Abstract
We describe construction of a novel modification, “6C,” of chromatin looping assays that allows specific proteins that may mediate long-range chromatin interactions to be defined. This approach combines the standard looping approaches previously defined with an immunoprecipitation step to investigate involvement of the specific protein. The efficacy of this approach is demonstrated by using a Polycomb group (PcG) protein, Enhancer of Zeste (EZH2), as an example of how our assay might be used. EZH2, as a protein of the PcG complex, PRC2, has an important role in the propagation of epigenetic memory through deposition of the repressive mark, histone H3, lysine 27, tri-methylation (H3K27me3). Using our new 6C assay, we show how EZH2 is a direct mediator of long-range intra- and interchromosomal interactions that can regulate transcriptional down-regulation of multiple genes by facilitating physical proximities between distant chromatin regions, thus targeting sites within to PcG machinery.
The past few years have seen an exciting development in technologies to address long-range chromatin interactions in vivo (3C, 4C, 5C, 3C-chip, ACT, open-ended 3C to name a few) (Dekker et al. 2002; Dostie et al. 2006; Ling et al. 2006; Lomvardas et al. 2006; Simonis et al. 2006; Wurtele and Chartrand 2006; Zhao et al. 2006). These techniques have revolutionized our concepts about key aspects of transcriptional regulation. However, all of these methodologies, to date, investigate physical proximities between chromatin elements without specifically identifying the protein components that may bridge them. Separate assays must then be carried out in order to address the potential role of specific regulatory factor(s) in mediating such long-range chromatin interactions. Moreover, when it comes to knowing whether a specific protein has the property to mediate physical pairing between distant chromatin elements, there have not yet been tools to specifically monitor this question.
In the present work, we combine multiple techniques to construct a novel 6C (combined 3C-ChIP-cloning) assay to address the question of long-range chromatin interactions mediated by specific proteins. To demonstrate how this methodology can work, we focus the assay on the protein Enhancer of Zeste (EZH2). This protein is a key member of the Polycomb protein complex (PcG) that regulates long-term gene silencing in multiple organisms. PcG proteins play crucial roles during early embryonic development (Sparmann and van Lohuizen 2006; Ringrose 2007) and are important for the self-renewal and pluripotency of embryonic stem cells (Bernstein et al. 2006; Boyer et al. 2006; Sparmann and van Lohuizen 2006). Among the PcG proteins, EZH2 is particularly important since it contains the histone methyltransferase (HMTase) activity that catalyzes trimethylation of histone H3 at Lys 27 (H3-K27) (Cao et al. 2002; Cao and Zhang 2004). This mark is required for attracting key components of PcG-mediated gene repression, and this activity is shown to require two other components of PcG machinery, SUZ12 and EED (Cao et al. 2002; Muller et al. 2002; Kirmizis et al. 2004; Pasini et al. 2007). Using our new assay, we now show that EZH2, and/or its H3K27me3 catalyzed mark, can play an important role in mediating long-range chromosomal interactions, in cis and trans, that modulate transcription of genes encompassed in the involved connected regions.
Results
Construction of the 6C assay to study whether EZH2 mediates inter- and intrachromosomal interactions
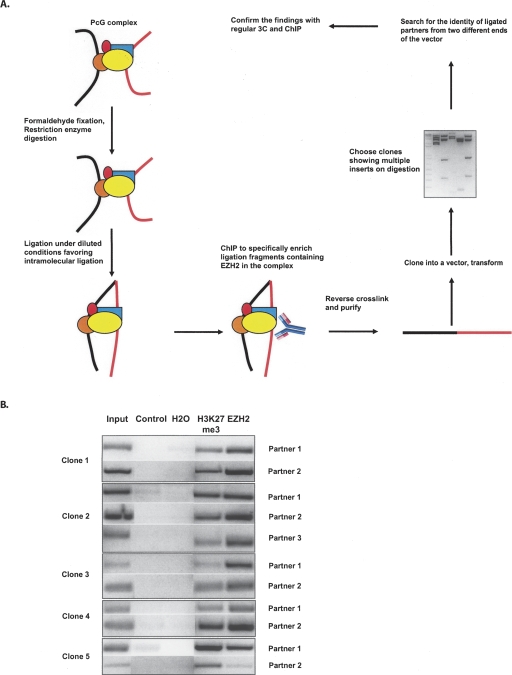
In order to address possible long-range interactions mediated by, or in which EZH2 participates, we combined three different methodologies, namely chromosome conformation capture (3C) (Dekker et al. 2002), chromatin immunoprecipitation (ChIP), and cloning to build a novel 6C assay (Fig. 1A). The beginning steps involve the usual 3C procedure, where the crosslinked chromatin is subjected to restriction enzyme-mediated digestion followed by intramolecular ligation. However, at the end of the 3C procedure, instead of de-crosslinking, the ligated chromatin is subjected to ChIP using an antibody against a protein of interest (the suspected “bridging protein”). This is followed, to facilitate insert cloning, by reversal of the crosslinking, purification of the immunoprecipitated DNA, and ligation into a vector bearing the sequence overhangs generated in the enzyme digestion used in the 3C assay. This directly allows the cloning of 3C ligated fragments pulled down by the antibody. The ligation mix is then transformed, and the clones are screened by digestion with the same restriction enzyme that was used for the 3C assay (Fig. 1A). The clones showing multiple fragments upon digestion (multiple inserts ideally mean 3C ligated partners) were subjected to sequencing from both ends of the vector to discover the identity of the interacting partners. Each step in the procedure was monitored via controls run in parallel for 3C, ChIP, and cloning steps (Supplemental Fig. 1A–C).
