Abstract
To understand the mechanism regulating the effector function of memory CD8 T cells, we examined expression and chromatin state of a key transcription factor (eomesodermin, EOMES) and two of its targets: perforin (PRF1) and granzyme B (GZMB). Accessible chromatin associated histone 3 lysine 9 acetylation (H3K9Ac) was found significantly higher at the proximal promoter and the first exon region of all three genes in memory CD8 T cells than in naive CD8 T cells. Correspondingly, EOMES and PRF1 were constitutively higher expressed in memory CD8 T cells than in naive CD8 T cells at resting and activated states. In contrast, higher expression of GZMB was induced in memory CD8 T cells than in naive CD8 T cells only after activation. Regardless of their constitutive or inducible expression, decreased H3K9Ac levels after treatment with a histone acetyl-transferase inhibitor (Curcumin) led to decreased expression of all three genes in activated memory CD8 T cells. These findings suggest that H3K9Ac associated accessible chromatin state serves as a corner stone for the differentially high expression of these effector genes in memory CD8 T cells. Thus, epigenetic changes mediated via histone acetylation may provide a chromatin “memory” for the rapid and robust transcriptional response of memory CD8 T cells.
During the process of naive cell to memory cell differentiation, memory CD8 T cells acquire two unique properties: long life and enhanced effector function (1–3). This enhanced effector function is associated with a low threshold for activation and a rapid production of effector molecules of memory CD8 T cells, and is controlled in part by transcription factors such as Eomesodermin (EOMES) and T-box transcription factor 21 (TBX21) (4–6). It has been shown that EOMES directly regulates expression of perforin (PRF1), granzyme B (GZMB), and IFN γ (IFNG), and promotes effector function in CD8 T cells (5). However, the mechanisms that differentially regulate the expression of EOMES and its targets in memory CD8 T cells are not fully understood.
Activation of gene transcription is controlled by the availability of transcription factors and the accessibility of chromatin structure at the gene locus. Recent studies suggest that an “accessible” or “repressed” chromatin structure is regulated in part by the modifications of histone amino-terminal tails such as acetylation, methylation, phosphorylation, and other posttranslational modifications (7–9). Histone acetylation occurs at a lysine residue of histone tails and is modified by two opposing enzymes: histone acetyltransferases (HATs)3 and histone deacetylases (HDACs) (10, 11). An enhanced level of histone acetylation is generally correlated with accessible chromatin structure, which is seen not only in active gene transcription but also in nontranscribed genes poised for response (12). Furthermore, a decrease in histone acetylation using an inhibitor of HAT activity, such as curcumin, results in a decrease in gene expression (13). Thus, histone acetylation could provide a means of rapid activation of specific gene expression in a cell in response to stimulation.
Accumulating evidence suggests that acetylation of histone H3 lysine 9 (H3K9Ac) and lysine 14 (H3K14Ac) are markers for accessible chromatin or chromatin that is transcriptionally active while methylation of H3 lysine 9 (H3K9me) is associated with gene silencing (14 –18). In naive CD4 T cells, histones H3 and H4 in the Il4 and Ifng loci were unacetylated but were rapidly acetylated after activation (19). In activated CD8 T cells, hyperacety-lation of histone H3 at the Ifng promoter and enhancer was maintained through memory T cells (20). Our recent findings demonstrate that the differentially higher expressed genes have high levels of H3K9Ac in their gene loci in memory CD8 T cells and increased H3K9Ac levels after an HDAC inhibitor (Trichostatin A) treatment increases gene expression in naive CD8 T cells (21), providing evidence that histone acetylation may regulate gene expression and function of memory CD8 T cells. In addition, changes in DNA methylation in some of these gene loci have also been reported (20, 22, 23). There is an inverse correlation between the methylation of the 5′UTR and EOMES expression (24) and DNA hypomethylation in the PRF1 promoter increases PRF1 expression (25). However, it is unknown whether epigenetic changes such as histone modifications play a role in the regulation of the effector function of memory CD8 T cells.
In this study, we examined the expression and H3K9Ac of EOMES, PRF1, and GZMB in human naive and memory CD8 T cells before and after stimulation. EOMES and PRF1 were expressed at higher levels in memory cells than in naive cells at rest and after activation while GZMB was expressed at higher levels in memory cells than in naive cells only after activation. The levels of H3K9Ac in the EOMES, PRF1, and GZMB loci were higher, particularly at the transcription start site, in memory cells than in naive cells. Induced hypoacetylation in these gene loci by a HAT inhibitor (curcumin) resulted in decreased expressions of EOMES, PRF1, and GZMB in memory cells in response to in vitro stimulation. These findings suggest that histone acetylation is essential in determining the level of EOMES, PRF1, and GZMB expressions in memory CD8 T cells.
