Abstract
Comparative transcriptomics studies in hominoids are difficult because of lack of EST information in the great apes. Nevertheless, processed pseudogenes (PPGs), which are reverse-transcribed ancient transcripts present in the current genome, can be regarded as a virtual transcript resource that may compensate for the paucity of ESTs in non-human hominoids. Here we show that chimpanzee PPGs can be applied to identification of novel human exons/alternatively spliced variants (ASVs) and inference of the ancestral hominoid transcriptome and chimpanzee exon loss events. We develop a method for comparatively extracting novel transcripts from PPGs (designated “CENTP”) and identify 643 novel human exons/ASVs. RT-PCR-sequencing experiments confirmed >50% of the tested exons/ASVs, supporting the effectiveness of the CENTP pipeline. With reference to the ancestral transcriptome inferred by CENTP, 47 chimpanzee exon loss events are identified. Furthermore, by combining out-group and PPG information, we identify 20 chimpanzee-specific exon loss and 10 human-specific exon gain events. We also demonstrate that the ancestral transcriptome and exon loss/gain events inferred based on comparisons of current transcripts may be incomplete (or occasionally inappropriate) because ancestral transcripts may not be represented in the ESTs of existing species. Finally, functional analysis reveals that the novel exons identified based on chimpanzee transcripts are significantly enriched in genes related to translation regulatory activity and viral life cycle, suggesting different expression levels of the associated transcripts, and thus divergent splicing isoform composition between human and chimpanzee in these functional categories.
The complexity of a transcriptome is directly related to the proteome size and functional versatility of organisms (Graveley 2001; Maniatis and Tasic 2002; Black and Grabowski 2003; Bracco and Kearsey 2003). In humans, 40%–70% of genes have more than one transcript (Brett et al. 2000; Kan et al. 2001; Modrek et al. 2001; Johnson et al. 2003), and different transcript isoforms of the same genes can serve very different or even opposite functions (Bracco and Kearsey 2003; Akey et al. 2004; Li and Manley 2006; Scotlandi et al. 2007). A considerable number of disease-state-specific transcripts have also been identified (Cooper and Mattox 1997; Caceres and Kornblihtt 2002; Faustino and Cooper 2003; Musunuru 2003; Garcia-Blanco et al. 2004; Buratti et al. 2006). The correlation between transcript variants and cancers particularly comes into widespread notice (Venables 2004). Transcript isoforms (and the resulting protein isoforms) may differ in functional domains (Goodman et al. 2003), post-translational modifications (Duma et al. 2006; Lim and Cao 2006), and protein–protein interactions (Lee et al. 2007), which, in turn, affect a wide range of biological functions. Considering the functional importance of transcriptome diversity, it is of great interest to investigate transcriptome evolution (Boue et al. 2003).
For genetically close but phenotypically divergent species, such as human and the common chimpanzee (Pan troglodytes), transcriptome evolution is considered relevant to interspecies functional divergence. However, comparative studies of transcriptomes in hominoids have been hampered by the paucity of expressed sequence tag (EST) information and experimentally validated transcripts in the great apes. Recently, Shemesh et al. (2006) have suggested that processed pseudogenes (PPGs) can be regarded as a “virtual cDNA library” when ESTs are unavailable. PPGs are intronless “dead-on-arrival” genes that derive from reverse transcription and subsequent re-insertion and pseudogenization of spliced mRNA transcripts (Vanin 1985; Carlton et al. 1995; Esnault et al. 2000; Goncalves et al. 2000; Graur and Li 2000; Mighell et al. 2000). They represent the mature forms of transcripts that were present in the ancestors of currently living organisms. In other words, PPGs are “genomic fossils” that may record the expressions of ancestral genes (Shemesh et al. 2006). Therefore, PPGs are a good alternative resource in identifying new exons and studying hominoid transcriptome evolution.
