding specificity. In contrast, 10 subfamilies (including subfamily E) completely lack conserved sugar-binding motifs. There are six subfamilies with a total of 40 models encoding collectin-like structures (COL-CTLD). In addition, at least 26 collectin-like models belong to other subfamilies. In vertebrates, collectins sense pathogenic carbohydrates and activate the complement system (Gupta and Surolia 2007).
Figure 3.
The ME tree of CTLD of amphioxus CTLs, including 879 amphioxus CTLDs. Subfamilies are colored. Five large amphioxus-specific CTL families are labeled A, B, C, D, and E.
We have cloned six cDNAs of small CTLs from Chinese amphioxus (Supplemental Table S2), including one from subfamily D (GenBank accession no. EU183370), three from subfamily E (GenBank accession nos. EU183373–EU183375), and two from two small subfamilies (GenBank accession nos. EU183371 and EU183372). Expression analysis indicates that EU183373–EU183375 are highly concentrated in the gut and the gills, while EU183370–EU183372 are mainly expressed in the gut and the skin, and are up-regulated by immune challenge (Yu et al. 2007a). The recombinant protein of EU183370 also shows strong microbial agglutination and anti-fungal activity (Yu et al. 2007a).
Scavenger receptors
Vertebrate scavenger receptors (SR) are usually surface proteins on macrophages and are able to recognize endogenous or bacterial lipoproteins (Mukhopadhyay and Gordon 2004). There are two types of scavenger receptors, namely, CD36 and the SRCR-containing receptor. Human has one CD36 and 16 SRCR-containing genes, while the sea urchin genome contains five CD36-like genes and 218 SRCR-containing models with a total of 1095 SRCR domains. The amphioxus genome possesses three CD36-like genes and 270 SRCR-containing models with a total of 497 SRCR domains (Table 2). Since a large proportion of the SRCR models are complex or problematic, 497 SRCR domains provide more accurate estimation of the size of this family.
RIG-I-like helicases
Besides NLRs, RIG-I-like helicases (RLHs) represent another crucial family of intracellular immune receptors, which use C-terminal RNA helicases to recognize viral dsRNA and N-terminal domains for signaling. Vertebrate RLHs use the CARD–CARD structure and the downstream CARD-containing adapter VISA for signal transduction, while sea urchin RLHs use DEATH and the DEATH-containing VISA-like adapters (Hibino et al. 2006). Seven RLH genes are present in amphioxus (Table 2). Four of them contain CARD-TIR domains, one contains CARD, one contains DEATH, and the last one contains DED. This represents the first report of CARD-TIR-containing RLHs and DED-containing RLHs, which will require further verification. As for adapters, only a CARD-containing VISA-like gene was identified in amphioxus so far.
Other PAMP recognition receptors
The amphioxus genome also encodes other conserved PRRs, including one galectin (Yu et al. 2007b), 16 PGRPs, and five GNBPs. In addition to the conserved PRRs, a set of amphioxus genes encoding novel protein architectures may have potential pathogen-sensing ability. These architectures include LRR-TM-DEATH (three genes) (Supplemental Material A), LRR-DEATH or LRR-DEATH-kinase (13 models) (Supplemental Material A), LRR-GTPase or LRR-GTPase-DEATH (37 models) (Supplemental Material A), LRR-DD-TIR (two genes), LRR-CCP-TM-TIR (three genes), CARD-TIR-LRR (two genes), and DEATH-glycosyl transferase (38 models; glycosyl transferase is capable of carbohydrate binding). Since few of these architectures have EST evidence, they require further verification.
The complement system
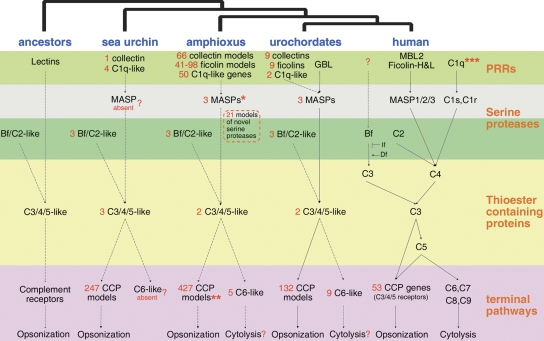
The vertebrate complement system is a sophisticated proteolysis system that has two major functions in humoral immunity: one is to lyse pathogenic cells; the other is to opsonize pathogens for phagocytosis and to recruit immunocytes to the reaction focus. The mammalian complement system has three activation pathways (lectin, classical, and alternative) and two terminal pathways (opsonic and cytolytic) (Fig. 4). Mammal C1q proteins can recognize antibodies (Igs) and activate the classical pathway, while the lamprey C1q acts as a carbohydrate-binding lectin like collectins and ficolins (Matsushita et al. 2004). C1qs, collectins, and ficolins can recruit MASP/C1r/C1s through their COL domain and activate complement pathways. The core pathway of the complement system consists of three protein families: MASP/C1r/C1s, Bf/C2, and C3/C4/C5 (Nonaka and Kimura 2006). The opsonic pathway has been identified in the sea urchin (Clow et al. 2004), but the C6-like proteins required for cytolysis are absent (Hibino et al. 2006). Urochordates and jawless vertebrates have multiple C6-like proteins, but their functions are awaiting further study. Taken together, the lectin pathway and the opsonic pathway seem to constitute the most primitive complement pathway in deuterostomes (Fig. 4).