Figure 1.
Construction of the 6C assay for EZH2. (A) Outline of the 6C assay. Briefly, cells are subjected to the conventional 3C procedure as described previously (Tolhuis et al. 2002). However, subsequent to the ligation step, the chromatin is subjected to chromatin immunoprecipitation (ChIP) using an antibody against the protein of interest (EZH2, in the present study), followed by ligation of the purified DNA into a cloning vector bearing the sequence overhangs generated in the enzyme digestion used in the 3C assay (EcoRI, in our case) to facilitate insert cloning and further screening. Clones are screened by digestion with the restriction enzyme that was used for the 3C assay, and the ones showing multiple inserts were subjected to sequencing from both directions in the vector to reveal the identity of the partners. (B) EZH2 is bound to the chromatin regions discovered in the 6C assay and is associated with the trimethylation of H3K27 in undifferentiated Tera-2 cells. The ChIP was performed using antibodies against EZH2 and H3K27me3 followed by amplification of the precipitated DNA using primers spanning the EcoRI enzyme sites (from each of the partners) used for 3C assay. The gel shows the results of semiquantitative PCR reactions for each of the partner regions in the five clones under investigation.
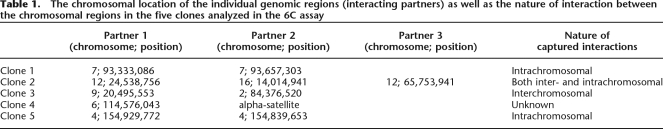
We performed our assay in Tera-2 teratocarcinoma cells, using an antibody against EZH2, to identify novel long-range chromatin interactions involving EZH2 as an integral component (Fig. 1A). We took five clones that were obtained with evidence for multiple inserts for further analysis. Two had intrachromosomal ligations (clones 1 and 5), although we cannot fully exclude the possibility of interchromosomal interactions between the homologous chromosomes. One clone had ligation of one fragment from chromosome 6 to alpha-satellite sequences of an unknown chromosomal origin (clone 4), one had ligated interchromosomal regions (clone 3), and one had both inter- and intrachromosomal ligations (clone 2) (Table 1; Supplemental Table 1; Supplemental Fig. 2).
Table 1.
The chromosomal location of the individual genomic regions (interacting partners) as well as the nature of interaction between the chromosomal regions in the five clones analyzed in the 6C assay
Next, we attempted to perform a deeper analysis with these five clones. Since the above observations left us with clues that EZH2 might be involved in the physical pairing of newly discovered chromatin regions, it was important to establish that the interacting regions are truly occupied by EZH2 in vivo using independent methods. Toward this end, we carried out ChIP analysis using antibodies specific for EZH2 and H3K27me3, and the immunoprecipitated DNA was amplified using primers spanning the EcoRI sites found to be involved in 3C ligation with other partner(s) in each of these clones. This semiquantitative PCR analysis again revealed that all of our defined chromatin interacting regions are bound by EZH2 proteins and have corresponding H3K27me3 marks (Fig. 1B).
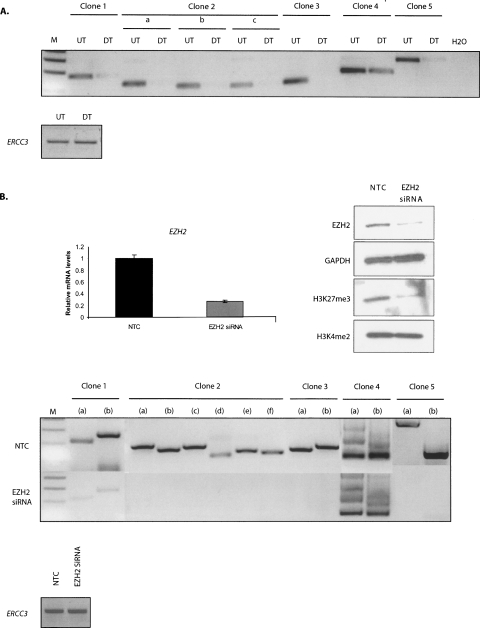
In order to further confirm that these interactions indeed exist in the nucleus, we went back to the formaldehyde-crosslinking-based 3C technique, which only detects intimate interactions as formaldehyde bridges relatively short distances (2 Å) and is reliable when it comes to finding a specific, functional chromatin interaction present in the interphase nucleus (Solomon and Varshavsky 1985; Jackson 1999; Splinter et al. 2004). Moreover, 3C generates a population-average measurement of interaction frequency between given genomic loci (Dekker et al. 2002), providing reliable and consistent observations across different experiments. We carried out 3C PCRs using primers to probe our above candidate physical interactions and confirmed that the identified partners in each of these five 6C clones (clone 1 to clone 5) are in close physical proximity in the nucleus of undifferentiated Tera-2 cells (Fig. 2A). In addition, very strikingly, each of these interactions were either severely reduced or lost when these cells were differentiated, toward a neuronal lineage, using ATRA (Fig. 2A). Importantly, this was an indirect indication that Polycomb proteins such as EZH2 might hold these regions together as the levels of polycomb components are down-regulated as cells differentiate (Supplemental Fig. 3; Boyer et al. 2006; Lee et al. 2006; Bracken et al. 2007; Ohm et al. 2007).