Materials and Methods
Abs and reagents
The Abs and reagents used in this study are as follows: FITC-conjugated anti-CD45RA mAb was purchased from eBioscience. Tri color-conjugated anti-CD8 mAb was purchased from Invitrogen. Anti-acetyl-Histone H3 (Lys9) and purified rabbit IgG were purchased from Upstate Biotechnology. Anti-GZMB mAb (2C5/F5) was purchased from BD Biosciences. Anti-perforin mAb (P1– 8) was purchased from Kamiya Biomedical. Anti-ZAP70 Ab was a gift from Dr. Ronald Wange (Food and Drug Administration, Rockville, MD). HRP-linked anti-mouse, anti-rabbit, or anti-rat Abs were purchased from GE Healthcare. Curcumin was purchased from Sigma-Aldrich.
Isolation and stimulation of naive and memory CD8 T cells
Peripheral blood was obtained from healthy adults by the National Institute on Aging Pheresis Unit (Institutional Review Board-approved protocol MRI2003–054). The procedure for isolation and stimulation of naive and memory CD8 T cells was previously described (26). In brief, PBMC were isolated by Ficoll (GE Healthcare) gradient centrifugation. Naive and memory CD8 T cells were then enriched by removing other types of cells through incubation with a panel of mouse mAbs against human CD4, CD19, CD11b, CD14, CD16, MHC class II, erythrocytes, platelets, and CD45RO (for the enrichment of naive cells) or CD45RA (for memory cells). Ab-bound cells were subsequently removed by incubation with anti-mouse IgG-conjugated magnetic beads (Qiagen). These enriched naive and memory CD8 T cells were further purified into CD8+CD45RA+ naive T cells and CD8+CD45RA− memory T cells by a cell sorter (MoFlo; Dako). The purity of sorted naive and memory CD8 T cells was over 96%. The sorted naive and memory CD8 T cells were either used for experiments right away or incubated with anti-CD3 plus anti-CD28 Ab (anti-CD3/CD28) coupled magnetic beads (Invitrogen) at the cell:bead ratio of 1:1 in RPMI 1640 with 10% FBS and penicillin (10 U/ml)/streptomycin (10 µg/ml) (Invitrogen). The stimulated cells were harvested at the specified time for analyses of mRNA, protein, and chromatin immunoprecipitation (ChIP) assay.
Real-time quantitative RT-PCR
The procedure was standard as previously described (21). In brief, total RNA was extracted from fresh and stimulated naive and memory CD8 T cells by Stat 60 (Tel-Test). The quantity of total RNA was measured by NanoDrop (Agilent) and 500 ng of total RNA was used to synthesize cDNA by reverse transcriptase (SuperScript II; Invitrogen). PCR was conducted in 20 µl of total volume with 0.125 µM primers using a SYBR GREEN kit (Applied Biosystems) for 40 cycles on ABI Prism 7500 (Applied Biosystems). The specific amplification of RT-PCR products was confirmed by agarose (2%) gel electrophoresis. The threshold cycle (Ct) of genes was normalized with that of ACOX1. Primers of PCR were designed by using the Primer Express software (Applied Biosystems) and made by Integrated DNA Technologies. The following are their sequences. EOMES: forward- 5′-AGGCGCAAATAACAACAACACC-3′ and reverse- 5′-ATTCAAGTCCTCCACGCCATC-3′; PRF1: forward- 5′-GTGGAGTGCCGCTTCTACAGTT-3′ and reverse- 5′-TGCCGTAGTTGGAGATAAGCCT-3′; GZMB: forward- 5′-GGTGGCTTCCTGATACAAGACG-3′ and reverse- 5′-GGTCGGCTCCTGTTCTTTGAT-3′;GAPDH: forward- 5′-GGAGTCAACGGATTTGGTCGTA-3′ and reverse- 5′-GCAACAATATCCACTTTACCAGAGTTAA-3′; ACOX-1: forward- 5′-TGCTTTGGTTGATGCATTTGA-3′ and reverse- 5′-CATAGCGGCCAAGCACAGA-3′.
Treatment of memory CD8 T cells with curcumin
Curcumin (Sigma-Aldrich), a HAT inhibitor, was dissolved in ethanol at 10 mM. Memory CD8 T cells were cultured in the presence or absence of 20 µM curcumin for 2 h, and followed by stimulation with anti-CD3/28 Abs for the specified time. Cells were then harvested for the analyses of mRNA, protein, and ChIP assay.