Figure 4.
A schematic of the evolution of the complement system. (Solid line) The pathway has experimental evidence; (dashed lines) no experimental support; (?) the existence of the item or pathway is not verified; (*) amphiMASP1/3 gene can produce two proteins, MASP1 and MASP3; (**) amphioxus contains the most abundant CCP domains, see Supplemental Table S1; (***) the human C1q proteins recognize antibody, while the lamprey C1q serves as a lectin.
While no authentic C1q proteins have been identified, the amphioxus genome contains 50 C1q-domain-containing genes compared to 29 in human and four in the sea urchin (Supplemental Fig. S7). There are 42 amphioxus C1q-like genes adopting the typical COL-C1q architecture, 33 of which even encode COL and C1q in the same exon. Since no antibody is present in amphioxus and the lamprey C1q acts as a lectin, amphioxus C1q-like proteins may also function as lectins. We have cloned a cDNA from B. japonicum encoding a C1q-like protein that can inhibit platelet agglutination similarly to the mammalian C1q proteins (Y. Yu, H. Huang, Y. Wang, S. Yuan, S. Huang, Y. Yu, M. Pan, K. Feng, A. Xu, in prep.). Both vertebrate collectin MBL2 (mannose-binding lectin, COL-CTLD) and urochordate GBL (glucose-binding C-type lectin, no COL) can recruit MASPs and activate C3 (Sekine et al. 2001). Although no clear orthologs for MBL2 or GBL have been identified in amphioxus, there are 1215 CTL models in amphioxus, at least 66 of which have COL-CTLD structures (Supplemental Table S2). Vertebrate ficolins are also humoral lectins capable of recruiting MASPs to activate C3. There are 347 FBG models in amphioxus, compared to 26 in human. At least 41 amphioxus FBG models have the ficolin-like structure (COL-FBG), compared to three in human. All amphioxus ficolin-like models belong to two distinct families, which comprise 98 FBG models.
In activation pathways, C1q/MBL2/ficolin recruit serine proteases, including MASP/C1r/C1s, Bf/C2, factor D (Df), and factor I (If). The sea urchin has no MASP/C1r/C1s-like proteins, but mammals have five (MASP1/2/3, C1r, and C1s), and all of them have the CUB-EGF-CUB-(CCP)2-serine_protease structure. It has been reported that the amphioxus MASP1/3 gene can produce two proteins (MASP1 and MASP3) by alternative splicing (Endo et al. 2003). Here we found another gene (Bf288621) that is similar to amphioxus MASP1/3 proteins but encodes no CUBs [EGF-(CCP)6-serine_protease]. As for Bf/C2-like, three homologs are present in amphioxus. They adopt the conserved (CCP)n-VWA-serine_protease structure (n = 3 in human, n = 3, 4, 7 in amphioxus). As for Df and If, there is no clear homolog. In addition, the amphioxus genome contains another 21 models encoding novel CUB or CCP-containing serine proteases. In vertebrates, only those complement-related serine proteases carry CUB and CCP domains.
C3 is a thioester-containing protein that plays a central role in the complement system, whose origin can be dated back before the split of cnidaria and bilateria (Miller et al. 2007). In the early evolution of vertebrates, C3 was separated into C3, C4, and C5 by duplications. The emergence of C4 separates activation pathways into the alternative pathway and the lectin/classical pathway, while C5 bridges the C3 and the cytolytic pathway. Human has seven thioester-containing genes, including C3/C4A/C4B/C5, CD109, A2M, and a novel thioester gene. A search of the amphioxus genome identified six thioester genes, including two C3/4/5-like (one ortholog and one distant homolog), two A2M-like, one CD109-like genes, and one gene close to both A2M and CD109. The amphioxus ortholog of C3/4/5 has previously been characterized (Suzuki et al. 2002).
In vertebrates the cytolytic pathway includes at least four C6-like proteins (C6/C7/C8/C9), which assemble the Membrane Attack Complex (MAC) on the targeted cells. These proteins feature a MACPF domain required for membrane perforation. C6 is the longest protein containing the “prototypic” structure (TSP1)2-LDLa-MACPF-EGF-TSP1-(CCP)2-(FIMAC)2, while other genes contain fewer domains. In particular, C9 adopts the shortest structure, TSP1-LDLa-MACPF-EGF. The amphioxus genome has 29 MACPF genes, five of which encode C6-like proteins. They all adopt the same structure, (TSP1)2-LDLa-MACPF-EGF-TSP1, and lack the C-terminal CCP or FIMAC domains, which in vertebrates are required for interacting with C5. There are four amphioxus gene models carrying FIMAC, three encoding FIMAC and CCP domains, while the remaining one encodes FIMAC, EGF, and TSP1 domains.