Meanwhile, it has been demonstrated that cross-species EST-to-genome comparisons are suitable for identification of uncharacterized exons/alternatively spliced variants (ASVs) (Chuang et al. 2004; Kan et al. 2004; Chen and Chuang 2005; Chen et al. 2006, 2007d) and exploration of transcriptome evolution (Chen et al. 2006, 2007d). The power of novel exon/ASV detection of this comparative approach is negatively related to the genetic distances between the compared species (Chen et al. 2006). Since the chimpanzee is human’s closest relative in nature, chimpanzee ESTs are very suitable for detecting novel human exons or ASVs, and vice versa. However, considering the limited availability of chimpanzee ESTs, we propose that chimpanzee PPGs can serve as a surrogate of full-length transcripts in this regard. Chimpanzee PPGs have another advantage in human–chimpanzee comparative studies. Since the alignable sequences between the human and chimpanzee genomes are almost 99% identical (Chimpanzee Sequencing and Analysis Consortium 2005), genomic sequence conservation is ubiquitous, and very little information is provided for distinguishing coding regions from non-coding regions (Nekrutenko et al. 2002). Comparative analyses between chimpanzee PPGs and the human genome can surmount this obstacle and enable us to extract functional features from the conserved information between these two genomes.
In this study, we develop a method for comparatively extracting novel transcripts from PPGs (designated “CENTP”). By cross-species PPG-to-genome mapping, we cannot only detect unannotated human exons/ASVs, but also infer the transcriptome in the Homo–Pan common ancestor. With reference to the ancestral transcriptome, we can identify chimpanzee exon loss events without having to reference out-group information. In addition, we demonstrate that inference of exon loss events based on comparisons with out-group sequences may be inappropriate if PPGs are not considered. Finally, we functionally analyze the ASVs that are lost in chimpanzee, and briefly discuss the possible impacts of these events in Homo–Pan functional divergence.
Results and Discussion
More than 600 novel human exons/ASVs are identified by CENTP
Table 1 lists the 643 CENTP-identified novel human exons (named “CENTP exons”) that are absent in current annotation databases or EST libraries (see Fig. 1 and Methods). These novel exons also represent novel human ASVs because no transcripts that include the CENTP exons have been characterized. For simplicity, we term CENTP exons identified based on human PPGs, chimpanzee PPGs, and chimpanzee transcripts (collectively called “CENTP cDNAs”) as CENTPH_PPG, CENTPC_PPG, and CENTPC_gene exons, respectively. As expected, the number of CENTPH_PPG exons (121) is much larger than that of CENTPC_PPG exons (29). This is understandable because the number of the extracted human PPGs is larger than that of chimpanzee PPGs, and human PPGs may have preserved more human expression information. As well, CENTP identifies a much larger number of human PPG-based novel exons as compared with a recent study that applied human PPGs to ASV detection (Shemesh et al. 2006) (see Supplemental material for details).
Table 1.
Novel cassette-on exons and retained introns identified by CENTP
Figure 1.
Flowchart of the CENTP pipeline.
Notably, a large number of potentially novel human exons (469) are inferred from chimpanzee transcripts, lending solid support for the power of novel exon/ASV detection based on cross-species EST-to-genome comparisons. Previously, we have demonstrated that the power of such a comparative approach is negatively related to the interspecies divergence level (Chen et al. 2006). Considering the small genetic distance between human and chimpanzee, the number of CENTP-identified novel human exons can grow rapidly with the increase of available EST data from chimpanzee and other primate species.