ChIP assay
The procedure of ChIP was previously described (21). In brief, cross-linking of chromosomal DNA of naive or memory CD8 T cells were conducted by incubating with 1% formaldehyde at 37°C for 10 min, quenching by 125 mM glycine, and washing twice with cold PBS containing a mixture of protease inhibitors (Roche). The treated cells were then lysed in three sequential lysis buffers 1–3 containing protease inhibitors (lysis buffer 1: 50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, and 0.25% Triton X-100; lysis buffer 2: 10 mM Tris-HCl (pH 8), 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA; lysis buffer 3: 10 mM Tris-HCl (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, and 0.5% N-lauroylsacrosine). The chromosomal DNA was then sheared with sonication (Sonic Dismembrator Model 100; Fisher Scientific), and the average length of sonicated chromosomal DNA was around 200 bp. A total of 1/10 volume of sonicated lysates was used for DNA isolation as INPUT while the remaining lysates (9/10 volumes) were used for ChIP with anti-H3K9Ac and control Abs. For the ChIP, cell lysates were first incubated with protein-A conjugated agarose (GE Healthcare) and 3 µg anti-acetyl-H3K9Ac antiserum (Upstate Biotechnology) or 3 µg control rabbit IgG overnight at 4°C, followed by washing with RIPA buffer (50 mM HEPES-KOH (pH 7.5), 500 mM LiCl, 1 mM EDTA, 1% Nonidet P-40, and 0.7% sodium deoxycholate) and TE buffer containing 50 mM NaCl sequentially, and then immunoprecipitated histone complexes were eluted at 65°C for 15 min. DNA was reverse-crosslinked by incubating at 65°C for 16 h and RNA and proteins were removed by treatment of RNaseA and proteinase K sequentially. Immunoprecipitated DNA was purified by phenol/chloroform extraction and ethanol precipitation, and was used for analysis by real-time PCR. The same PCR reagents, instrument, and verification of the amplification product were used here as described in above section. The formula for quantitation of ChIP results is INPUT % = 1/9 × 2Ct of INPUT − Ct of ChIP × 100. The primer sequences are for EOMES: forward_9- 5′-TGAAAAAGGGCAGAAAGGCG-3′ and backward_9 –5′-GAAAGCAGGAGGTGGAAACTAACC-3′; forward_10- 5′-TCATTACGAAACAGGGCAGGTG-3′ and backward _10- 5′-CGGTGTCTACGGAGATTTATTGCG-3′; PRF1: forward_9- 5′-TGTGTGCCCTGAGTCCCCG-3′ and backward_9- 5′-TCCTTTGTGCTCTCCCCTCC-3′; forward_10- 5′-TGGGAGGGGAGAGCACAAAG-3′ and backward_10- 5′-CACCAGCCACCACTCACATCAC-3′; GZMB: forward_8- 5′-CACTTCATAGGCTTGGGTTCCTG-3′ and backward_ 8- 5′-CTCTGGGTGCTTGTGTGAGAATC-3′; forward_9- 5′-AAGAGAGCAAGGAGGAAACAACAG-3′ and backward_9- 5′-AATGTGGAGAAAGGGCAGGAGACC-3′;GAPDH:forward-5′-GCGTGCCCAGTTGAACCAG-3′ and backward- 5′-TCAGCCAGTCCCAGCCCAAG-3′.
Western blot
Cells were washed once in PBS, resuspended in lysis buffer (50 mM Tris-HCl (pH 8), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) with protease inhibitors (Roche), and incubated on ice for 30 min. Lysates were spun for 15 min at 14,000 rpm at 4°C, mixed with NuPage LDS sample buffer (Invitrogen), and loaded onto a NuPAGE 4–12% gradient Bis-Tris gel for GZMB or a 7.5% SDS-PAGE gel (non-reducing condition) for PRF1 detection. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane (Immobiolon-P; Millipore) using an XCell II Blot Module (Novex) in Transfer buffer (25 mM bicine, 25 mM Bis-Tris, 1 mM EDTA, 8% methanol, and 0.0375% SDS). The membranes were first incubated with a blocking buffer (PBS, 5% BSA, and 0.1% Tween 20) at room temperature for 1 h, and followed by the incubation with the primary Ab for 1 h at room temperature. Anti-GZMB mAb (2C5/F5) was used at a 1/1000 dilution and anti-perforin mAb (P1–8) at a 1/1000 dilution. The membranes were washed three times in PBS, 0.1% Tween 20, incubated with a HRP-labeled anti-Ig secondary Ab at a 1/5000 dilution, washed three times, developed for 1 min in ECL Western Blotting Detection reagents (GE Healthcare) and exposed between 10 s and 5 min to Hyperfilm (GE Healthcare). The film images were digitized by AlphaImager (Alpha Innotech) and quantified by AlphaEase FC software (Alpha Innotech).
Statistical analysis
Two-tailed Student’s t test was used in analysis of the levels of EOMES, PRF1, and GZMB mRNA expression and the levels of H3K9Ac in these gene loci between naive and memory CD8 T cells. The significance was defined by a value of p < 0.05.
