Abstract
The emergence of the neural crest has been proposed to play a key role in early vertebrate evolution by remodeling the chordate head into a “new head” that enabled early vertebrates to shift from filter feeding to active predation. Here we show that the genome of the basal chordate, amphioxus, contains homologs of most vertebrate genes implicated in a putative neural crest gene regulatory network (NC-GRN) for neural crest development. Our survey of gene expression shows that early inducing signals, neural plate border patterning genes, and melanocyte differentiation genes appear conserved. Furthermore, exogenous BMP affects expression of amphioxus neural plate border genes as in vertebrates, suggesting that conserved signals specify the neural plate border throughout chordates. In contrast to this core conservation, many neural crest specifier genes are not expressed at the amphioxus neural plate/tube border, raising the intriguing possibility that this level of the network was co-opted during vertebrate evolution. Consistent with this, the regulatory region of AmphiFoxD, homologous to the vertebrate neural crest specifier FoxD3, drives tissue-specific reporter expression in chick mesoderm, but not neural crest. Thus, evolution of a new regulatory element may have allowed co-option of this gene to the NC-GRN.
In vertebrate embryos, neural crest cells arise at the neural plate border, undergo an epithelial-to-mesenchymal transition, and become a migratory cell population that forms defining features of vertebrates including craniofacial skeleton, peripheral nervous system, and pigment cells (Le Douarin and Kalcheim 1999). Recent comparative developmental and molecular studies on the sister groups of vertebrates (e.g., amphioxus and tunicates) have provided important insights into the evolutionary origin of the neural crest (Gans and Northcutt 1983; Shimeld and Holland 2000; Northcutt 2005). While no migrating neural crest cells have been discovered in amphioxus (Holland and Holland 2001; Meulemans and Bronner-Fraser 2004), some cells with neural crest-like properties such as the ability to migrate and form pigment cells, have been reported in the ascidian tunicate Ecteinascidia turbinate (Jeffery et al. 2004). Recently, similar cells were also reported in diverse ascidian species, suggesting that they might be a common character in this clade (Jeffery 2006). This discovery suggests that the evolutionary origin of neural crest in chordates may be more deeply rooted than previously thought. Given that amphioxus is now considered a basal chordate rather than the immediate sister-taxon of vertebrates (Bourlat et al. 2006), it occupies a key position for understanding the origin of the neural crest. To address this important question, the draft genome sequence of amphioxus Branchiostoma floridae (Putnam et al. 2008) enables a global comparison between amphioxus and vertebrates of signaling and gene regulatory interactions operating at the neural plate border.
Studies of several vertebrate models have led to the hypothesis that a common set of genes with apparently conserved functions operate in a genetic network at the vertebrate neural plate border to specify and maintain the neural crest cells (Mayor et al. 1999; Meulemans and Bronner-Fraser 2004; Steventon et al. 2005). By comparing the gene expression patterns and experimental results from gain- or loss-of-function studies, a putative neural crest gene regulatory network (NC-GRN) has been proposed to mediate events leading from initial induction to final differentiation of the neural crest (Supplemental Fig. S1A). Despite the current paucity of cis-regulatory analysis required to define direct versus indirect interactions, this putative NC-GRN provides a useful conceptual framework to understand neural crest development and its evolution. According to this NC-GRN, the neural crest regulatory state is initiated in this border region in response to intercellular signals like BMP, Wnt, FGF, and Notch. These segregate the neural plate border from neuroectoderm and epidermis by activating a suite of transcription factors whose expression defines a territory competent to form the neural crest at the juncture between neural plate and epidermis. Next, inductive signals and neural plate border specifiers cooperate to activate another set of transcriptional regulators (neural crest specifiers) that specify neural crest fate and migratory state. Finally, the neural crest specifiers control a variety of downstream effector genes involved in generating migratory cells and lineage decisions for pigment cells, neurons, glia, and craniofacial elements. We present here the identification of amphioxus homologs of vertebrate NC-GRN genes, and the comparison of gene deployment for neural plate border development between amphioxus and vertebrate embryos.