In terms of ASV types, CENTP totally identifies 434 cassette-on exons (Fig. 2A) and 209 retained introns (Fig. 2B). Note that two types of cassette-on exons are identified here: simple and complex (see Fig. 2A and Methods). Of these exons, 387 are located in coding sequences (CDSs). The remaining 256 are located in untranslated regions (UTRs). It is worth noting that the majority of CENTP cassette-on exons are located in CDSs rather than in UTRs. This is because UTRs in most cases are the initial/terminal exons in the transcripts in which they reside, and such exons cannot pass the CENTP filters (for accuracy, CENTP only identifies novel exons located between two well-known exons; see Methods). On the other hand, more CENTP retained introns are located in UTRs than in CDSs, which is consistent with Galante et al.’s report (Galante et al. 2004). This bias may be due to nonsense-mediated decay, which triggers the degradation of transcripts that include premature stop codons, allowing less time for the last intron to be spliced out (Black 2003; Galante et al. 2004). Another reason is that UTR events do not need to satisfy our rule that the identified exons must neither disrupt the reading frame nor interrupt the coding protein, whereas CDS events do (see Methods).
Figure 2.
Examples of simple and complex cassette-on exons (A) and retained introns (B). Simple cassette-on exons (Case 1) do not alter the boundaries of their flanking exons when they are included in transcripts, while complex ones (Case 2) do.
To validate the CENTP-identified exons/ASVs, three subsets from CENTPC_PPG, CENTPH_PPG, and CENTPC_gene (21, 29, and 28 events, respectively) are selected for RT-PCR-sequencing verification (Supplemental Table 1). More than 50% (40/78) of the tested exons are experimentally confirmed. These include simple/complex cassette-on exons and retained introns. The RT-PCR results of the confirmed exons are given in Supplemental Figures 1–3. Our results indicate that a considerable proportion of the ASVs identified based on human/chimpanzee PPGs or chimpanzee transcripts are still active in the human transcriptome.
Ancestral hominoid transcriptomes and chimpanzee exon loss events inferred from PPGs
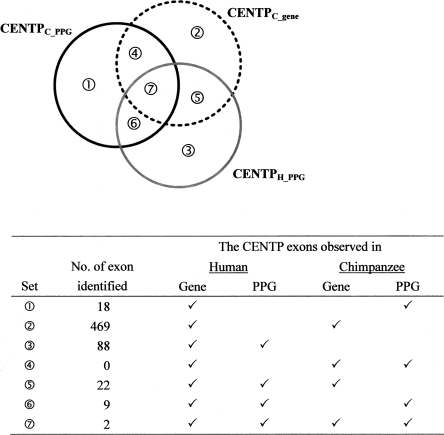
Figure 3 shows the Venn diagram of the numbers of CENTPH_PPG, CENTPC_PPG, and CENTPC_gene exons, which are supported by different CENTP cDNAs and may have different evolutionary implications. Among these 643 potentially novel human exons, only 33 (∼5%) are supported by at least two CENTP cDNA resources (i.e., Sets 4–7), and only two (<0.5%) are supported by all three resources (i.e., Set 7). Figure 3 also shows that Set 2 exons (i.e., exons supported only by chimpanzee genes) account for the majority (73%) of the collective total of CENTP exons, whereas Set 1 (supported only by chimpanzee PPGs) and Set 3 (supported only by human PPGs) exons account for 3% and 14%, respectively. By definition, the CENTPC_GENE exons (the dotted circle) and CENTPC_PPG exons (the black circle) might have existed in the Homo–Pan common ancestral transcriptome. Therefore, if the exons are supported by chimpanzee PPGs but not by chimpanzee transcripts (i.e., Sets 1 and 6 exons), we may infer that these exons (27 exons) represent chimpanzee exon loss events without having to reference out-group information.
Figure 3.
Venn diagram of the CENTP exons inferred from chimpanzee PPGs, human PPGs, and chimpanzee genes.