Results
Memory CD8 T cells express higher levels of EOMES than do naive CD8 T cells before and after in vitro stimulation
As memory cells exhibit a faster and more robust effector response than do naive cells and EOMES regulates expression of several key cytolytic effector molecule genes (4), we compared EOMES expression in human naive and memory CD8 T cells from peripheral blood of normal donors before and after activation in vitro with anti-CD3/CD28 Abs. Resting memory CD8 T cells expressed significantly higher levels of EOMES than did resting naive cells (2.5-fold, p < 0.01) (Fig. 1A). After activation, EOMES mRNA level peaked around 4–8 h and then gradually decreased in both naive and memory cells. Although the overall kinetic pattern of EOMES expression was similar between naive and memory cells, memory cells consistently expressed higher levels of EOMES mRNA (from 2.1–3.0-fold) than did naive cells throughout the course of 72-h stimulation (Fig. 1B). The higher expression of EOMES in resting and activated memory CD8 T cells provides a plausible mechanism for their rapid and robust effector response compared with naive cells.
FIGURE 1.
EOMES is expressed at higher levels in memory CD8 T cells than in naive CD8 T cells. A, The levels of EOMES expression in naive and memory CD8 T cells before and after stimulation in vitro. Sorted naive and memory CD8 T cells were stimulated with anti-CD3/CD28 Abs for 0, 4, 8, 16, 32, and 72 h. The level of EOMES expression was determined by quantitative real-time PCR at each time point. The data are presented as mean ± SEM (n = 9). B, The relative expression levels of EOMES in naive and memory CD8 T cells. The data are presented as the ratio of each time point to freshly isolated naive cells in mean ± SEM (n = 9). Values of p are indicated by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001) used in all figures.
The EOMES gene locus contains higher levels of H3K9Ac in memory CD8 T cells than in naive CD8 T cells
To determine the mechanism underlying the differential expression of EOMES in memory CD8 T cells, we analyzed the levels of H3K9Ac in the promoter, the coding region, and the 3′UTR of the EOMES gene locus in naive and memory CD8 T cells by ChIP and quantitative real-time PCR assays. To provide good spatial resolution, we designed nine pairs of primers covering the promoter (2000 bp of upstream region from the transcription start site), six pairs of primers in the coding region, and one pair of primers at the 3′UTR (data not shown). As high levels of H3K9Ac are found in the proximal promoter and the first exon, we focused on the quantitative analysis of these regions. Resting memory cells had significantly higher levels of H3K9Ac ( p < 0.05) than did resting naive cells (Fig. 2). After in vitro stimulation (at 6, 24, and 72 h), increase of H3K9Ac levels was observed mainly in memory cells but did not reach statistical significance (Fig. 2), suggesting that H3K9Ac levels in the EOMES locus may not be strongly regulated by activation. Together, the high levels of H3K9Ac in the EOMES locus are associated with high EOMES expression in resting and activated memory CD8 T cells.
FIGURE 2.
High levels of H3K9Ac are found in the EOMES locus of memory CD8 T cells. The relative levels of H3K9Ac in the proximal promoter and the first exon in the EOMES locus in naive and memory CD8 T cells at rest and after stimulation were presented. Sorted naive and memory CD8 T cells were stimulated with anti-CD3/CD28 Abs and were used for ChIP assay. The primer pairs 9 and 10 were used for quantitative real-time PCR (same conditions used in the Fig. 3), and the data are presented as the average ratio of the sum of these products at each time point to freshly isolated naive cells in mean ± SEM (n = 7).
Reduced histone acetylation (H3K9Ac) levels decrease EOMES expression in memory CD8 T cells
To further determine the role of H3K9Ac levels in EOMES expression, we pretreated memory CD8 T cells with a HAT inhibitor, curcumin (20 µM), for 2 h before stimulation with anti-CD3/CD28 Abs. A significant decrease of the H3K9Ac levels in the EOMES locus but not in a control gene (GAPDH) locus was observed in curcumin-treated memory CD8 T cells after stimulation for 6 h compared with curcumin-untreated activated memory T cells (Fig. 3A). The H3K9Ac level of the EOMES locus in curcumin-treated memory CD8 T cells after stimulation was only slightly higher than in resting naive CD8 T cells (Fig. 3A). Consequently, a significant decrease of EOMES mRNA levels (63% reduction) but not a control gene GAPDH mRNA levels was observed in curcumin-treated activated memory cells compared with curcumin-untreated activated memory T cells ( p < 0.001) (Fig. 3B). These results suggest that the level of H3K9Ac in the EOMES locus may regulate the expression of EOMES.
FIGURE 3.