Results and Discussion
To identify amphioxus homologs of vertebrate neural crest network genes, we searched the newly assembled amphioxus genome. Despite the lack of definitive neural crest in amphioxus, its genome contains most homologs of vertebrate NC-GRN genes (Supplemental Table S1). We have identified amphioxus genes encoding inductive signaling molecules in the BMP, Wnt, FGF, and Notch pathways. The amphioxus genome also contains all of the transcription factors that are deployed in the vertebrate neural crest regulatory network, including genes encoding neural plate border specifiers and neural crest specifiers. In addition, we identified homologs of downstream mediators of neural crest migration and differentiation, such as Rho, cRet, Erbb3, Mitf, tyrosinase, and tyrosinase-related proteins. In contrast, we failed to identify amphioxus homologs of c-Kit, a receptor tyrosine kinase essential for migration, survival, and differentiation of neural crest-derived melanocytes (Wehrle-Haller et al. 2001), or of myelin protein P0, consistent with the notion that the glial myelin sheath is a vertebrate innovation (Gould et al. 2005). The presence of all of the major signaling molecules and transcription factors for NC-GRN in amphioxus suggests that the evolution of neural crest is likely due to a change of network architecture (Davidson and Erwin 2006), perhaps by novel deployment of existing genes or network subcircuits at the neural plate border.
As a first step in identifying evolutionary changes in NC-GRN architecture, we compared the expression pattern of amphioxus homologs to those of traditional vertebrate models. For reference, a diagram showing the amphioxus neurulation process and domains within the neural plate border is provided in Supplemental Figure S1B. Carefully staged embryos were examined from mid-gastrulation through neurulation to give an approximate picture of dynamic changes in gene expression during early development. Prior to neurulation, genes encoding inductive signals are expressed in amphioxus similarly to their vertebrate homologs (Fig. 1), suggesting that they have conserved roles in patterning the neural ectoderm. In vertebrates, high BMP signaling levels activate epidermal fate via activation of transcription factors Msx, Dlx, and AP2; conversely, inhibition of BMP signaling induces expression of the neural markers, Sox2 (SoxB) and Zic, in the neural plate (Meulemans and Bronner-Fraser 2004). In addition, the combined input of inductive signals up-regulates the border specifiers Dlx, Msx, Zic, and Pax3/7 at the neural plate border. To determine whether the second level of the NC-GRN is conserved between amphioxus and vertebrate, we examined the expression of homologs of vertebrate epidermal, neural plate, and neural plate border marker genes in amphioxus. Our expression analysis in carefully stage-matched embryos during neurulation allows direct and unequivocal comparisons between these genes relative to the neural plate border (Fig. 2A). Amphioxus AP2 (tfap2) is expressed only in the epidermal ectoderm. The Dlx expression boundary is more medial than that of AP2and strongly demarcates the epidermal border near the neural ectoderm, whereas Msx is expressed not only in the epidermal ectoderm, but extends more medially into the neural ectoderm at the edges of the neural plate. Ectodermal Zic and Pax3/7 expression marks the neural plate border, and amphioxus SoxB1-a expression labels the entire neural plate. These results suggest that amphioxus uses similar inductive signaling input and transcription factors to specify epidermis, neural plate, and the border between them.
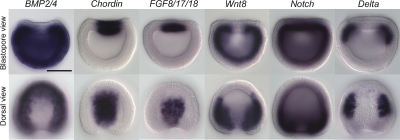
Figure 1.
Expression of amphioxus genes of inductive signals during late gastrula stage. Anterior is perpendicular to the plane of the page in the blastopore view and at the top in the dorsal view. Dorsal is up in the blastopore view. Scale bar, 50 μm. Amphioxus BMP2/4 is expressed throughout the ectoderm and mesendoderm, except the dorsal axial mesoderm and ectoderm, and the BMP antagonist Chordin is expressed in complementary domain in the most dorsal-medial mesendorem and ectoderm. Fgf8/17/18 is expressed in the dorsal axial mesendoderm underlying the prospective neural plate, and Wnt8 is expressed in the paraxial mesoderm and ventral mesendoderm. Notch is expressed throughout the entire mesendoderm and in the prospective neural plate, and its ligand Delta is expressed in the paraxial dorsal mesoderm.
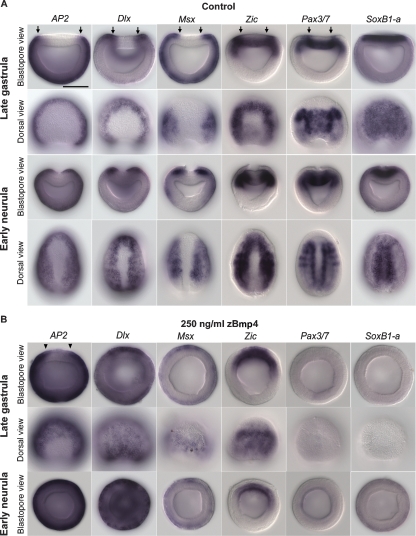
Figure 2.