We then examine these 27 exons using the CENTP pipeline to determine whether these exons are really lost or simply unannotated in chimpanzee. We align the chimpanzee PPGs that include these exons against the introns of their parent genes and examine the matches using the CENTP exon-checking rules stated in Methods. If novel exons are identified, they are considered as currently unannotated chimpanzee exons and being included in the active transcripts of both human and chimpanzee. Otherwise, exon loss events are thought to have occurred in the chimpanzee lineage. Two types of exon loss events are expected: exon deletion and pseudogenization. In the former case, the PPG-derived exons will be non-alignable against the introns of their parent genes. In the latter case, the exons should be conserved in the introns of the parent genes but have incurred frameshift or nonsense mutations, or loss of splicing signals. In fact (also see Table 2), both cases are observed for Set 1 and Set 6 exons. We therefore identify 16 chimpanzee exon loss events, five potentially novel chimpanzee exons (newly annotated by CENTP), and six exons of uncertain status for lack of information. For the chimpanzee exon loss events, we further estimate the time of pseudogenization by calculating the genetic distances between PPGs that support these 16 exons and their parent genes. All except one of the PPG–parent gene pairs have distances much larger than 2.6% (Supplemental Table 2), which is the largest background human–chimpanzee sequence divergence (in 1-Mb windows across the autosomes; ranging from ∼0.4 to ∼2.6%) (Chimpanzee Sequencing and Analysis Consortium 2005). The only exception occurs in Set 1, where one gene pair has a genetic distance of 2.7%. Therefore, our results indicate that most (probably all) of the PPGs that support these 16 CENTP exons were present in the Homo–Pan common ancestor. Interestingly, some of the PPGs are, in fact, very ancient, having genetic distances as large as >20% from their parent genes. Such large distances imply a long evolutionary history that can be traced back further than the common ancestor of current primates.
Table 2.
Classification of absent-in-chimpanzee exons from Sets 1, 3, and 6 in Figure 3
aThe parent genes of chimpanzee PPGs are not found in the Ensembl annotation.
bIncluding five chimpanzee-specific exon loss events.
cSeven chimpanzee-specific exon loss events are inferred based on the macaque/mouse genomes. The other 46 exons are not found (or identified) in the macaque/mouse genomes.
dThe 26 exons include four very short exons (<12 bp) and 22 exons of which orthologous chimpanzee genes are not found in the Ensemble annotation.
To investigate whether these exon loss events are actually specific to chimpanzee, we retrieved the macaque/mouse orthologous genes and examined whether these 16 exons were present in these genes. In fact, 13 exons are well-annotated or can be identified by CENTP in the macaque or mouse genome (Supplemental Table 2), implying that they represent chimpanzee-specific exon loss events. Although the lineage-specificity of the other three exons remains unclear, current PPG evidence appears to suggest that they represent chimpanzee exon loss events (rather than human gain of exons). This example demonstrates the usability of PPGs as an indicator to distinguish between exon losses and gains when out-group information is unavailable.
Meanwhile, Set 3 exons (88 exons) are observed in neither Ensembl-annotated chimpanzee transcripts nor chimpanzee PPGs (Fig. 3). Again, these exons may be either lost or not yet annotated in chimpanzee. Among the 88 exons, 22 have no chimpanzee orthologs, and four are too short (<12 bp) for BLAST alignments. We examined the remaining 62 exons using the CENTP pipeline and identified nine potentially novel chimpanzee exons. With reference to the macaque/mouse orthologous genes, seven of the remaining 53 exons are found to result from chimpanzee-specific exon loss events (Table 2). Meanwhile, the other 46 exons, which are not found in the macaque/mouse genomes, may represent human exon gain events. However, considering the incompleteness of the macaque genomic sequences and annotations, more evidence is required before any conclusions can be drawn. We therefore calculated the genetic distances between PPGs that support the 46 exons and their parent genes. We find that 12 PPG–parent gene pairs (covering 15 exons) have distances <2.6% (Supplemental Table 3). Ten out of the 12 gene pairs (covering 10 exons) have distances even ≤0.7%, which is much smaller than the genome-wide human–chimpanzee nucleotide divergence (1.23%) (Chimpanzee Sequencing and Analysis Consortium 2005). The result indicates that these pseudogenization events may have occurred after the Homo–Pan divergence and these 10 exons very likely represent human-specific exon gain events. On the other hand, the other 31 (46 minus 15) exons are probably chimpanzee exon loss events (with PPG–parent gene distances ≥2.6%), for the pseudogenization of the supporting PPGs obviously predate the Homo–Pan divergence (Supplemental Table 3). This result demonstrates that the inadequateness of out-group information in discrimination between exon gain and loss events can be compensated by PPGs.