Inducing hypoacetylation of H3K9 in the EOMES locus decreases EOMES expression in memory CD8 T cells. A, The relative levels of H3K9Ac in the proximal promoter and the first exon in the EOMES locus in curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation, and resting naive CD8 T cells. The data are presented as the ratio to curcumin-untreated memory CD8 T cells in mean ± SEM (n = 4). The relative levels of H3K9Ac in the proximal promoter of the GAPDH locus in curcumin-treated and curcumin-untreated memory CD8 T cells are also shown. B, The relative levels of EOMES expression of curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation, and resting naive CD8 T cells. The levels of EOMES and GAPDH expression are presented as the ratio to curcumin-untreated memory CD8 T cells in mean ± SEM (n = 5).
Memory CD8 T cells express high levels of perforin (PRF1) and contain high levels of H3K9Ac in the PRF1 locus
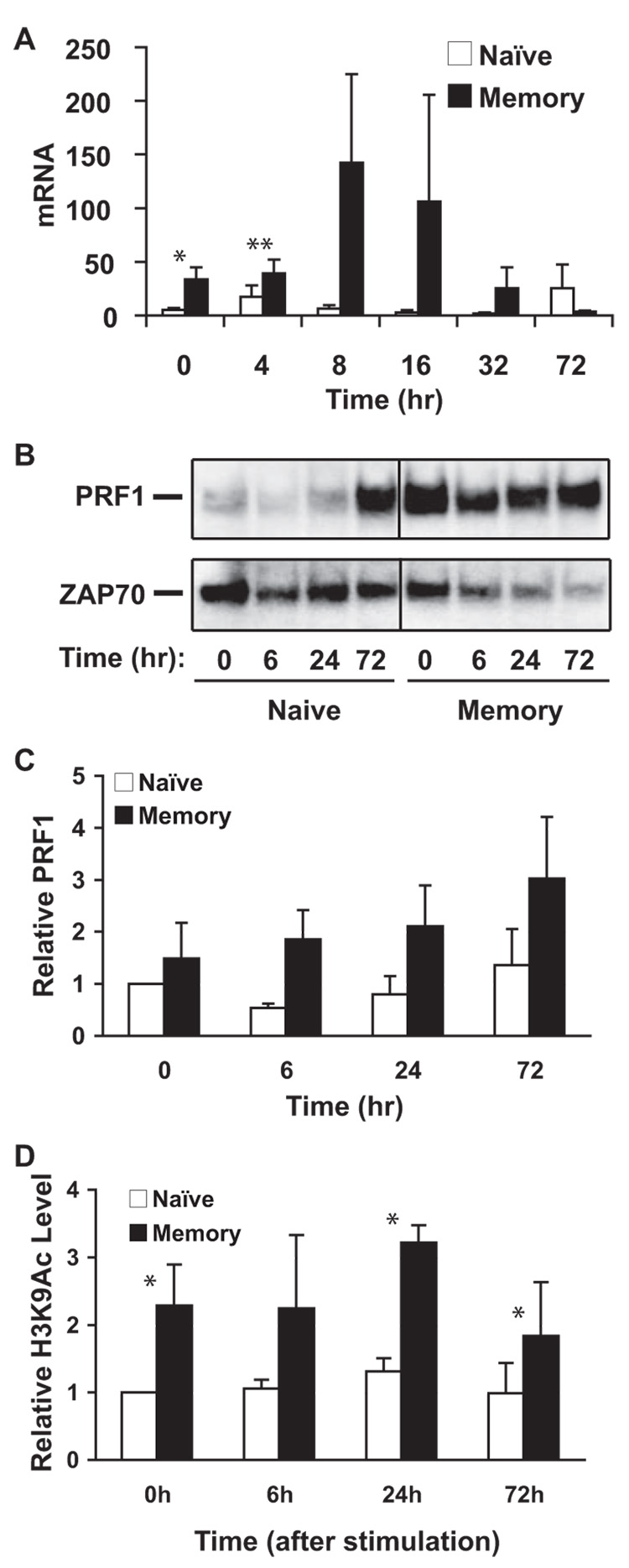
Next, we wanted to determine whether the target gene of EOMES, PRF1, is expressed and regulated in a similar manner to EOMES in memory CD8 T cells. The mRNA levels of PRF1 were significantly higher in resting memory CD8 T cells than in resting naive cells (6.1-fold, p < 0.05) (Fig. 4A). After activation, mRNA levels were up-regulated consistently higher in memory cells than naive cells until 72 h after stimulation. Corresponding to the mRNA levels, perforin protein levels were also higher in memory cells than in naive cells before and after activation (Fig. 4, B and C). We then examined H3K9Ac levels and found significant higher levels of H3K9Ac at the PRF1 locus in resting memory CD8 T cells than in resting naive cells (Fig. 4D). After in vitro stimulation (at 6, 24, and 72 h), increase of H3K9Ac levels was observed mainly in memory cells but their differences did not reach statistical significance (Fig. 4D). Thus, high levels of H3K9Ac in the PRF1 locus are also associated with high PRF1 expression in resting and activated memory CD8 T cells.