Figure 2.
EZH2 mediates long-range inter- and intrachromosomal interactions in undifferentiated Tera-2 cells. (A) 6C interactions exist in vivo in undifferentiated Tera-2 cells and are lost on ATRA-induced cellular differentiation toward neuronal lineage. Semiquantitative 3C PCRs were performed using one primer from each partner in the clone to detect interactions between 6C partners in undifferentiated Tera-2 (UT) and differentiated Tera-2 (DT) cells. For clone 2, “a” refers to the interaction between partner 1 and partner 2; “b,” the interaction between partner 1 and partner 3; and “c,” the interaction between partner 2 and partner 3. In the lower panel; interaction between two fragments at the ERCC3 locus, separated by ∼10 kb, was used to normalize crosslinking frequency between UT and DT samples. (B) siRNA-mediated depletion of EZH2 leads to loss of 6C interactions. The upper panel shows the elimination of the target mRNA (left panel) and protein (right panel) by siRNA against EZH2. As a control, a random-sequence nontargeting control siRNA was used (NTC). The undifferentiated Tera-2 cells were transfected with 25 nM EZH2 siRNA for two consecutive days using Lipo 2000 reagent and were processed 72 h later for real-time RT-PCR analysis using EZH2 and GAPDH-specific primers (as controls) (upper left panel) and for Western analysis using anti-EZH2 and anti-H3K27me3 antibodies (upper right panel). The blot was subsequently stripped and reprobed with anti-GAPDH and H3K4me2 antibodies as controls. The H3K4me2 marks remain unaffected, indicating that the targeting is specific for EZH2, which catalyzes the H3K27me3 marks. 3C assay was carried out in undifferentiated Tera-2 cells 72 h post-transfection with either siRNA specific for EZH2 or with a NTC (lower panel). Semiquantitative 3C PCRs were performed using multiple primer combinations to detect interactions between 6C partners. For the 3C PCRs, following combinations of primers (FP indicates forward primer; RP, reverse primer) were used. For clone 1, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; for clone 2, (a) FP of partner 1 and FP of partner 2, (b) FP of partner 1 and FP of partner 3, (c) RP of partner 1 and RP of partner 2, (d) RP of partner 1 and RP of partner 3, (e) FP of partner 2 and FP of partner 3, (f) RP of partner 2 and RP of partner 3; for clone 3, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; for clone 4, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2; and for clone 5, (a) FP of partner 1 and FP of partner 2, (b) RP of partner 1 and RP of partner 2. In the lower panel; interaction between two fragments at ERCC3 locus, separated by ∼10 kb, was used to normalize crosslinking frequency between NTC and EZH2 siRNA-treated samples. The figure represents the results of one experiment, which were further confirmed with samples generated in independent experiments.
EZH2 knockdown leads to loss of physical proximity between the paired partners
Our observations that the interactions between several remote chromatin regions are possibly mediated by EZH2, prompted us to investigate the effect of depletion of cellular levels of EZH2 on the interactions between the 6C partners in each of our five clones. We transiently transfected Tera-2 cells with small interfering RNA (siRNA) specific for EZH2 or a nontargeting control (NTC), and both Western blot and real-time RT-PCR confirmed efficient knockdown of EZH2, as shown in Figure 2B (upper panels). Moreover, the EZH2 knockdown led to a very efficient and global reduction in the associated mark, H3K27me3, compared with a random NTC, without affecting another histone mark H3K4me2 (Fig. 2B, upper right panel). Interestingly, 3C analysis using multiple primer combinations revealed that siRNA against EZH2 led to reduction or complete loss of interactions between 6C partners in each of these clones, while NTC did not interfere with any of these physical proximities (Fig. 2B, lower panels). This provided strong evidence that EZH2 is a direct mediator of this pairing between different chromosomal segments, both inter- and intrachromosomally.