Expression of amphioxus neural plate border specifier genes is effected by exogenous zebrafish Bmp4 (zBmp4) protein. Anterior is perpendicular to the plane of the page in the blastopore view and at the top in the dorsal view. The medial boundaries of gene expression in dorsal ectoderm are indicated by arrows (A) or arrowheads (B). Scale bar, 50 μm. (A) In normal late gastrula stage embryos, AP2, Dlx, and Msx are expressed throughout the epidermal ectoderm. Msx is also strongly expressed at the neural plate border. Zic and Pax3/7 expression in the dorsal ectoderm marks the neural plate border, and SoxB1-a expression is in the entire neural plate. During the early neurula stage, the edges of the epidermal ectoderm dissociate from the neuroectoderm and migrate toward the midline. This process is shown in the blastopore view of early neurula embryos. (B) In zBmp4 treated embryos, expression of epidermal markers AP2 and Dlx is greatly expanded, suggesting that the entire dorsal ectoderm is transformed to epidermal fate. The expression of neural plate border markers Msx and Zic is converged to the dorsal midline; expression of another neural plate border marker Pax3/7 and neural plate marker SoxB1-a is lost. In addition, no neurulation process can be observed in zBmp4 treated embryos.
During gastrulation, expression of amphioxus BMP2/4 and its antagonist Chordin appear to set up a BMP signaling gradient in the dorsal ectoderm (high laterally and low medially) (Fig. 1). This is consistent with the possibility that amphioxus BMP signaling might function similarly in patterning dorsal ectoderm in amphioxus as in vertebrates. Furthermore, administration of exogenous BMP protein ventralizes amphioxus embryos (Yu et al. 2007). To address whether BMP signaling specifies the neural plate boundary in amphioxus, we treated developing amphioxus embryos with exogenous zebrafish Bmp4 protein (zBmp4) and examined the subsequent effects on gene expression (Fig. 2B; Supplemental Fig. S2). In Bmp4-treated embryos, the expression of the epidermal marker AP2 was expanded medially; the epidermal domains of Dlx and Msx also were expanded and the strong expression that originally marked the neural plate border converged toward the midline of dorsal ectoderm. Ectodermal expression of Zic also converged toward the midline of the treated late gastrula stage, but disappeared by the early neurula stage. The neural plate border marker Pax3/7 and neural plate marker SoxB1-a were either greatly reduced or completely absent in BMP-treated embryos, indicating that the entire dorsal ectoderm was transformed to an epidermal fate. Cumulatively, these results suggest that BMP signaling patterns the neural ectoderm and regulates the mediolateral expression boundaries of neural plate border specifier genes in the dorsal ectoderm of amphioxus. This is consistent with the data in vertebrates showing that neural plate specifier genes (Dlx, Msx, Zic, and Pax3/7) function to mediate the effects of inductive signals at the neural plate border (Meulemans and Bronner-Fraser 2004). This group of genes performs the general function of establishing the neural plate border and acts genetically upstream of neural crest markers like Snail/Slug and FoxD3 (Meulemans and Bronner-Fraser 2004). Whereas the neural crest genes are general markers of premigratory, migrating, and/or post-migratory neural crest cells, neural plate border specifiers have a broader function in the development of other cell types derived from neural plate border including roof plate, dorsal interneurons, placodes, and Rohon-Beard cells. From our experimental data, we hypothesize that these neural plate border specifiers function downstream of inductive signals to specify the neural plate border in amphioxus and may also be involved in formation of dorsal cells in the developing neural tube of amphioxus. Together with studies showing the conserved role of BMP signaling in neural induction and mediolateral patterning of neuroectoderm among vertebrates, fly Drosophila, and annelid Platynereis (De Robertis and Kuroda 2004; Mizutani et al. 2006; Denes et al. 2007), our findings provide strong support that the upper two levels of the NC-GRN represent an ancient pan-bilaterian subcircuit for setting up the boundary between neuroectoderm and epidermis.