FIGURE 4.
High levels of perforin expression are associated with high levels of H3K9Ac in the PRF1 locus in memory CD8 T cells. A, The mRNA levels of PRF1 expression in naive and memory CD8 T cells before and after stimulation. The levels of PRF1 expression are presented as mean ± SEM (n = 9). B, The protein levels of perforin in naive and memory CD8 T cells before and after stimulation. ZAP70 levels were used as the loading control. C, The quanitified data of B are presented as mean ± SEM (n = 3). The intensity of perforin was normalized based on ZAP70 (same as in Fig. 5). D, The relative levels of H3K9Ac in the PRF1 locus in naive and memory CD8 T cells before and after stimulation. The primer pairs 9 and 10 were used for quantitative real-time PCR (same conditions used in the Fig. 6) and the data are presented as the average ratio of the sum of these two products at each time point to freshly isolated naive cells (n = 7).
Memory CD8 T cells express high levels of GZMB and contain high levels of H3K9Ac in the GZMB locus
GZMB is another key target of EOMES in regulation of effector function. We found low levels of GZMB mRNA (Fig. 5A) and no detectable levels of granzyme B protein (Fig. 5, B and C) in resting naive or memory CD8 T cells. In vitro stimulation increased the mRNA levels of GZMB in both naive and memory cells. Activation-induced GZMB expression increased significantly faster and higher in memory cells than in naive cells for the first 24 –32 h (Fig. 5, A–C). At 72 h after stimulation, naive cells expressed higher levels of GZMB than did memory cells. Unlike the differential expression of EOMES and PRF1 in resting memory cells, the differential expression of GZMB in memory cells occurred only after activation. We then examined H3K9Ac levels and found that the resting memory CD8 T cells had significant higher levels of H3K9Ac than did resting naive cells (Fig. 5D). Like the EOMES and PRF1 loci, H3K9Ac levels increased in the GZMB locus mainly in memory CD8 T cells after stimulation (Fig. 5D). However, the control of the GZMB expression differs from that of the EOMES and PRF1 expression in resting memory cells.
FIGURE 5.
Rapid increase of GZMB expression after stimulation is associated with high levels of H3K9Ac in the GZMB locus in memory CD8 T cells. A, The mRNA levels of GZMB expression in naive and memory CD8 T cells before and after stimulation. The levels of GZMB expression are presented as mean ± SEM (n = 9). B, The protein levels of GZMB expression in naive and memory CD8 T cells before and after stimulation. C, The quanitified data of B are presented as mean ± SEM (n = 4). D, The relative levels of H3K9Ac in the GZMB locus in naive and memory CD8 T cells at rest and after stimulation. The primer pairs 8 and 9 were used for quantitative real-time PCR (same conditions used in the Fig. 7) and the data are presented as the average ratio of the sum of these two products at each time point to freshly isolated naive cells (n = 5).
Decreasing acetylation (H3K9Ac) in the PRF1 and GZMB loci in memory CD8 T cells decreases PRF1 and GZMB expression
To determine whether the H3K9Ac levels play an essential role in activation-induced rapid up-regulation of PRF1 and GZMB in memory CD8 T cells, we pretreated resting memory CD8 T cells with curcumin (20 µM) for 2 h followed by stimulation with anti-CD3/CD28 Abs. Like the EOMES gene, H3K9Ac levels in the PRF1 and GZMB loci significantly decreased after curcumin treatment in memory cells (Fig. 6A and 7A). The levels of H3K9Ac in the PRF1 and GZMB loci in curcumin-treated memory cells after stimulation were close to the H3K9Ac levels in resting naive cells. Strikingly, significant decreases of PRF1 and GZMB mRNA and protein were observed in curcumin-treated activated memory cells (Fig. 6, B and C, and 7, B and C). Interestingly, the levels of mRNA of PRF1 and GZMB in curcumin-treated memory cells after activation were comparable with their levels in resting naive cells while the control gene GAPDH did not change significantly between curcumin-treated and curcumin untreated memory cells. These findings support the notion that higher levels of histone acetylation (H3K9Ac) in memory CD8 T cells is responsible for their activation-induced rapid expression of PRF1 and GZMB and rapid effector function.
FIGURE 6.