Many proteins involved in complement regulation contain multiple CCPs, such as Factor H, DAF, and complement receptor Types 1 and 2. In spite of the absence of clear orthologs for these genes, the amphioxus genome contains 427 CCP-containing models, thereby comprising the biggest CCP repertoire seen in any species (Fig. 4; Supplemental Table S1). In actual fact, those domains involved in the complement system like CCP, CUB, TSP1, MACPF, C1q, FBG, VWA, and LDLa are all greatly expanded in amphioxus (Supplemental Table S1). Taken together, amphioxus has a very complicated and novel complement system, despite having only two C3-like genes.
The TNF system
It is perhaps because of the rapid and direct delivery of extrinsic signals by tumor necrosis factors (TNF) and their receptors (TNFR) that the TNF system is thriving in vertebrates, and extensively engaged in the immune system (Locksley et al. 2001). In vertebrates, the TNF system mediates activation, proliferation, differentiation, and homeostasis of immunocytes; participates in the development and maintenance of long-lived or evanescent lymphoid tissues; and implements the clearance of cancerous, aged, and diseased cells (Hehlgans and Pfeffer 2005). Vertebrates have approximately 20 TNF and 20–30 TNF receptor genes. The prominent among these genes include TNF, FASLG, TNFSF10, TNFSF11, LTA, TNFSF15, CD40LG, TNFSF13, TNFSF13B, and EDA. In contrast, only one TNF (called eiger) and one TNFR have been identified in D. melanogaster, and four TNFs and seven TNFRs in the sea urchin (Robertson et al. 2006). However, to our surprise, the amphioxus genome contains 24 TNF and 36 TNFR genes, numbers comparable to those of vertebrates (Table 2).
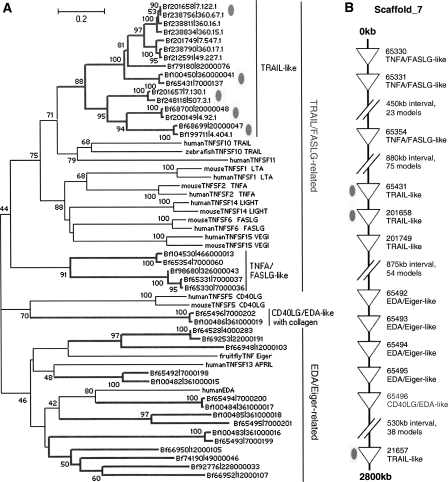
Phylogenetic analysis of the TNF domain indicates that amphioxus TNF proteins can be separated into two major lineages, the TRAIL/FASLG-related and the EDA/Eiger-related lineages (Fig. 5A; Supplemental Fig. S8). The TRAIL/FASLG-related lineage includes 10 TRAIL genes and three FASLG/TNFA-like genes, whereas the EDA/Eiger-related lineage contains 11 TNF genes homologous to insect Eiger, vertebrate EDA, and TNFSF13. Amphioxus TRAIL proteins share 45%–58% amino acid identity with vertebrate TRAILs, whereas the FASLG/TNFA-like proteins have little amino acid similarity with vertebrate FASLG or TNFA. In addition, an amphioxus TNF is clustered with vertebrate CD40LG, but the sequence similarity is rather weak. There is a TNF cluster of 12 genes located on the Scaffold_7, comprising five EDA/Eiger-related TNFs in the middle, six TRAIL/FASLG-related TNFs on one side, and one on the other side (Fig. 5B). Together with the phylogenetic analysis, this genomic configuration provides an update to the evolutionary story of vertebrate TNFs (Collette et al. 2003). The TRAIL/FASLG-related lineage was first created by tandem duplication from the ancient EDA/Eiger-related lineage and then underwent substantial functional divergence before whole-genome duplications; later, in the early evolution of vertebrates, whole-genome duplications produced TNFSF11, LTA, and TNFSF15 from the ancient TRAIL/FASLG-related lineage, and produced TNFSF13 and TNFSF13B from the ancient EDA/Eiger-related lineage. We have cloned cDNAs for five TRAILs from B. japonicum (Fig. 5A,B, marked by gray ovals). Initial analysis indicates that they assume disparate expression patterns in different tissues, suggesting that these TRAIL proteins may have achieved substantial functional divergence after rapid lineage-specific duplications (Supplemental Fig. S9).
Figure 5.
(A) The ME tree of the amphioxus TNF family. Both alleles are included in the analysis. (Bf) B. floridae. (B) The configuration of the TNF cluster on Scaffold_7. Gray ovals indicate genes with expression data (Supplemental Fig. S9).
TNFR can be divided into two types, TNFR with (death receptor, DR) and without a cytoplasmic DEATH domain (TNFR-noDD). DRs can activate caspase-dependent apoptosis, while TNFR-noDDs can act as DR antagonists or activate NF-κB and JNK pathways. The amphioxus genome contains 14 DRs and 22 TNFR-noDDs. Sequence analysis of the DEATH domain indicates that only two amphioxus DRs have some similarity to the vertebrate DR genes NGFR and EDAR. However, the rest of the amphioxus DRs are more similar to each other than to vertebrate DRs, suggesting that most amphioxus DRs have undergone a lineage-specific expansion. As for amphioxus TNFR-noDDs, their TNFR repeats are too divergent to be used for reliable phylogenetic analysis. We have cloned two TNFR-noDDs with weak similarity to the human TNFR-noDD genes TNFRSF1B or TNFRSF14 and have demonstrated their immune relevance (Yuan et al. 2007).