Overall, 69 potential absent-in-chimpanzee exons are identified from Sets 1, 3, and 6, of which 20 and 10 represent possible chimpanzee-specific exon loss and human-specific exon gain events, respectively. Note that 47 (16 plus 31) chimpanzee exon loss events are identified without referring to out-group information.
The implications of PPGs in comparative and evolutionary studies
Figure 4, A and B, respectively, illustrate possible evolutionary scenarios of Set 1 and Set 6 CENTP exons, both of which are supported by chimpanzee PPGs but not by chimpanzee genes. Since PPGs must have been expressed at the time of pseudogenization, they represent part of the ancient transcriptome. Therefore, we suggest that the human ASVs supported by chimpanzee PPGs were present in the common ancestor of human and chimpanzee (Fig. 4), regardless of whether such ASVs are observed in chimpanzee functional genes or not. Meanwhile, the PPGs that lack the CENTP exons (ASV2) are either present or absent in the chimpanzee genome. (Note that in either case, a chimpanzee exon loss event is thought to have occurred.) If the ASV2 PPG does exist, three scenarios are possible. Firstly, ASV2 was present in the Homo–Pan common ancestral transcriptome, and it resulted in this PPG. Secondly, chimpanzee had lost the CENTP exon after Homo–Pan divergence, and subsequently this lost-exon ASV2 formed a PPG. Thirdly, a PPG had included the CENTP exon (i.e., it had resulted from ASV1) but somehow lost it because of random sequence losses and/or nucleotide substitutions. Anyhow, this example demonstrates that PPGs can bring a new vista for inferences of ancestral transcriptomes. If PPGs are not considered, interpretations of transcriptome evolution may sometimes be incorrect.
Figure 4.
Possible evolutionary scenarios of CENTP exons supported by chimpanzee PPGs but not by chimpanzee genes. In both scenarios, a chimpanzee exon loss event is inferred. (A) The corresponding human PPG is absent (i.e., Set 1 exons); (B) the corresponding human PPG is present (i.e., Set 6 exons).
In another case (see Fig. 5) when both the active transcript and PPG of ASV 1 are absent in chimpanzee, one may consider ASV 2 as the Homo–Pan ancestral form. In this case, out-group species (e.g., macaque) information may be used to distinguish between chimpanzee exon loss and human exon gain events. If the active transcript of ASV 1 is observed in macaque, most likely an exon loss event has occurred in chimpanzee and ASV 1 was present in the human–chimpanzee–macaque common ancestor. On the other hand, if ASV 2 rather than ASV 1 is active in macaque, one may speculate that ASV 2 was the ancestral form of these three primates, and an exon gain event has occurred in human. Nevertheless, if a macaque ASV 1 PPG is found, a second scenario is also likely, that an exon loss event has occurred in both chimpanzee and macaque, and ASV 1 represents the human–chimpanzee–macaque ancestral form.
Figure 5.
An example that shows how out-group species PPGs can help distinguish between a chimpanzee exon loss event and a human exon gain event.
In sum, these examples illustrate that PPGs may significantly affect our inference of the ancestral state of transcriptome. PPGs are therefore a valuable resource in view of evolutionary transcriptomics studies.