Inducing hypoacetylation of H3K9 in the PRF1 locus results in decreased PRF1 expression in memory CD8 T cells. A, The relative levels of H3K9Ac in the PRF1 locus in curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation, and resting naive CD8 T cells. The data are presented as the average ratio to curcumin-untreated memory CD8 T cells (n = 4). B, The relative levels of PRF1 expression of curcumin-untreated and curcumin-treated memory CD8 T cells, and resting naive CD8 T cells. The levels of PRF1 and GAPDH expression are presented as the average ratio to curcuminun-treated memory CD8 T cells (n = 5). C, Perforin protein levels in curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation.
FIGURE 7.
Inducing hypoacetylation of H3K9 in the GZMB locus results in decreased GZMB expression in memory CD8 T cells. A, The relative levels of H3K9Ac in the GZMB locus in curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation, and resting naive CD8 T cells. The data are presented as the average ratio to curcumin-untreated memory CD8 T cells (n = 4). The relative levels of H3K9Ac in the proximal promoter in the GAPDH locus are also shown. B, The relative levels of GZMB expression of curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation, and resting naive CD8 T cells. The levels of GZMB and GAPDH expressions are presented as the average ratio to curcumin-untreated memory CD8 T cells (n = 5). C, GZMB protein levels in curcumin-treated and curcumin-untreated memory CD8 T cells after stimulation.
Discussion
In this study, we showed that the effector function regulator (EOMES) and two of its targets (PRF1 and GZMB) are expressed at a higher level in memory CD8 T cells than in naive CD8 T cells in the resting stage and/or after activation. Furthermore, the significant higher levels of H3K9Ac were observed in the EOMES, PRF1, and GZMB loci in memory CD8 T cells compared with naive CD8 T cells. Although the levels of EOMES and PRF1 expression were correlated with the levels of H3K9Ac in resting memory CD8 T cells, GZMB expression not in resting memory CD8 T cells but in activated memory CD8 T cells was correlated with the level of H3K9Ac in resting memory CD8 T cells. Finally, we demonstrated that reduced levels of H3K9Ac in the EOMES, PRF1, and GZMB loci by a HAT inhibitor (curcumin) resulted in decreased expression of all three genes in activated memory CD8 T cells. Together, these results indicate that histone acetylation is tightly correlated with the rapid and robust increase in expression of effector function genes in memory CD8 T cells.
The analysis of expression of EOMES, PRF1, and GZMB in naive and memory CD8 T cells before and after activation showed that memory cells rapidly and greatly increased the levels of EOMES, PRF1 and GZMB regardless of their initial expression levels in the first 32 h after activation. Interestingly, EOMES expression peaked at 4–8 h and declined at 32 h after stimulation, slightly ahead of the peak of expression of PRF1 and GZMB. Although the kinetic pattern of PRF1 expression was similar to EOMES in memory CD8 T cells before and after stimulation with PRF1 levels slightly behind EOMES, GZMB expression exhibited a different pattern before and after stimulation. GZMB mRNA level was similar between naive and memory CD8 T cells at rest and remained stably high through 72-h stimulation in memory CD8 T cells. This difference in PRF1 and GZMB expression suggests that the regulation of PRF1 and GZMB expression is different, even though similarly high levels of H3K9Ac are present at these genes in memory CD8 T cells. It is intriguing that the expression of these two key effector molecules functioning in the same cytotoxic pathway is regulated differently. Further study will be needed to determine how PRF1 and GZMB expression are precisely regulated in naive and memory CD8 T cells and the relative contributions of EOMES and other transcription factors as well as other histone modifications associated with accessible chromatin structure.
Although histone acetylation at the cytokine gene loci facilitates their expression in effector T cells (19, 20), its role in effector function-related genes in memory T cells has not been reported. Our analysis showed that the acetylation levels of one specific residue on histone H3 (H3K9Ac) were higher in resting memory CD8 T cells than in resting naive CD8 T cells in the EOMES, PRF1, and GZMB loci, providing a chromatin basis for the high level of expression of EOMES, PRF1, and GZMB in memory CD8 T cells. Equally interesting is that the levels of H3K9Ac in these gene loci do not appear to increase dramatically after activation. Thus, activation-induced changes in gene expression may be mediated by acetylation of other histone residues and/or by different types of histone modification.