EZH2 siRNA directed loss of physical pairing leads to up-regulation of flanking genes
Our findings that EZH2 brings distinct chromatin domains together led us to speculate that such proximity might strengthen PcG-dependent silencing by increasing the concentration of constituent PcG proteins and other associated repressors around genes in a silencing hub. This kind of pairing might promote efficiency of PcG function by allowing delimited numbers of PcG complexes to regulate large numbers of genes that are subject to silencing at a given time in the EC cells. Similar suggestions have been put forth previously for transcriptional activation in the case where multiple genes were found to share the same transcription factory (RNA pol II machinery) in a transcription-dependent manner (Osborne et al. 2004). There is also evidence that transitions between active and silent transcriptional states involve shuttling between euchromatic and heterochromatic nuclear compartments (Brown et al. 1997; Francastel et al. 1999).
To help study the potential gene expression implications of our demonstrated long-range interactions of EZH2, we first searched for genes to examine by combining siRNA-mediated depletion of EZH2, expression microarray technology, and quantitative pathway analysis to determine the effects of EZH2 knockdown in undifferentiated human EC (Tera-2) cells. We transfected these cells in a proliferative phase with siRNA oligonucleotides specific for EZH2. Real-time RT-PCR and Western blot analysis of lysates prepared 72 h after transfection confirmed, as was shown in Figure 2B, that the siRNA efficiently inhibited the RNA and protein synthesis of its specific target, which accompanied a very efficient and global reduction in the associated heterochromatic mark, H3K27me3.
RNA was extracted at this point, labeled, and hybridized to Agilent 4x44K gene expression arrays. After image processing, the data were normalized as described (Zahurak et al. 2007), and fold changes were calculated to compare expression after siRNA-mediated knockdown of EZH2 to the untreated control. Results obtained on dye-swapped replicate arrays were averaged to produce reliable measures of differential expression. A total of 253 genes were identified as potential targets of EZH2-mediated transcriptional repression (at least twofold up-regulated upon down-regulation of EZH2) (Supplemental Fig. 4A). Moreover, as many as 79 genes were found to be down-regulated in these siRNA-treated cells. We randomly chose 16 genes from this list of up-regulated genes for further validation by ChIP analysis for EZH2 occupancy, and we found that 13 out of 16 genes are indeed occupied by EZH2 at their promoters (Supplemental Fig. 4B). This gives a one-sided 95% confidence interval for the true proportion of (0.58, 1.0); i.e., unless EZH2 occupies nearly 60% of all genes, this represents significant enrichment at the 0.05 level.
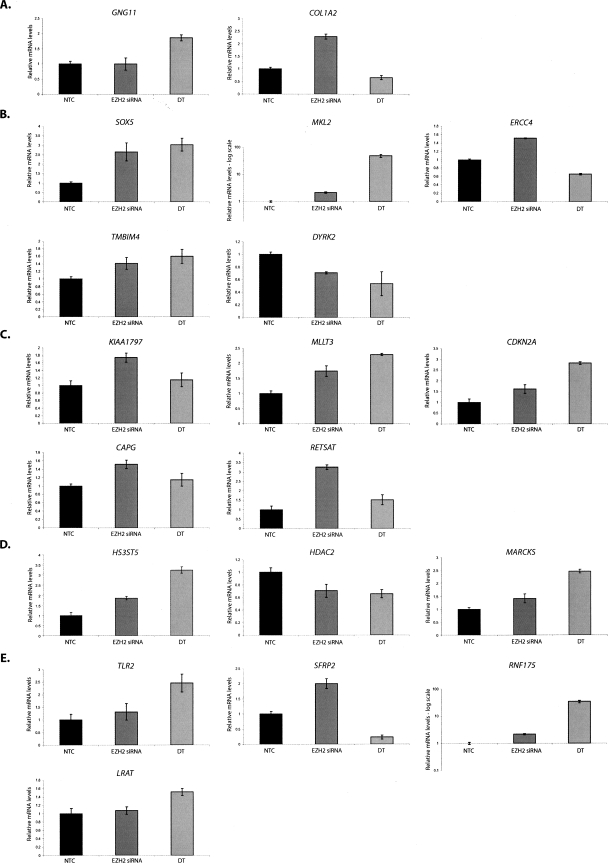
We then used some of the genes identified above to investigate our EZH2-dependent long-range interactions. We randomly chose and analyzed the transcript levels of a total of 19 genes from our five analyzed clones, based on their residing in the flanking chromatin regions around the interaction sites in each of the partners in the clones captured in our 6C assay. Interestingly, real-time RT-PCR discovered that 14 out of 19 genes are up-regulated to varying degrees (∼40%–350% of the NTC) after depletion of EZH2 (Fig. 3). While, the up-regulation seen in each of these cases solely by EZH2 depletion is not very dramatic for some genes, this may be consistent with a requirement for specific differentiation signals, such as those induced by ATRA treatment of the Tera-2 cells, which pushes them toward neuronal differentiation. We have previously shown (Ohm et al. 2007), as have others (Bernstein et al. 2006; Boyer et al. 2006; Lee et al. 2006), that this differentiation is associated with gene loss of the EZH2 catalyzed H3K27me3 mark in the setting of full transcriptional activation. Indeed, we found that 10 of our 19 genes were up-regulated to different degrees above the EZH2 siRNA-induced levels (Fig. 3) upon ATRA-induced differentiation of the Tera-2 cells, which also accompanies severe down-regulation of EZH2 (Supplemental Fig. 3). Thus, it appears likely that our broad domains of linked chromatin regions may link multiple genes over megabases of distance through PcG-dependent associations that, in turn, modulate gene regulation.