Functional influences of transcriptome evolution
By performing the Gene Ontology (Gene Ontology Consortium 2001) analyses, we find that the over-representation of transcripts that contain both CENTPC_gene exons and absent-in-chimpanzee exons occurs in the same functional categories: translation regulation and vial life cycle (see Supplemental Fig. 4 and Supplemental Table 4, respectively). There are several possible implications for the enrichment (which are not mutually exclusive). Firstly, since the relative abundance of PPGs is directly related to the expression levels of their parent genes, it is likely that such enrichment actually results from the high expression levels of the parent genes in these functional categories. Secondly, the enrichment may indicate remarkable disparity in ASV compositions in the related functional categories and possibly functional divergence between human and chimpanzee. Thirdly, a considerable number of transcripts in these two categories have not been discovered in human. Since the human transcriptome has been extensively studied, the newly identified human ASVs very likely are expressed in a restricted pattern (in terms of tissue specificity, developmental stage, or expression level). Meanwhile, the currently available chimpanzee ESTs are most likely highly expressed, implying that inclusion of these CENTP exons may be associated with the human–chimpanzee divergence in expression regulation of genes involved in these two categories. Interestingly, these categories are also enriched with human-specific insertions/deletions (Chen et al. 2007b, c). Whether such co-occurrence of different types of genetic changes in the same functional categories is merely accidental or evolutionarily meaningful awaits further investigation.
Methods
The CENTP pipeline
CENTP makes use of the “CENTP cDNAs,” including chimpanzee PPGs, human PPGs, and chimpanzee genes, to identify potentially novel human exonic sequences. As shown in Figure 1, we first retrieved 6932 chimpanzee PPGs (5904 from Yale [Zhang et al. 2003, 2006; Karro et al. 2007] and 1028 from Ensembl), 8242 human PPGs (7069 from Yale and 1173 from Ensembl), and 33,880 chimpanzee transcripts (from Ensembl), respectively. These CENTP cDNAs were then BLAST-aligned to the human genic regions (including exons and introns) annotated by UCSC, Ensembl, and NCBI. By doing so, CENTP cDNAs conserved in human well-annotated genic regions were identified. Using the SIM4 package (Florea et al. 1998), we further identified CENTP cDNA segments conserved in human introns (the “meta-CENTP exons”), which might also be potentially novel human AS events, including cassette-on exons (Fig. 2A) and retained introns (Fig. 2B). Note that the human introns used here were “pure introns,” which did not overlap with any well-annotated transcripts.
Subsequently, four exon-checking filters were used to eliminate potential false positives (Fig. 1). The meta-CENTP exons that passed all of these four rules were regarded as novel human exons (termed “CENTP exons”): Rule 1, For each identified exon, both of its flanking exonic regions must overlap with a well-annotated human transcript to avoid accidental matches; Rule 2, the identified cassette-on exons must be flanked by legal splicing sites (i.e., GT-AG/GC-AG); Rule 3, the identified exons that were located in CDSs must not disrupt the reading frame or contain any premature stop codons; Rule 4, the meta-CENTP exons that overlapped with human ESTs (GenBank UniGene) were discarded to ensure the novelty of the CENTP exons. Through these filtering processes, the majority of meta-CENTP exons were removed (from >4 million to 643 exons). Some of the CENTP exons were examined for validity using RT-PCR-sequencing (see Supplemental material for details).
In addition, CENTP can identify simple and complex cassette-on exons (Fig. 2). These two exon types are defined in the European Bioinformatics Institute Alternative Splicing Database (EBI-ASD) (Stamm et al. 2006). Identification of complex cassette-on exons is a unique feature of the CENTP system. In the case of simple cassette-on exons, if the addition of the potentially novel exons caused reading frame disruption or premature stop codons, they were discarded by CENTP. However, it was sometimes possible to identify complex cassette-on exons, whereby the boundaries of one or two of their flanking exons were slightly shifted to restore the reading frames that were otherwise disrupted by their insertion. To identify a complex cassette-on exon, we sought canonical splicing sites within a 20-bp window at boundaries of a meta-CENTP exon and its two flanking exons. The splicing sites that enabled the identified transcript to pass the CENTP exon-checking rules stated above and generate the longest transcript were then chosen. Note that, in the CENTP process, if a meta-CENTP exon had been identified as a simple cassette-on exon, it was not further examined for consideration of a complex exon, for simple exons constitute the majority of cassette exons (>80%) (Chen and Chuang 2007; Chen et al. 2007a). For accuracy, we only identified complex cassette-on exons that were located in CDSs.