Transduction of TNF signals requires the interaction between TNFR cytoplasmic tails and the downstream adapters. Human has six TRAFs and four DFD adapters (FADD, TRADD, CRADD, EDARADD) for this purpose, while the sea urchin draft genome contains only one FADD, one CRADD, and four TRAF adapters (Robertson et al. 2006). The amphioxus genome contains a set of homologs of FADD, CRADD, and EDARADD, a family of 24 TRAFs, and a total of 541 DFD-containing models (discussed below). If a substantial proportion of these genes participate in the TNF system, it would represent the most complicated TNF signaling network ever known.
Expansion and reshuffling of the death-fold domains
The signal transduction of PRRs and cytokine receptors requires a cytosolic protein interaction network composed of various adapters or intermediate transducers. Death-fold domains (DFD), including DEATH, CARD, and DED, are basic building blocks for homotypic interactions. They are widely present in NLRs, RLHs, DRs, apoptotic proteins, and other signal transducers. As such, they broadly participate in TLR/IL-1R, NLR, TNF, RLH, and apoptosis pathways, as well as cross-talk among them. The human genome contains about 60 DFD genes, while the sea urchin genome contains 116 DFD genes (Robertson et al. 2006). It has also been reported that in the sea urchin genome, more than 200 DFDs coexist with NLRs (Hibino et al. 2006). A search of the amphioxus genome yields 541 DFD-containing models (NLRs and DRs excluded). Notably, this number is sensitive to the cutoff E-value of the homology search. For instance, if the E-value is relaxed to 0.01 or 0.1, the DFD number increases to 781 or 861, respectively. Moreover, many DFD sequences in the amphioxus genome fail to be captured by gene prediction (data not shown). In other words, a great expansion of the amphioxus DFD repertoire has occurred. These DFD models can be classified into 53 groups each consisting of similar domain structures (Supplemental Table S2).
Human DFD proteins consist of 16 distinct architectures, of which amphioxus has at least 14. In contrast, the amphioxus DFD repertoire has at least 40 domain combinations not seen in human, provided that most structures listed in Supplemental Table S2 are real. Since a novel domain combination may create a novel signaling pathway, increased architectural complexity may lead to increased complexity of the signaling network. However, novel architectures can either be inherited from ancient ancestors or result from recent domain reshuffling. The analysis of the DFD sequence similarity shows that identity between cognate domains from different DFD architectures ranges from 30% to more than 70%, where higher identity indicates more recent domain reshuffling events (Supplemental Fig. S10). This analysis suggests that dynamic domain reshuffling contributes to the architectural complexity of the amphioxus DFD repertoire.
Expansion of TIR adapters, TRAFs, and initiator caspases
As mentioned above, amphioxus has 95 vertebrate TIR, four plant-TIR-like, and 15 bacterial-TIR-like genes. In 95 vertebrate TIR genes, 40 are potential TIR adapters. By comparison, human has five TIR adapters and the sea urchin has approximately 26.
TRAFs compose a family of signal transducers capable of interacting with non-TRAF proteins through the TRAF domain. D. melanogaster, the sea urchin, and vertebrates possess three, four, and six TRAFs (TRAF1∼6), respectively. In vertebrates, TRAFs are involved in TLR pathways, TNF pathways, apoptosis, antiviral response, and NF-κB activation (Chung et al. 2002). A search of the amphioxus genome identifies 24 TRAF genes, including one corresponding to vertebrate TRAF6, one to TRAF4, four to TRAF3, and 16 to TRAF1/2 (Supplemental Fig. S11). Notably, 12 amphioxus TRAFs corresponding to the vertebrate TRAF1/2 lineage are encoded on Scaffold_558.
Vertebrate caspases can be separated into two classes, the ICE-like and the apoptotic CED-3-like. The ICE-like caspases mediate inflammation and NLR pathways, while the apoptotic caspases are downstream of TNF- or mitochondria-mediated apoptosis (Fig. 2; Riedl and Shi 2004). The apoptotic caspases can be further divided into “effector caspases” and “initiator caspases.” Caspase-3, caspase-6, and caspase-7 are effector caspases that can be activated by initiator caspases and execute the final suicidal process. Caspase-8 and caspase-10 are initiator caspases for delivery of extrinsic “death” signals from TNF receptors. Caspase-9 is responsible for intrinsic “death” signals from mitochondria, whereas caspase-2 relays “death” signals from both TNFs and mitochondria. Caspase-3/6/7 has no DFD domains, while caspase-8/10 contains a pair of DED domains and caspase-2 and caspase-9 each carries a CARD domain. At least 45 caspase genes have been identified from the amphioxus genome. Phylogenetic analysis reveals eight amphioxus caspase genes loosely related to both caspase-9 and caspase-2, 15 genes corresponding to caspase-8/10, 10 genes to caspase-2, five genes to caspase-3/6/7, and seven unknown caspases (Supplemental Fig. S11). Therefore, amphioxus “initiator caspases” have undergone sevenfold to 10-fold expansions compared to vertebrates. Such expansion is not evident in the sea urchin, although its genome encodes 31 caspases (Robertson et al. 2006). Notably, some amphioxus caspases contain DEATH instead of DED and CARD domains (Supplemental Fig. S12). This domain combination occurs several times in amphioxus and is also present in sea urchin, but not in vertebrates, and hence deserves further study.