Figure 3.
The genes flanking the interacting chromatin regions are up-regulated in Tera-2 cells depleted for EZH2 and in the ATRA differentiated cells. The loss of pairing between 6C partners induced by EZH2 knockdown, or by ATRA-induced cellular differentiation of the Tera-2 cells toward a neuronal lineage, is concomitant with the up-regulation of flanking genes residing in the interacting chromatin regions. Total RNA was prepared from NTC and EZH2 siRNA-treated Tera-2 cells 72 h post-transfection using RNeasy Kit (QIAGEN), followed by cDNA synthesis and real-time RT-PCR for a total of 19 genes flanking the interacting chromatin region in each 6C partner for the five clones that we studied. The relative mRNA levels of the chosen genes in NTC vs. EZH2 siRNA-treated Tera-2 cells are shown for clone 1 (A), clone 2 (B), clone 3 (C), clone 4 (D), and clone 5 (E). PCR of GAPDH mRNA was used to normalize values across different samples.
Previous findings that PcG occupancy and associated H3K27 trimethylation marks are distributed over large chromosomal domains might help explain our observations (Boyer et al. 2006; Lee et al. 2006; Squazzo et al. 2006; Bracken et al. 2007; Rinn et al. 2007). There have been observations in Drosophila showing that PcG proteins are localized primarily to the polycomb response elements (PREs) while me3K27 is spread over the entire gene (Kahn et al. 2006). By extrapolation, the distinct chromatin regions that we captured in our 6C assay might represent the docking sites for the PcG proteins, from where the heterochromatin mark H3K27me3 self-propagates and spreads in both-directions, thereby silencing a number of genes on the way. We studied this possibility for clone 4, where we found that the PcG-dependent association of a chromatin element to alpha-satellite sequences accompanied silencing of a number of genes flanking this region. We performed ChIP using antibodies against EZH2 and H3K27me3 for the clone 4 partner 1, clone 4 partner 2 (alpha-satellite), and three gene promoters (HS3ST5, HDAC2, and MARCKS) flanking partner 1. Semiquantitative PCRs followed by gel-based quantiation indicated that while the levels of H3K27me3 are similarly high on all of the three promoters as well the two partners, the EZH2 occupancy was substantially reduced as we moved away (genes HDAC2 and MARCKS) from the interacting region (clone 4 partner 1) (Supplemental Figure 5).
Discussion
In this article, we describe our construction of an assay to determine how specific proteins may mediate and/or participate in complexes that mediate long-range chromatin interactions that, in turn, can regulate the transcriptional status of multiple genes simultaneously. We use the protein EZH2, as our first example, to show how the assay might work and show an illustrative body of initial data to demonstrate that this is the case. Thus, we provide the first demonstration that the PcG protein EZH2 is able to bring distant chromatin regions, both intra- and interchromosomal, together in the three-dimensional space of the mammalian nucleus (Fig. 4). This is ultimately verified by the fact that siRNA-mediated knockdown of EZH2 leads to loss of pairing between the 6C partners, concomitant with the up-regulation of multiple flanking genes around each of the interacting chromatin regions.
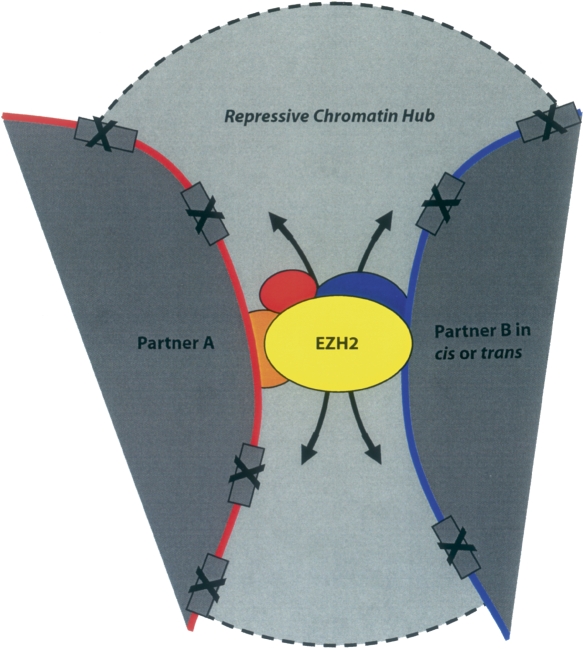
Figure 4.