Based on these findings, we propose a transcription and chromatin model for the effector functions of memory CD8 T cells (Fig. 8). In naive CD8 T cells, the EOMES, PRF1, and GZMB loci are in a repressed chromatin state as shown by low levels of H3K9Ac and of their mRNA levels. By contrast, in memory CD8 T cells, these three gene loci are in an active chromatin state as shown by high levels of H3K9Ac with either high levels of EOMES and PRF1 mRNA (“primed”) or no increase of GZMB mRNA (“poised”). Upon activation, both naive and memory CD8 T cells increase expression of EOMES, PRF1 and GZMB. However, the accessible chromatin structure of these gene loci in memory CD8 T cells leads to a much more rapid and vigorous transcriptional response than in naive cells. Thus, the memory CD8 T cell response at the transcription level is controlled at two stages: 1) differential expression of the key effector regulators such as EOMES (through regulation of the chromatin structure of the gene locus and of transcription activators); and 2) differential expression of the effector targets (PRF1 and GZMB) through two modes. PRF1 has accessible chromatin at its locus and differential transcription (“primed” status), whereas GZMB has accessible chromatin at its locus without increased expression (“poised” status) until receiving an activation signal. This sequential induction of these two main effectors allows memory CD8 T cells to deliver a rapid response and at the same time to prevent these weapons of destruction from potential off-target killing. These epigenetic changes provide a chromatin basis for differential expression of regulators and their targets, which may well be a general mechanism for memory T cell function.
FIGURE 8.
Model for epigenetic regulation of differential gene expression associated with effector functions of CD8 T cells. In naive CD8 T cells, the EOMES, PRF1, and GZMB loci contain repressive (“not primed”) chromatin with low level of H3K9Ac in naive CD8 T cells corresponding to low levels of mRNA. In memory CD8 T cells, the EOMES, PRF1, and GZMB loci contain accessible chromatin as shown by the high levels of H3K9Ac corresponding to either high levels of mRNA (EOMES and PRF1) or low levels of mRNA (GZMB) until activation. Due to the differences of chromatin structure and transcription activator(s), memory CD8 T cells have a much more rapid and strong transcription response than do naive cells upon antigenic stimulation.
The level of histone acetylation is established and maintained by the interplay between HATs and HDACs. It is clear that inhibition of HATs led to decreased levels of H3K9Ac in the EOMES, PRF1, and GZMB loci resulting in a decrease in their mRNA levels in memory CD8 T cells. As curcumin inhibits p300/CBP but not PCAF and p300/CBP is capable of acetylation several lysine residues of histone (13), the relative importance of H3K9Ac and histone acetylating of other residues in the regulation of expression of these three genes is not known. Nevertheless, our data suggest that p300/CBP may play a critical role in regulating EOMES, PRF1, and GZMB expression in memory CD8 T cells. In addition to histone acetylation, histone methylation, regulated by histone methyltransferases, histone demethylases, and histone replacement can also influence chromatin structure. How these different modifications of histone tails are regulated and coordinated remains to be determined. Further elucidation of the histone modifications and other epigenetic changes in memory cells will not only shed new light on the molecular nature of memory T cells but also provide new targets and strategies for improving vaccines.
Acknowledgments
We thank Drs. Dan Longo and Mike Pazin for critical reading of the manuscript. We thank Francis J. Chrest and Cuong Nguyen of the Flow Cytometry Unit for sorting cells, and Karen Madara and her staff at the National Institute on Aging Apheresis Unit for collecting blood samples.
Footnotes
This work was supported by the Intramural Research Program of the National Institute on Aging and National Cancer Institute, National Institutes of Health.
Abbrevations used in this paper: HAT, histone acetyltransferase; PRF1, perforin 1; GZMB, granzyme B; HDAC, histone deacetylase; ChIP, chromatin immunoprecipitation.
Disclosures
The authors have no financial conflict of interest.
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- 1.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu. Rev. Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 3.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 4.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 5.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T. Chromatin modifications and their function 3. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Roth SY, Denu JM, Allis CD. Histone acetyltransferases 1. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 11.Nusinzon I, Horvath CM. Histone deacetylases as transcriptional activators: role reversal in inducible gene regulation 4. Sci. STKE. 2005;2005:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- 12.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective 1. Mol. Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription 1. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 14.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 15.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 16.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc. Natl. Acad. Sci. USA. 2004;101:7398–7403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields PE, Kim ST, Flavell RA. Changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 20.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-γ loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 2006;177:1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 21.Fann M, Godlove JM, Catalfamo M, Wood Iii WH, Chrest FJ, Chun N, Granger L, Wersto R, Madara K, Becker K, et al. Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8+ T cell response. Blood. 2006;108:3363–3370. doi: 10.1182/blood-2006-02-005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 23.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics 2. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Ivascu C, Wasserkort R, Lesche R, Dong J, Stein H, Thiel A, Eckhardt F. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas 1. Int. J. Biochem. Cell Biol. 2007;39:1523–1538. doi: 10.1016/j.biocel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression 17. J. Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- 26.Chiu WK, Fann M, Weng NP. Generation and growth of CD28nullCD8+ memory T cells mediated by IL-15 and its induced cytokines. J. Immunol. 2006;177:7802–7810. doi: 10.4049/jimmunol.177.11.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]