Immune-related cytokines and transcription factors
Although homologs for IL-1 receptors, TNFs, IL-17, and macrophage migration inhibitory factors (MIF) are present in the amphioxus genome (Table 1), most of the vertebrate cytokines are absent, including most interleukins, all interferons, chemokines, colony-stimulating factors (CSFs), and their cognate receptors. A similar situation is observed in the sea urchin and other invertebrates (Hibino et al. 2006). The reason for this can be either a true absence of these genes or the inability of similarity searches to identify such fast-evolving genes.
In contrast, most of the immune-related transcription factors and their direct upstream kinases are present in amphioxus, including NF-κB, NFAT, IRF, Ikaros, PU.1/Spi, IKK/TBK, MAPK/JNK, and the like (Table 1). Unlike upstream signal transducers (such as TRAFs and TIR adapters), these transcription factors and kinases have not undergone expansion and maintain typical numbers. Notably, in vertebrates, interferon-regulatory factors (IRFs) can induce type I interferons and play a crucial role in the regulation of TLRs, NLRs, RLHs, and other PRRs (Honda and Taniguchi 2006). Although interferon is absent, there are 11 IRF genes in amphioxus, compared to nine in human and only two in the sea urchin. Phylogenetic analysis indicates that seven amphioxus IRFs are closely related to human IRFs, especially to human IRF1/2 and IRF4/8/9 lineages (Supplemental Fig. S13).
Rudiments of the adaptive immunity related to innate molecules
The amphioxus genome contains some basic components of the VAIS, such as the protoMHC region (Abi-Rached et al. 2002), recombination activating gene 1-like (RAG1-like) genes (Kapitonov and Jurka 2005), terminal deoxynucleotidyl transferase-like (TdT/polμ-like) genes, and enzymes/factors involved in DNA rearrangement and DNA repair (Holland et al. 2008). However, all molecular hallmarks of the VAIS, including VLR, BCR, TCR, and MHC class I and II molecules, are absent in amphioxus.
Vertebrates generate two forms of somatically diversified receptors by DNA rearrangement, the VLR of jawless vertebrates and the BCR/TCR/Ig of jawed vertebrates. Thus far no somatically rearranged receptor has been found in amphioxus. However, as mentioned previously, there is a huge arsenal of LRR-containing genes in amphioxus. Moreover, the previously identified amphiLRR1 genes (from B. japonicum) have been found to have high polymorphism at the population level (G. Huang and A. Xu, unpubl.). The amphioxus draft genome also contains approximately 1000 IgSF domains, of which two types have been documented. One is the VCBP family, which produces secreted proteins with highly polymorphic IgV-like domains (Cannon et al. 2002). The other is the VCP family, which contains IgV-like-IgC ectodomains (Yu et al. 2005). The latest study indicates that both IgV-like and IgC domains of VCP proteins have significantly high polymorphism at the population level and that the phosphorylated cytoplasmic ITIM-like motifs of VCP can induce extensive intracellular phosphorylation in B-cell lines (C. Yu and A. Xu, unpubl.). Taken together, these data indicate that some basic elements of the vertebrate BCR/TCR/VLR systems, including molecular structures, high levels of polymorphism, and signaling mechanisms, already exist in amphioxus.
Conclusions
This survey shows that amphioxus and vertebrates share an overall conserved framework of the innate immune system in terms of sequence homology, domain composition, receptor repertoire, and core pathways. When sea urchin and other non-chordates are used as references, some conservation can be reinterpreted as chordate innovation, including an expanded TNF system, a more complex complement system, an expanded IRF family, and the like.
The expansion of innate receptors differentiates sea urchin and amphioxus from vertebrates. Both sea urchin and amphioxus exhibit expansion of TLR, NLR, and SR families, while amphioxus further expands CTL, LRRIG, LRR-only, C1q-like, ficolin-like, and even genes containing complement-related domains like CCP, CUB, and TSP1. Since sea urchin and amphioxus represent basal deuterostome lineages, it is possible that increased innate diversity is the prevalent strategy in chordate ancestors. However, in vertebrates, innate diversity apparently has been reduced concurrently with the rise of the VAIS. A plausible hypothesis for this transition is that somatically diversified receptors may provide equivalent (or larger) recognition capacity and, more importantly, may allow more plasticity for increasing developmental and morphological complexity during vertebrate evolution (Pancer and Cooper 2006). Innate diversity may not be comparable to the somatic diversity of the VAIS, but considering that amphioxus is a small and apparently simple animal with a short life span, high polymorphism, and high reproductive capacity, innate diversity at the population level may offset the advantage of those somatic mechanisms.
The expansion of adapter-like TIR genes, TRAFs, initiator caspases, and the DFD gene repertoire distinguishes amphioxus from vertebrates. In a “funnel” model of signaling such as the vertebrate BCR/TCR and perhaps the sea urchin TLR, the specificity of downstream cellular responses for receptors arises in the context of selective cellular expression of receptors (cell-type-specific or clonal expression) (Rast et al. 2006). As in amphioxus, since expansion occurs at both the receptor level and the adapter level, it is possible that the specificity of downstream cellular responses may be modulated at both levels through selective expression.