In vertebrates, inductive signals and neural plate border specifiers cooperate to activate the third level of the NC-GRN by regulating Snail/Slug, AP-2, FoxD3, Twist, Id, cMyc, and Sox9/10 (SoxE) transcription factors at the neural plate border to specify neural crest fate (Meulemans and Bronner-Fraser 2004). In comparing the expression of amphioxus homologs of these genes, we found that they were expressed in broad and diverse patterns, but were absent from the amphioxus neural plate border (Langeland et al. 1998; Yasui et al. 1998; Meulemans and Bronner-Fraser 2002; Yu et al. 2002; Meulemans et al. 2003) (Fig. 3A). The single exception was amphioxus Snail, which is transiently expressed at the neural plate border in the early neurula stage. This suggests that in vertebrates neural crest specifier genes may have been co-opted for new roles at this level of the network (Meulemans and Bronner-Fraser 2002, 2005; Yu et al. 2002; Meulemans et al. 2003). Interestingly, many of these neural crest specifier genes have a single copy in amphioxus, whereas in vertebrates they have multiple paralogs, presumably due to whole-genome duplication events (Supplemental Figs. S6, S9) (Meulemans and Bronner-Fraser 2002; Yu et al. 2002; Meulemans et al. 2003). This is consistent with the possibility that gene duplication may have facilitated co-option of genes into the NC-GRN. To test the possible role of co-option in the evolution of the neural crest, we introduced into vertebrate embryos a regulatory region located immediately 5′ of the coding region of amphioxus FoxD (AmphiFoxD) (Yu et al. 2004), homologous to the vertebrate neural crest specifier FoxD3. A similar approach previously was used to assay the regulatory activity of genomic DNA from amphioxus Hox genes (Manzanares et al. 2000). To achieve cross-species transgenesis, we electroporated an EGFP reporter construct containing the AmphiFoxD regulatory region into the early chick gastrula together with a ubiquitously expressed RFP reporter construct. Interestingly, we found that the EGFP reporter was expressed in a pattern highly reminiscent of endogenous AmphiFoxD, with numerous EGFP-expressing cells observed in the chick somites and notochord (Fig. 3B–E; Supplemental Table S2). Also consistent with the amphioxus pattern, a few EGFP+ cells were observed in the central nervous system, particularly at hindbrain levels. Although numerous neural crest cells were RFP positive, indicating efficient transfection, no EGFP+/HNK1+ cells were observed within the prominent neural crest migratory streams. Even when the reporter construct was electroporated into the neural tube at stages 8–12 in order to selectively target neural tube and neural crest, EGFP was not observed in migrating crest cells, with the exception of one to two cells immediately external to the dorsal hindbrain levels in 2/15 embryos, perhaps reflecting perdurance of EGFP from neural tube cells. These results suggest that the regulation of FoxD gene expression in somites, notochord, and anterior neural tube is conserved between amphioxus and vertebrates, but that the expression in neural crest is a vertebrate innovation. We hypothesize that, after duplication of FoxD genes at the emergence of the vertebrate lineage, the FoxD3 paralog was co-opted to the neural plate border/neural crest via introduction of new regulatory elements.
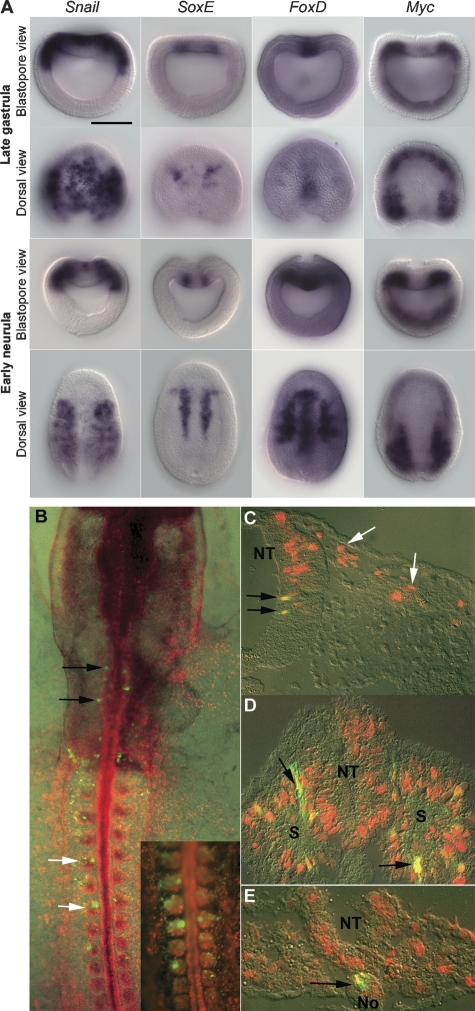
Figure 3.