A hypothetical model showing that EZH2 mediates long-range chromatin interactions, both in cis and in trans and brings about silencing of multiple genes in a broad chromatin domain flanking the interacting regions. Such proximity might help achieve physiologically significant levels of PcG-dependent transcriptional repression of the target genes simply by increasing the concentration of Polycomb proteins and associated repressors in the vicinity of the interaction, by virtue of having multiple PcG occupied sites together. Alternatively, this kind of pairing might also result simply from the sharing of Polycomb factories (or PcG bodies) by multiple genes possibly due to less number of polycomb proteins available compared with the large number of genes that are subject to silencing at a given time in these stem-cell like cells where PcGs are crucial regulators of transcription. The distinct chromatin regions that we captured in our 6C assay might represent the docking sites for the PRC2 complex proteins (containing EZH2), from where the heterochromatin mark H3K27me3 self-propagates and spreads in both directions, thereby silencing a number of genes on the way, or the complex might contact the promoter directly by establishing small loop(s) between these docking sites and the site of action (promoter). Some genes might be protected from such reaction due to their promoter-specific protection mechanism(s), against such marks.
It is important to stress, given recent studies of the dynamics for the rapid “on-off” interactions between transcriptional factors and proteins to the chromatin (Cheutin et al. 2003; Kosak and Groudine 2004; Meshorer and Misteli 2006) that we probably have captured only a glimpse of some of the more stable interactions out of the plethora of dynamic associations that might be ongoing in the nucleus with regard to PcG functions and structures. The chromatin domains localized within the PcG bodies may only temporarily associate with these structures and then leave one PcG body to incorporate into another one and may not necessarily affect other partners in the complex. Recently, using 3C-chip technology, it was discovered that one inactive gene tends to cluster with other nontranscribed genes in the nucleus, located both in cis and in trans (Simonis et al. 2006). Such preferential clustering of inactive genes might be explained by our findings relating the PcG-dependent physical proximity of multiple chromatin domains with the silenced status of the associated genes (Fig. 4).
From a technical standpoint, it is important to mention that for our first construction of the 6C assay, while screening the clones by restriction digestion, we found a very low frequency of clones having multiple inserts (five out of 352). The rest of the clones had a single insert and may represent distinct genomic sites that are bound by PcG proteins but are not engaged in any long-range associations. Very importantly, in essence, such findings may also provide an indirect indication of the frequency at which EZH2-dependent long-range associations take place in the nucleus. On the other hand, the number of clones with multiple inserts might also get reduced due to difficulty in cloning (ligation and transformation) and sequencing of large size fragments that might result from the ligation of EcoRI digestion generated fragments in this first iteration of our 6C assay. Moreover, intramolecular ligation after crosslinking and digestion will lead to a fraction of circular DNA that cannot be cloned and may further reduce the number of clones in such an assay. Finally, we could have also missed some interactions involving partners that bear DNA methylation at the EcoRI site as EcoRI is a DNA methylation–sensitive enzyme. Thus, it may give partial digestion when the last C of its recognition site is part of a methylated CpG dinucleotide. We also analyzed clones from the control plate, which all had single sequences and may represent nonspecifically precipitated sequences during the ChIP step. In addition, the sequencing of five such clones revealed no similarity to any of the clones obtained in the EZH2 6C assay.
Although, our 6C assay partly resembles the ChIP-loop assay previously described (Horike et al. 2005), in terms of combining 3C and ChIP assays, it differs from ChIP-loop assay in the order in which these steps were performed and the other additional steps we have added in order to make it more useful for a global analysis. 6C offers a promising tool when the aim is to discover novel interactions mediated by specific protein(s) of interest without any presumptive knowledge of the interacting partners. Previously performed ChIP-loop assays impose certain technical concerns, as also discussed recently in a review by Simonis et al. (2007), where they also point out for a need of techniques very similar to our 6C assay. In a ChIP-loop assay, the ligation is done after the ChIP procedure when the fragments are bound and concentrated on the beads. Such a ligation step might facilitate the ligation between all the fragments that are juxtaposed due to their association with beads and not necessarily due to formaldehyde crosslinking, and thus may not reflect associations that take place in the three-dimensional space of the mammalian nucleus. In addition, the fragments generated at the end of the ChIP-loop assay do not have any restriction enzyme ends and thus are not easy to clone for further characterization and therefore are only suited for assaying interaction between known partners, unlike our 6C assay. This limitation would also inhibit the ChIP-loop assay from being combined with other assays (3C-chip, 4C, etc.) for high-throughput global analysis (discussed below) (Supplemental Fig. 6). This may be especially so when reproducibility is concerned given the sonication procedure utilized in the ChIP-loop assay vs. the use of restriction enzyme digestion in the 6C assay.