We have identified many novel domain combinations in amphioxus NLRs, CTLs, SRs (data not shown), TIR proteins, DFD proteins, and complement-related proteins. Although the vertebrate immune system also contains many protein architectures absent in amphioxus, this does not compare to the diversity and complexity seen in amphioxus. By having analyzed the DFD genes, we propose that an ongoing domain-reshuffling mechanism may contribute to this increased complexity of immune protein architectures. Since domain reshuffling can lead to changes in the signaling network, it would be very interesting to determine/examine whether an immune strategy for amphioxus is to shape its immune gene repertoire by employing a dynamic domain-reshuffling mechanism.
The expanded receptor and adapter repertoires and the increased architectural variety produce extraordinary innate complexity and diversity in amphioxus. It might seem like quite a burden to dedicate about 1/10 of its genes (>5000 of 50,718 models) to host defense, but there is no question about the success of this immune system, considering that amphioxus is widely distributed around the world and has been thriving for more than 500 million yr. Extensive experimentation is still required to verify that these genes are functional and working together as a system. Nevertheless, since its immune gene repertoire contains myriad innovations and conservations, amphioxus is invaluable for comparative immunological study.
Methods
The draft genome (release V1.0) of B. floridae and corresponding analysis tools can be accessed at http://genome.jgi-psf.org/Brafl1/Brafl1.home.html. The NCBI gene sets of human, mouse, zebrafish, sea urchin, and D. melanogaster; the Gnomon-predicted gene set of the sea urchin; and the trace archive of the jawless vertebrate lamprey were all downloaded from the NCBI FTP site.
Domain and gene identification were mainly performed with the HMMER2.0 plus SMART data set (Letunic et al. 2004). The RPSBLAST program and PFAM data set were also used in some cases. For genes without conserved domains or sequences, homology searches were performed with the BLASTALL program combined with various known genes. For domain comparison (Supplemental Table S1), the cutoff E-value was set to 0.01 to achieve an upper bound of domain hits. For the analysis of protein architecture, the E-value was set to 1e−5 to acquire reliable predictions. To search for the “missing” domains or to correct the poorly predicted models (such as TLR with missing LRRs, NLR with missing NACHT, DFD, or LRRs), the E-value was set to 10. Intronic sequences and 20-kb genomic sequences flanking the models were also examined for “missing” domains. Finally, manual inspection was performed.
The novel protein architectures were validated by two means. First, we searched for EST and cDNA evidence from the NCBI B. floridae EST data set, as well as our EST data set and cDNA cloning project of Chinese amphioxus (B. japonicum and B. belcheri). Second, a novel architecture without cDNA evidence was considered relatively reliable when it met the following three criteria: (1) presence in multiple copies dispersed throughout the genome; (2) a compact and neat exon structure; and (3) no other uncaptured domains in its genomic vicinity. If not specified, the novel domain combinations mentioned in the paper have been verified by one of the abovementioned means.
Phylogenetic analysis was based on protein sequences (plus 5–10 extra amino acids flanking the domain) and performed as described previously (Huang et al. 2005). Briefly, alignment was produced with CLUSTAL V1.83, sequences of poor quality (diverged, frame shifts, large gaps, etc.) were reciprocally deleted, and manual improvement was conducted with GENEDOC software (using LogOdds score as criteria). Phylogenetic analyses using the Neighbor-Joining (NJ) and Minimum-Evolution (ME) methods were performed with MEGA3.1 software (Kumar et al. 2004), with 1000 bootstrap tests, Poisson correction, and pairwise deletion of gaps. The obtained tree was improved by reciprocal deletion of diverged or unstable sequences. The major topology of all trees was verified by using PHYLIP software (Maximum-Likelihood (ML) method, pairwise deletion, 100 bootstrap tests, and Poisson correction). Here only bootstrapped consensus ME trees were presented. All trees were unrooted trees.
Expression analysis was conducted with adult Chinese amphioxus. If not specified, real-time quantitative RT-PCR was used for expression analysis with four tissues (skin, muscle, gut, and gill). If not specified, immune challenge was carried out with a cocktail of Vibrio vulnificus (G−), Staphylococcus aureus (G+), and bacterial cell wall components lipopolysaccharide (LPS) and lipoteichoic acid (LTA). The rationale and practice of these methods have been detailed in our previous studies (Yu et al. 2005; Huang et al. 2007a, b; Yuan et al. 2007).
Acknowledgments
We thank Linda Z. Holland for her comments on this manuscript. We also thank Dr. Ildiko Somorjai for her kindness in proofreading the manuscript. This work was supported by Project 2007CB815800 of the National Basic Research Program (973) and Project 2006AA09Z433 of the State High-Tech Development Project (863), Project 2007DFA30840 of International S&T Cooperation Program of China from the Ministry of Science and Technology of China, Key Project (0107) from the Ministry of Education, and Key Projects of Commission of Science and Technology of Guangdong Province and Guangzhou City, and Project of Sun Yet-sen University Science Foundation. A.X. is a recipient of the ′Outstanding Young Scientist Award′ from the National Natural Science Foundation of China. The genome of Branchiostoma floridae were sequenced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). Children's Hospital & Research Center Oakland (CHORI; Pieter de Jong and Kazutoyu Osoegawa) provided the DNA and the BAC library. Linda Z. Holland provided the animals for the DNA and the money for the BAC library and Asao Fujiyama (NIG, Japan) provided BAC end sequences. NSF funded part of the BAC sequencing, performed by TIGR.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. EU183366–EU183375.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.069674.107.