Expression of amphioxus homologues of neural crest specifier genes and reporter gene expression mediated by amphioxus FoxD (AmphiFoxD) regulatory region in chick embryos. (A) Anterior is perpendicular to the plane of the page in the blastopore view and at the top in the dorsal view. Dorsal is up in the blastopore view. Scale bar, 50 μm. Amphioxus Snail is expressed in the entire prospective neural plate and paraxial dorsal mesoderm in late gastrula stage. During the early neurula stage, Snail is down-regulated in the neural plate and transiently expressed at the neural plate border. SoxE is expressed in the dorsal mesendoderm at the border of future axial and paraxial mesoderm. FoxD is expressed in the dorsal axial mesoderm, paraxial mesoderm, and the anterior neural plate. Amphioxus Myc is expressed in the dorsal paraxial mesoderm and ventral mesendoderm. (B–E) Reporter gene expression in mesoderm derivatives and hindbrain directed by AmphiFoxD in chick embryos. Rostral is to the top in B, and dorsal to the top in C–E. EGFP+ cells have yellow or orange nuclei due to coexpression of RFP. (B) Whole-mount image of stage 11 chick embryo expressing RFP in all electroporated cells and EGFP under the control of the AmphiFoxD regulatory region; EGFP+ cells are seen in somites (white arrows, shown at higher power in inset), paraxial mesoderm, and hindbrain (black arrows). (C–E) Cross-sections showing expression of EGFP in the hindbrain (C, black arrows), somites (D, arrows), and notochord (E, arrow). No expression of EGFP is seen in premigratory or migratory neural crest cells (C, white arrows). (NT) neural tube; (S) somite; (No) notochord.
The distal portion of the vertebrate NC-GRN contains effector genes encoding proteins involved in cell migration and differentiation. Among the identified homologs of neural crest effector genes, we found a group of genes that are involved in pigment cell development. These include genes encoding the transcription factor Mitf, along with melanin synthesizing enzymes tyrosinase and tyrosinase-related proteins. These have been implicated as a conserved differentiation gene battery for melanocyte development (Widlund and Fisher 2003; Martinez-Morales et al. 2004). Notably, these genes are coexpressed during amphioxus embryogenesis and eventually become localized in the rudiment of the primary pigment spot located in the amphioxus neural tube (Fig. 4A; Supplemental Fig. S3). This pigment spot becomes the first pigment cell of the amphioxus photoreceptor organs of Hesse (Conklin 1932; Lacalli et al. 1994; Lacalli 2002). Thus, this pigment cell lineage within the amphioxus neural tube appears to utilize the same transcription factor input and downstream differentiation gene battery as do neural crest-derived pigment cells and retinal pigment epithelial cells in vertebrates. Unlike neural crest cells, these cells never migrate out of the neural tube.
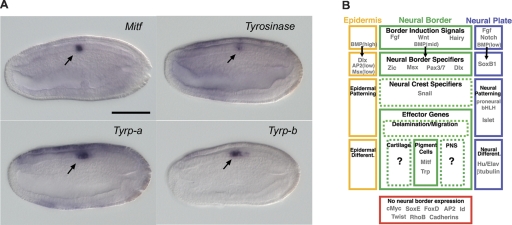
Figure 4.
Expression of amphioxus homologs of neural crest effector genes and summary for putative neural border gene network in amphioxus. (A) Expression of Mitf, tyrosinase, Tyrp-a, and Tyrp-b in late neurula stage embryos. Side views are shown here; anterior is at left and dorsal is up. Scale bar, 50 μm. All four of these genes are coexpressed in the neural tube at the level where the first primary pigment spot will form. (B) In amphioxus, gene expression and BMP signaling perturbation suggest that the processes of dorsal ectoderm patterning and neural border specification are conserved between vertebrates and amphioxus. In addition, the differentiation gene battery for pigment cell development appears conserved in both vertebrates and amphioxus. However, at intermediate levels of their respective neural border networks, the two groups diverge. Amphioxus lacks neural border expression of most neural crest specifiers, as well as the effector subcircuit controlling neural crest delamination and migration, consistent with a lack of bona fide neural crest cells in this lineage.