Computation of substitution rates
The substitution rates between human/chimpanzee PPGs and their parent genes were calculated using the TN93 model implemented in the Baseml program of the PAML package (Yang 1997; Yang and Nielsen 2000).
Data retrieval and availability
The human/chimpanzee PPGs were downloaded from the Yale Pseudogene Database (http://www.pseudogene.org) and Ensembl (http://www.ensembl.org). The chimpanzee, macaque, and mouse annotated genes/transcripts were downloaded from Ensemble (release 45). The human annotated genes/transcripts were downloaded from the Ensemble genome browser, the UCSC genome browser (http://hgdownload.cse.ucsc.edu/downloads.html), and the NCBI RefSeq database (ftp://ftp.ncbi.nih.gov/refseq/). The original human, chimpanzee, macaque, and mouse genomic data are versions hg18 (or NCBI Build 36.1), panTro2 (or NCBI Build 2), rheMac2 (or NCBI Build 1), and mm8 (or NCBI Build 36), respectively. These genomic sequences and the human EST-to-genome alignments were all downloaded from the UCSC genome browser. The CENTP-identified exons/ASVs are available at http://www.sinica.edu.tw/~trees/CENTP/CENTP.html.
Acknowledgments
We thank Wen-Hsiung Li for experimental assistance. This work was supported by the Genomics Research Center, Academia Sinica, Taiwan (T.J.C.); the National Health Research Institutes (NHRI), Taiwan (under contract NHRI-EX97-9408PC) (T.J.C.); the National Science Council, Taiwan (under contract NSC 96-2628-B-001-005-MY3) (T.J.C.); and NHRI intramural funding (F.C.C.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075556.107.
References
- Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Black D.L., Grabowski P.J. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- Boue S., Letunic I., Bork P. Alternative splicing and evolution. BioEssays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kearsey J. The relevance of alternative RNA splicing to pharmacogenomics. Trends Biotechnol. 2003;21:346–353. doi: 10.1016/S0167-7799(03)00146-X. [DOI] [PubMed] [Google Scholar]
- Brett D., Hanke J., Lehmann G., Haase S., Delbruck S., Krueger S., Reich J., Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- Buratti E., Baralle M., Baralle F.E. Defective splicing, disease and therapy: Searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Kornblihtt A.R. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Carlton M.B., Colledge W.H., Evans M.J. Generation of a pseudogene during retroviral infection. Mamm. Genome. 1995;6:90–95. doi: 10.1007/BF00303250. [DOI] [PubMed] [Google Scholar]
- Chen F.C., Chuang T.J. ESTviewer: A web interface for visualizing mouse, rat, cattle, pig and chicken conserved ESTs in human genes and human alternatively spliced variants. Bioinformatics. 2005;21:2510–2513. doi: 10.1093/bioinformatics/bti332. [DOI] [PubMed] [Google Scholar]
- Chen F.C., Chuang T.J. Different alternative splicing patterns are subject to opposite selection pressure for protein reading frame preservation. BMC Evol. Biol. 2007;7:179. doi: 10.1186/1471-2148-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Chen C.J., Ho J.Y., Chuang T.J. Identification and evolutionary analysis of novel exons and alternative splicing events using cross-species EST-to-genome comparisons in human, mouse and rat. BMC Bioinformatics. 2006;7:136. doi: 10.1186/1471-2105-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Chaw S.M., Tzeng Y.H., Wang S.S., Chuang T.J. Opposite evolutionary effects between different alternative splicing patterns. Mol. Biol. Evol. 2007a;24:1443–1446. doi: 10.1093/molbev/msm072. [DOI] [PubMed] [Google Scholar]