References
- Abi-Rached L., Gilles A., Shiina T., Pontarotti P., Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat. Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- Azumi K., De Santis R., De Tomaso A., Rigoutsos I., Yoshizaki F., Pinto M.R., Marino R., Shida K., Ikeda M., Ikeda M., et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “Waiting for Godot.”. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- Cannon J.P., Haire R.N., Litman G.W. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat. Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- Carty M., Goodbody R., Schroder M., Stack J., Moynagh P.N., Bowie A.G. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Chen Y., Aulia S., Li L., Tang B.L. AMIGO and friends: An emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res. Brain Res. Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Chung J.Y., Park Y.C., Ye H., Wu H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- Clow L.A., Raftos D.A., Gross P.S., Smith L.C. The sea urchin complement homologue, SpC3, functions as an opsonin. J. Exp. Biol. 2004;207:2147–2155. doi: 10.1242/jeb.01001. [DOI] [PubMed] [Google Scholar]
- Collette Y., Gilles A., Pontarotti P., Olive D. A co-evolution perspective of the TNFSF and TNFRSF families in the immune system. Trends Immunol. 2003;24:387–394. doi: 10.1016/s1471-4906(03)00166-2. [DOI] [PubMed] [Google Scholar]
- Couillault C., Pujol N., Reboul J., Sabatier L., Guichou J.F., Kohara Y., Ewbank J.J. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dodd R.B., Drickamer K. Lectin-like proteins in model organisms: Implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:71R–79R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- Dong M., Fu Y., Yu C., Su J., Huang S., Wu X., Wei J., Yuan S., Shen Y., Xu A. Identification and characterisation of a homolog of an activation gene for the recombination activating gene 1 (RAG 1) in amphioxus. Fish Shellfish Immunol. 2005;19:165–174. doi: 10.1016/j.fsi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Endo Y., Nonaka M., Saiga H., Kakinuma Y., Matsushita A., Takahashi M., Matsushita M., Fujita T. Origin of mannose-binding lectin-associated serine protease (MASP)-1 and MASP-3 involved in the lectin complement pathway traced back to the invertebrate, amphioxus. J. Immunol. 2003;170:4701–4707. doi: 10.4049/jimmunol.170.9.4701. [DOI] [PubMed] [Google Scholar]
- Fritz J.H., Ferrero R.L., Philpott D.J., Girardin S.E. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Fuentes M., Benito E., Bertrand S., Paris M., Mignardot A., Godoy L., Jimenez-Delgado S., Oliveri D., Candiani S., Hirsinger E., et al. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum) J. Exp. Zoolog. B Mol. Dev. Evol. 2007;308:484–493. doi: 10.1002/jez.b.21179. [DOI] [PubMed] [Google Scholar]
- Gupta G., Surolia A. Collectins: Sentinels of innate immunity. Bioessays. 2007;29:452–464. doi: 10.1002/bies.20573. [DOI] [PubMed] [Google Scholar]
- Hehlgans T., Pfeffer K. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: Players, rules and the games. Immunology. 2005;115:1–20. doi: 10.1111/j.1365-2567.2005.02143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T., Loza-Coll M., Messier C., Majeske A.J., Cohen A.H., Terwilliger D.P., Buckley K.M., Brockton V., Nair S.V., Berney K., et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Holland L.Z., Yu J.K. Cephalochordate (amphioxus) embryos: Procurement, culture, and basic methods. Methods Cell Biol. 2004;74:195–215. doi: 10.1016/s0091-679x(04)74009-1. [DOI] [PubMed] [Google Scholar]
- Holland L.Z., Albalat R., Azumi K., Benito-Gutiérrez È., Blow M.J., Bronner-Fraser M., Brunet F., Butts T., Candiani S., Dishaw L.J., et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008 doi: 10.1101/gr073676.107. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Huang S., Yuan S., Dong M., Su J., Yu C., Shen Y., Xie X., Yu Y., Yu X., Chen S., et al. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics. 2005;86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Huang G., Liu H., Han Y., Fan L., Zhang Q., Liu J., Yu X., Zhang L., Chen S., Dong M., et al. Profile of acute immune response in Chinese amphioxus upon Staphylococcus aureus and Vibrio parahaemolyticus infection. Dev. Comp. Immunol. 2007a;31:1013–1023. doi: 10.1016/j.dci.