Recent study in lamprey has shown that the ground state of NC-GRN is already present in jawless vertebrates (Sauka-Spengler et al. 2007), suggesting that the emergence of a bona fide NC-GRN occurred during the transition from protochordates to vertebrates. Based on our examination of amphioxus homologs of NC-GRN genes, we propose that the initial step in assembling the vertebrate NC-GRN involved intercalation of genetic subnetworks for cell migration to precursor cells within the neural tube. We hypothesize that this intercalation was achieved by co-option of existing transcription factors to a new function as neural crest specifier genes into the network (Fig. 4B). This was inserted between the existing conserved neural plate border specification subnetwork and pigment cell differentiation gene battery. This hypothesis is consistent with data from vertebrate model systems demonstrating that several of the neural crest specifier genes are responsible for initiating cell migration by regulating expression of cell adhesion molecules (Dottori et al. 2001; Cheung et al. 2005). Furthermore, this idea meshes nicely with the discovery of migrating neural crest-like pigment cells in another invertebrate chordate lineage—the tunicates (Jeffery et al. 2004; Jeffery 2006), and the recent phylogenetic analysis placing tunicates as the sister group of vertebrates and amphioxus at the base of chordates. Taken together, these data suggest that migrating pigmented cells emerged after the split of amphioxus from tunicates and vertebrates, and this cell lineage might represent the first neural crest derivative in evolutionary history. In-depth examination of neural crest gene expression in urochordate pigment cells will help refine this scenario by establishing when neural crest specifiers were co-opted relative to the appearance of such cells. A recent comparative genomic analysis shows that many signaling receptor ligands associated with the cell lineage specification of various neural crest derivatives appear only in vertebrates (Martinez-Morales et al. 2007). These new signaling molecules and the expansion of neural crest-derived cell types might represent a special adaptation during evolution of the vertebrate lineage. Of particular interest is whether one can account for the evolutionary precursors of neural crest derivatives other than pigment cells in protochordates. Resolving this question will require comparative anatomy and developmental genetics in protochordates, and will shed light on the origin of neural crest and the vertebrate head.
Methods
Identification of vertebrate neural crest network genes in amphioxus
Vertebrate protein sequences for neural crest network genes were used as queries to TBLASTN search against the predicted gene models from the amphioxus draft genome (http://genome.jgi-psf.org/Brafl1/Brafl1.home.html). We retrieved the hit gene model sequences with E-value < 10−5 and used the peptide sequences deduced from the gene models to perform BLASTp searches against the International Protein Index proteome set released on May 2, 2006. When the best-hit sequence of the gene model corresponded to the starting vertebrate protein query, the gene model was assigned as the putative ortholog. Molecular phylogenetic analysis was used to confirm the orthology assignment of newly identified amphioxus genes (Supplemental Figs. S4–S13). Sequences of vertebrate neural crest network genes and other related sequences for molecular phylogenetic analysis were downloaded from the public database (http://www.ncbi.nlm.nih.gov/). The sequences were aligned using the Clustal X program, and the phylogenetic trees were calculated with MEGA program version 2.1 based on the Neighbor-joining method. Bootstrap support values were calculated by 1000 pseudoreplications. The sequences are designated by the accession number, gene name, and abbreviation of the species. For example, human FGF8 (accession no. P55075) is represented as “P55075 FGF8 Hs.” Species name abbreviations: Bf, Branchiostoma floridae; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hr, Halocynthia roretzi; Hs, Homo sapiens; Mm, Mus musculus; and Xl, Xenopus laevis. We subsequently searched in an amphioxus cDNA/EST database (Yu et al. 2007) to identify the corresponding cDNA cluster, and the longest clone from each cluster was picked for our gene expression survey.
In situ hybridization
We used a primer matching with the vector sequence adjacent to the 3′end of the cDNA insert with a T7 promoter sequence added to its 5′end as a reverse primer (pDONR222-T7-Reverse, 5′-TAA TACGACTCACTATAGGGAGGGGATATCAGCTGGATG-3′) and a primer matching with the vector sequence adjacent to the 5′end of the cDNA insert with a SP6 promoter sequence added as a forward primer (pDONR222-SP6-Forward, 5′-ATTTAGGTGAC ACTATAGAAGACGGCCAGTCTTAAGCTC-3′) to PCR amplify the full-length cDNA for antisense riboprobe synthesis. DIG-labeled antisense riboprobes were synthesized by T7 RNA polymerase, and the procedure of in situ hybridization for amphioxus embryos was performed as described (Yu et al. 2007). The expression pattern of genes was examined in embryos from mid-gastrula to late neurula stages (when the neural tube is formed) for all genes. Each of the developmental stages that we examined are ∼1.5 to 2 h apart under the culturing temperatures used in our study.