The 6C assay involves steps to directly investigate the potential of a candidate protein to be a chromatin looping mediator and to bridge multiple chromatin elements, both in cis and in trans in the nucleus. From a technical standpoint, the 6C technique could also be modified and incorporated with other recently published techniques to discover all the chromatin regions in the nucleus that interact with a given gene or a chromatin region of interest in a specific protein-dependent manner (Supplemental Fig. 6). In this regard, future work could involve construction of libraries of all the chromatin interactions mediated by protein complexes, such as PcG using the 6C assay—and comparison of such libraries from different cell types and diseased cells, such as cancer cells, to investigate potential alterations in the protein complex–dependent genome architecture and contribution of such differential chromatin organization to cell type–specific transcriptomes. The 6C assay is a potential development toward understanding the three-dimensional aspects of genome organization and should constitute an important future tool to unravel how specific proteins contribute to the spatial distribution of distinct genomic loci.
Examples of the above possibilities involve some of the 6C clones explored in this study that could shed light on the basics of transcription regulation in mammalian cells. One interesting example is clone 4, which harbors interactions wherein silencing of genes such as MARCKS and HS3ST5 was found to depend on PRC2 (EZH2)-dependent close physical proximity with alpha-satellite sequences (Figs. 2B, clone 4, 3D). It has been shown that the PcG proteins associate with pericentromeric heterochromatin in both primary and transformed cell lines (Saurin et al. 1998; Cmarko et al. 2003). Pericentromeric heterochromatin has been associated with gene silencing activity, and it is known that factors recruit genes destined for heritable inactivation to pericentromeric heterochromatin (Brown et al. 1999; Cobb et al. 2000; Lundgren et al. 2000). Our findings directly extend these observations and implicate that PcG proteins might target certain genomic loci to pericentromeric heterochromatin in order to silence them.
Methods
Cell culture and transient siRNA transfection
Tera-2 cells were maintained in McCoy’s 5A medium supplemented with 15% FBS and grown at 37°C under 5% CO2. The cells were transfected with either a NTC or EZH2 targeting siRNA (Dharmacon D-001210-01 and D-004218-01) using Lipofectamine 2000 (Invitrogen). Cells were transfected at a 25 nmol/L siRNA concentration and then transfected again 24 h later and continued to be passaged for up to 72 h.
Microarray and data analysis
Total RNA was harvested from Tera-2 cells depleted of EZH2 as well as control cells in log phase using the QIAGEN kit according to the manufacturer’s protocol. RNA was quantified by using the NanoDrop ND-100 followed by quality assessment with the 2100 Bioanalyzer (Agilent Technologies). The 28s/18s ratio was taken for all the samples and was found to be greater than 2.2. Sample amplification and labeling procedures were carried out using the low RNA input fluorescent linear amplification kit (Agilent Technologies) according to manufacturer’s suggestions. The labeled cRNA was purified using the RNA easy minikit (QIAGEN) and quantified. RNA spike-in controls (Agilent Technologies) were added to RNA samples before amplification. Samples (EZH2 siRNA) labeled with cy3 or cy5 were mixed with controls targets (NTC; Agilent Technologies), assembled on oligo microarray, hybridized, and processed according to the Agilent microarray protocols. Scanning was performed with the Agilent G25665BA microarray scanner using setting recommended by Agilent Technologies.
Agilent array data were preprocessed using a simple loess normalization without background subtraction, as described by Zahurak et al. (2007). Log fold changes were calculated for each array and averaged over the dye-swap replicates. A gene set enrichment analysis was performed for gene ontology (GO) terms and for involvement KEGG pathways (Ashburner et al. 2000). Genes were ranked by log fold change, and a nonparametric Wilcoxon rank sum test was used to determine whether the genes annotated to a given GO category (or given pathway) and which show more differential expression than would be expected by chance alone. P-values were adjusted using the Benjamini-Hochberg false discovery rate (Pawluk-Kolc et al. 2006).
ChIP
ChIP assays were performed as described previously (McGarvey et al. 2006), with only modification that immunoprecipitation was performed using Dynal Magnetic beads purchased from Invitrogen (Protein A beads [100-02D] and Protein G beads [100-04D]). Trimethylated H3K27 antibody was produced as previously described (McGarvey et al. 2006). Antibody to EZH2 (Upstate) was purchased from the commercial source. Primers and amplification conditions are available by request from the authors.
3C assay
3C assay was done essentially as discussed by Tolhuis et al. (2002) with minor modifications for human cells. The PCR analysis was optimized by mixing, digesting, and ligating the PCR products spanning the EcoRI sites from each of the partners in 6C clones. A novel control was designed and tested for human ERCC3 locus in order to be able to normalize crosslinking frequency across different samples. The primer sequences and PCR conditions used in the 3C assay are available on request.