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Huang G., Xie X., Han Y., Fan L., Chen J., Mou C., Guo L., Liu H., Zhang Q., Chen S., et al. The identification of lymphocyte-like cells and lymphoid-related genes in amphioxus indicates the twilight for the emergency of adaptive immune system. PLoS ONE. 2007b;2:e206. doi: 10.1371/journal.pone.0000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N., Nunez G. NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Inohara C., McDonald C., Nunez G. NOD-LRR proteins: Role in host–microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas E., Michaut L., Reichhart J.M., Hoffmann J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley R.M., Killeen N., Lenardo M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Matsushita M., Matsushita A., Endo Y., Nakata M., Kojima N., Mizuochi T., Fujita T. Origin of the classical complement pathway: Lamprey orthologue of mammalian C1q acts as a lectin. Proc. Natl. Acad. Sci. 2004;101:10127–10131. doi: 10.1073/pnas.0402180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.J., Hemmrich G., Ball E.E., Hayward D.C., Khalturin K., Funayama N., Agata K., Bosch T.C. The innate immune repertoire in Cnidaria—Ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Gordon S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology. 2004;209:39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Kishishita N., Shimizu K., Hirose S., Miyoshi M., Nezu J., Nishimura T., Nishizumi H., Takahashi Y., Hashimoto S., et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat. Immunol. 2007;8:206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- Neves G., Zucker J., Daly M., Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat. Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Nonaka M., Kimura A. Genomic view of the evolution of the complement system. Immunogenetics. 2006;58:701–713. doi: 10.1007/s00251-006-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z., Cooper M.D. The evolution of adaptive immunity. Annu. Rev. Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- Pancer Z., Amemiya C.T., Ehrhardt G.R., Ceitlin J., Gartland G.L., Cooper M.D. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Parker J.S., Mizuguchi K., Gay N.J. A family of proteins related to Spatzle, the toll receptor ligand, are encoded in the Drosophila genome. Proteins. 2001;45:71–80. doi: 10.1002/prot.1125. [DOI] [PubMed] [Google Scholar]
- Putnam N.H., Butts T., Ferrier D.E.K., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008 doi: 10.1038/nature06967. (in press) [DOI] [PubMed] [Google Scholar]
- Rast J.P., Smith L.C., Loza-Coll M., Hibino T., Litman G.W. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes C.P., Ratcliffe N.A., Rowley A.F. Presence of coelomocytes in the primitive chordate amphioxus (Branchiostoma lanceolatum) Science. 1982;217:263–265. doi: 10.1126/science.7089565. [DOI] [PubMed] [Google Scholar]
- Riedl S.J., Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Roach J.C., Glusman G., Rowen L., Kaur A., Purcell M.K., Smith K.D., Hood L.E., Aderem A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.J., Croce J., Carbonneau S., Voronina E., Miranda E., McClay D.R., Coffman J.A. The genomic underpinnings of apoptosis in Strongylocentrotus purpuratus. Dev. Biol. 2006;300:321–334. doi: 10.1016/j.ydbio.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Sansonetti P.J. The innate signaling of dangers and the dangers of innate signaling. Nat. Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- Schubert M., Escriva H., Xavier-Neto J., Laudet V. Amphioxus and tunicates as evolutionary model systems. Trends Ecol. Evol. 2006;21:269–277. doi: 10.1016/j.tree.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Sekine H., Kenjo A., Azumi K., Ohi G., Takahashi M., Kasukawa R., Ichikawa N., Nakata M., Mizuochi T., Matsushita M., et al. An ancient lectin-dependent complement system in an ascidian: Novel lectin isolated from the plasma of the solitary ascidian, Halocynthia roretzi. J. Immunol. 2001;167:4504–4510. doi: 10.4049/jimmunol.167.8.4504. [DOI] [PubMed] [Google Scholar]
- Suzuki M.M., Satoh N., Nonaka M. C6-like and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J. Mol. Evol. 2002;54:671–679. doi: 10.1007/s00239-001-0068-z. [DOI] [PubMed] [Google Scholar]
- Yu C., Dong M., Wu X., Li S., Huang S., Su J., Wei J., Shen Y., Mou C., Xie X., et al. Genes “waiting” for recruitment by the adaptive immune system: The insights from amphioxus. J. Immunol. 2005;174:3493–3500. doi: 10.4049/jimmunol.174.6.3493. [DOI] [PubMed] [Google Scholar]
- Yu Y., Yu Y., Huang H., Feng K., Pan M., Yuan S., Huang S., Wu T., Guo L., Dong M., et al. A Short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J. Immunol. 2007a;179:8425–8434. doi: 10.4049/jimmunol.179.12.8425. [DOI] [PubMed] [Google Scholar]
- Yu Y., Yuan S., Yu Y., Huang H., Feng K., Pan M., Huang S., Dong M., Chen S., Xu A. Molecular and biochemical characterization of galectin from amphioxus: Primitive galectin of chordates participated in the infection processes. Glycobiology. 2007b;17:774–783. doi: 10.1093/glycob/cwm044. [DOI] [PubMed] [Google Scholar]
- Yuan S., Yu Y., Huang S., Liu T., Wu T., Dong M., Chen S., Yu Y., Xu A. Bbt-TNFR1 and Bbt-TNFR2, two tumor necrosis factor receptors from Chinese amphioxus involve in host defense. Mol. Immunol. 2007;44:756–762. doi: 10.1016/j.molimm.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Zelensky A.N., Gready J.E. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zhang S.M., Adema C.M., Kepler T.B., Loker E.S. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]