Culture of embryos with zBmp4 protein
Cultures of amphioxus were treated with recombinant zebrafish Bmp4 (zBmp4, R&D systems) at a concentration of 250 ng/mL. For control experiments, the same concentration of bovine serum albumin (BSA) was applied separately to the same stage of culture. Proteins were added at the early blastula stage (2.5 h post-fertilization) and embryos were fixed for in situ hybridization for marker genes at late gastrula and early neurula stages.
Electroporation of AmphiFoxD regulatory region into chick embryos
The 6-kb regulatory region of AmphiFoxD (Yu et al. 2004) was cloned into the SmaI site of the reporter construct ptkEGFP (kindly donated by H. Kondoh, Osaka University, Japan). pRFP-H2B (kindly donated by T. Sauka-Spengler, Caltech, USA) was used as a tracer to mark electroporated cells. For Stage 3+ to 4 electroporations, embryos were collected on a ring of filter paper, placed into Ringer’s solution, and gently washed to remove the yolk. The constructs (1 mg/mL each in Ringer’s) were injected between the blastoderm and vitelline membrane. Embryos were immediately electroporated with five pulses of 7V for a duration of 50 msec with 100 msec between pulses, with the anodal electrode on the hypoblast side of the embryo. Embryos were cultured overnight at 37°C in New culture on a thin layer of liquid albumen with the hypoblast side up. Chick embryos from stages 8–12 were electroporated in ovo by injecting the constructs (2 mg/mL each) into the lumen of the neural tube and immediately electroporating the embryo with five pulses of 22 V (30 msec per pulse with 100 msec between pulses). Embryos were resealed and incubated at 37°C for 24 to 44 h and analyzed for expression of EGFP and RFP. Embryos expressing RFP were processed and cryosectioned at 14 mm. Some sections were labeled using an antibody for HNK-1 (diluted 1/50), and secondarily detected using goat anti-mouse IgM 350 (1/200; Molecular Probes). Images were captured using a Zeiss Axioskop 2 plus microscope with AxioVision 4.6 software, and compiled using Adobe Photoshop 7.0.
Acknowledgments
We thank Linda Holland, Noriyuki Satoh, Yutaka Satou, and Yuji Kohara for the amphioxus EST resources, and the Joint Genome Institute for the amphioxus genome sequence resources. We also thank John Lawrence, Susan Bell, Ray Martinez Jr., and James Swigart at the University of South Florida for providing laboratory facilities during the summer breeding season of amphioxus. We thank Hisato Kondoh for donating the ptkEGFP vector and Tatjana Sauka-Spengler for providing the pRFP-H2B plasmid. This work was funded by a grant from National Institutes of Health DE017911 (M.B.-F). J.-K.Y was supported by the Della Martin prize postdoctoral fellowship from the Division of Biology, California Institute of Technology. S.J.M. is supported by an Australian Government NH&MRC CJ Martin fellowship.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076208.108.
References
- Bourlat S.J., Juliusdottir T., Lowe C.J., Freeman R., Aronowicz J., Kirschner M., Lander E.S., Thorndyke M., Nakano H., Kohn A.B., et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Cheung M., Chaboissier M.C., Mynett A., Hirst E., Schedl A., Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Conklin E.G. The embryology of amphioxus. J. Morphol. 1932;54:69–151. [Google Scholar]
- Davidson E.H., Erwin D.H. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Denes A.S., Jekely G., Steinmetz P.R., Raible F., Snyman H., Prud'homme B., Ferrier D.E., Balavoine G., Arendt D. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- De Robertis E.M., Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M., Gross M.K., Labosky P., Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Gans C., Northcutt R.G. Neural crest and the origin of vertebrates—A new head. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Gould R.M., Morrison H.G., Gilland E., Campbell R.K. Myelin tetraspan family proteins but no non-tetraspan family proteins are present in the ascidian (Ciona intestinalis) genome. Biol. Bull. 2005;209:49–66. doi: 10.2307/3593141. [DOI] [PubMed] [Google Scholar]
- Holland L.Z., Holland N.D. Evolution of neural crest and placodes: Amphioxus as a model for the ancestral vertebrate? J. Anat. 2001;199:85–98. doi: 10.1046/j.1469-7580.2001.19910085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery W.R. Ascidian neural crest-like cells: Phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J. Exp. Zool. B Mol. Dev. Evol. 2006;306B:470–480. doi: 10.1002/jez.b.21109. [DOI] [PubMed] [Google Scholar]