6C assay
The 6C protocol was designed by combining 3C, ChIP, and cloning techniques with further modifications to optimize the efficiency of the experiment. The beginning steps involve the usual 3C procedure (Tolhuis et al. 2002), where the crosslinked chromatin is subjected to restriction enzyme-mediated digestion (EcoRI in our case) followed by intramolecular ligation. However, at the end of 3C procedure, instead of de-crosslinking, the ligated chromatin is subjected to ChIP as described above, using an antibody against the protein of interest (the suspected “looping mediator”; EZH2 in the present study) followed by reverse crosslinking, purification of the immunoprecipitated DNA, and ligation into pBluescript II RI Predigested Vector (Stratagene). This directly allows the cloning of 3C ligated fragments pulled down by the antibody. The ligation mix is transformed into XL10-Gold Ultracompetent Cells (Stratagene), and cells are subjected to blue-white screening. Plasmids are purified from white colonies and are screened by digestion with the same enzyme that was used in the 3C procedure (EcoRI in this study). The clones showing multiple fragments upon digestion (multiple inserts ideally mean 3C ligated partners) are subjected to sequencing from both ends of the vector (T3 and T7 promoter-specific primers for pBluescript II RI) to discover the identity of the interacting partners. Each step in the procedure was carefully monitored, and controls were run in parallel to ensure accuracy of the procedure (Supplemental Fig. 1), including controls for the 3C procedure by testing ERCC3 locus (Supplemental Fig. 1A), ChIP assay (GATA5 as a positive control in our case, shown previously by Ohm et al. 2007 as a PcG target in Tera-2 cells) (Supplemental Fig. 1B), and cloning method (calculation of the percentage of white colonies in experimental vs. control samples) (Supplemental Fig. 1C) during the corresponding steps in the 6C protocol.
RNA purification and real-time RT-PCR analysis
RNA was isolated with RNeasy kit (QIAGEN) according to the manufacturer’s instructions. For qPCR analysis, 1 μg of total RNA was reverse transcribed using the Superscript First-Strand Synthesis System (Invitrogen). Quantitative real-time RT-PCR using was performed using QuantiTect SYBR Green PCR Kit (QIAGEN). PCR primers and amplification conditions are available on request.
Acknowledgments
We thank all the members of the Baylin laboratory, members of Dr. James Herman’s laboratory, and Neha Tiwari in Dr. Barry Nelkin’s laboratory for their help and discussions. We thank Dr. Dirk Schübeler (FMI) for his support. We also thank Kathy Bender for administrative assistance. This work was supported by ES011858 (National Institute of Environmental Health Sciences) and CA116160 (National Institute of Health) grants to S.B.B. and P30 CA06973-44 (National Cancer Institute) grant to L.C.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073452.107.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Cao R., Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cheutin T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Cmarko D., Verschure P.J., Otte A.P., van Driel R., Fakan S. Polycomb group gene silencing proteins are concentrated in the perichromatin compartment of the mammalian nucleus. J. Cell Sci. 2003;116:335–343. doi: 10.1242/jcs.00225. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Morales-Alcelay S., Kleiger G., Brown K.E., Fisher A.G., Smale S.T. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C., Walters M.C., Groudine M., Martin D.I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Jackson V. Formaldehyde cross-linking for studying nucleosomal dynamics. Methods. 1999;17:125–139. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- Kahn T.G., Schwartz Y.B., Dellino G.I., Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- Kirmizis A., Bartley S.M., Kuzmichev A., Margueron R., Reinberg D., Green R., Farnham P.J. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes & Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Groudine M. Gene order and dynamic domains. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D.J., Mendelsohn M., Kirkland J., Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lundgren M., Chow C.M., Sabbattini P., Georgiou A., Minaee S., Dillon N. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell. 2000;103:733–743. doi: 10.1016/s0092-8674(00)00177-x. [DOI] [PubMed] [Google Scholar]
- McGarvey K.M., Fahrner J.A., Greene E., Martens J., Jenuwein T., Baylin S.B. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Ohm J.E., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk-Kolc M., Zieba-Palus J., Parczewski A. Application of false discovery rate procedure to pairwise comparisons of refractive index of glass fragments. Forensic Sci. Int. 2006;160:53–58. doi: 10.1016/j.forsciint.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Ringrose L. Polycomb comes of age: Genome-wide profiling of target sites. Curr. Opin. Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shiels C., Williamson J., Satijn D.P., Otte A.P., Sheer D., Freemont P.S. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Simonis M., Kooren J., de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: A probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A., van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Splinter E., Grosveld F., de Laat W. 3C technology: Analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- Squazzo S.L., O'Geen H., Komashko V.M., Krig S.R., Jin V.X., Jang S.W., Margueron R., Reinberg D., Green R., Farnham P.J. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Wurtele H., Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- Zahurak M., Parmigiani G., Yu W., Scharpf R.B., Berman D., Schaeffer E., Shabbeer S., Cope L. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjolinder M., Gondor A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]