- Chen F.C., Chen C.J., Chuang T.J. INDELSCAN: A web server for comparative identification of species-specific and non-species-specific insertion/deletion events. Nucleic Acids Res. 2007b;35:W633–W638. doi: 10.1093/nar/gkm350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Chen C.J., Li W.H., Chuang T.J. Human-specific insertions and deletions inferred from mammalian genome sequences. Genome Res. 2007c;17:16–22. doi: 10.1101/gr.5429606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Wang S.S., Chaw S.M., Huang Y.T., Chuang T.J. Plant gene and alternatively spliced variant annotator. A plant genome annotation pipeline for rice gene and alternatively spliced variant identification with cross-species expressed sequence tag conservation from seven plant species. Plant Physiol. 2007d;143:1086–1095. doi: 10.1104/pp.106.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chuang T.J., Chen F.C., Chou M.Y. A comparative method for identification of gene structures and alternatively spliced variants. Bioinformatics. 2004;20:3064–3079. doi: 10.1093/bioinformatics/bth368. [DOI] [PubMed] [Google Scholar]
- Cooper T.A., Mattox W. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma D., Jewell C.M., Cidlowski J.A. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J. Steroid Biochem. Mol. Biol. 2006;102:11–21. doi: 10.1016/j.jsbmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante P.A., Sakabe N.J., Kirschbaum-Slager N., de Souza S.J. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves I., Duret L., Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S.J., Branda C.S., Robinson M.K., Burdine R.D., Stern M.J. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130:3757–3766. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
- Graur D., Li W.-H. Fundamentals of molecular evolution. Sinauer Associates; Sunderland, MA: 2000. [Google Scholar]
- Graveley B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kan Z., Rouchka E.C., Gish W.R., States D.J. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z., Castle J., Johnson J.M., Tsinoremas N.F. Detection of novel splice forms in human and mouse using cross-species approach. Pac. Symp. Biocomput. 2004;2004:42–53. doi: 10.1142/9789812704856_0005. [DOI] [PubMed] [Google Scholar]
- Karro J.E., Yan Y., Zheng D., Zhang Z., Carriero N., Cayting P., Harrrison P., Gerstein M. Pseudogene.org: A comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007;35:D55–D60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Kwak H.Y., Hur J., Kim I.A., Yang J.S., Park M.W., Yu J., Jeong S. Beta-catenin regulates multiple steps of RNA metabolism as revealed by the RNA aptamer in colon cancer cells. Cancer Res. 2007;67:9315–9321. doi: 10.1158/0008-5472.CAN-07-1128. [DOI] [PubMed] [Google Scholar]
- Li X., Manley J.L. Alternative splicing and control of apoptotic DNA fragmentation. Cell Cycle. 2006;5:1286–1288. doi: 10.4161/cc.5.12.2832. [DOI] [PubMed] [Google Scholar]
- Lim C.P., Cao X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Mighell A.J., Smith N.R., Robinson P.A., Markham A.F. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Modrek B., Resch A., Grasso C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K. Cell-specific RNA-binding proteins in human disease. Trends Cardiovasc. Med. 2003;13:188–195. doi: 10.1016/s1050-1738(03)00075-6. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A., Makova K.D., Li W.H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002;12:198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotlandi K., Zuntini M., Manara M.C., Sciandra M., Rocchi A., Benini S., Nicoletti G., Bernard G., Nanni P., Lollini P.L., et al. CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. Oncogene. 2007;26:6604–6618. doi: 10.1038/sj.onc.1210481. [DOI] [PubMed] [Google Scholar]
- Shemesh R., Novik A., Edelheit S., Sorek R. Genomic fossils as a snapshot of the human transcriptome. Proc. Natl. Acad. Sci. 2006;103:1364–1369. doi: 10.1073/pnas.0509330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: A bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin E.F. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carriero N., Zheng D., Karro J., Harrison P.M., Gerstein M. PseudoPipe: An automated pseudogene identification pipeline. Bioinformatics. 2006;22:1437–1439. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]