- Jeffery W.R., Strickler A.G., Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–699. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- Lacalli T.C. The dorsal compartment locomotory control system in amphioxus larvae. J. Morphol. 2002;252:227–237. doi: 10.1002/jmor.1101. [DOI] [PubMed] [Google Scholar]
- Lacalli T.C., Holland N.D., West J.E. Landmarks in the anterior central-nervous-system of amphioxus larvae. Philos. T. Roy. Soc. B. 1994;344:165–185. [Google Scholar]
- Langeland J.A., Tomsa J.M., Jackman W.R., Kimmel C.B. An amphioxus snail gene: Expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the chordate embryo. Dev. Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- Le Douarin N., Kalcheim C. The neural crest. Cambridge University Press; Cambridge, UK: 1999. [Google Scholar]
- Manzanares M., Wada H., Itasaki N., Trainor P.A., Krumlauf R., Holland P.W.H. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature. 2000;408:854–857. doi: 10.1038/35048570. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales J.R., Rodrigo I., Bovolenta P. Eye development: A view from the retina pigmented epithelium. Bioessays. 2004;26:766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales J.-R., Henrich T., Ramialison M., Wittbrodt J. New genes in the evolution of the neural crest differentiation program. Genome Biol. 2007;8:R36. doi: 10.1186/gb-2007-8-3-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Young R., Vargas A. Development of neural crest in Xenopus. Curr. Top. Dev. Biol. 1999;43:85–113. doi: 10.1016/s0070-2153(08)60379-8. [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev. Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. Central role of gene cooption in neural crest evolution. J. Exp. Zool. B Mol. Dev. Evol. 2005;304:298–303. doi: 10.1002/jez.b.21047. [DOI] [PubMed] [Google Scholar]
- Meulemans D., McCauley D., Bronner-Fraser M. Id expression in amphioxus and lamprey highlights the role of gene cooption during neural crest evolution. Dev. Biol. 2003;264:430–442. doi: 10.1016/j.ydbio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Mizutani C.M., Meyer N., Roelink H., Bier E. Threshold-dependent BMP-mediated repression: A model for a conserved mechanism that patterns the neuroectoderm. PLoS Biol. 2006;4:e313. doi: 10.1371/journal.pbio.0040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt R.G. The new head hypothesis revisited. J. Exp. Zool. B Mol. Dev. Evol. 2005;304B:274–297. doi: 10.1002/jez.b.21063. [DOI] [PubMed] [Google Scholar]
- Putnam N., Butts T., Ferrier D.E.K., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008 doi: 10.1038/nature06967. in press. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Meulemans D., Jones M., Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Shimeld S.M., Holland P.W.H. Vertebrate innovations. Proc. Natl. Acad. Sci. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B., Carmona-Fontaine C., Mayor R. Genetic network during neural crest induction: From cell specification to cell survival. Semin. Cell Dev. Biol. 2005;16:647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Wehrle-Haller B., Meller M., Weston J.A. Analysis of melanocyte precursors in Nf1 mutants reveals that MGF/KIT signaling promotes directed cell migration independent of its function in cell survival. Dev. Biol. 2001;232:471–483. doi: 10.1006/dbio.2001.0167. [DOI] [PubMed] [Google Scholar]
- Widlund H.R., Fisher D.E. Microphthalamia-associated transcription factor: A critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- Yasui K., Zhang S.C., Uemura M., Aizawa S., Ueki T. Expression of a twist-related gene, Bbtwist, during the development of a lancelet species and its relation to cephalochordate anterior structures. Dev. Biol. 1998;195:49–59. doi: 10.1006/dbio.1997.8834. [DOI] [PubMed] [Google Scholar]
- Yu J.K., Holland N.D., Holland L.Z. An amphioxus winged helix/forkhead gene, AmphiFoxD: Insights into vertebrate neural crest evolution. Dev. Dyn. 2002;225:289–297. doi: 10.1002/dvdy.10173. [DOI] [PubMed] [Google Scholar]
- Yu J.K., Holland N.D., Holland L.Z. Tissue-specific expression of FoxD reporter constructs in amphioxus embryos. Dev. Biol. 2004;274:452–461. doi: 10.1016/j.ydbio.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Yu J.K., Satou Y., Holland N.D., Shin I.T., Kohara Y., Satoh N., Bronner-Fraser M., Holland L.Z. Axial patterning in cephalochordates and the evolution of the organizer. Nature. 2007;445:613–617. doi: 10.1038/nature05472. [DOI] [PubMed] [Google Scholar]