Abstract
The urochordate Ciona savignyi is an emerging model organism for the study of chordate evolution, development, and gene regulation. The extreme level of polymorphism in its population has inspired novel approaches in genome assembly, which we here continue to develop. Specifically, we present the reconstruction of all of C. savignyi’s chromosomes via the development of a comprehensive genetic map, without a physical map intermediate. The resulting genetic map is complete, having one linkage group for each one of the 14 chromosomes. Eighty-three percent of the reference genome sequence is covered. The chromosomal reconstruction allowed us to investigate the evolution of genome structure in highly polymorphic species, by comparing the genome of C. savignyi to its divergent sister species, Ciona intestinalis. Both genomes have been extensively reshaped by intrachromosomal rearrangements. Interchromosomal changes have been extremely rare. This is in striking contrast to what has been observed in vertebrates, where interchromosomal events are commonplace. These results, when considered in light of the neutral theory, suggest fundamentally different modes of evolution of animal species with large versus small population sizes.
Comparative analysis between the sister species Ciona savignyi and Ciona intestinalis offers a unique opportunity to understand the evolution of genomes. These ocean-dwelling broadcast spawners represent urochordates, the closest living sister group to the vertebrates (Delsuc et al. 2006). Urochordates have the basic set of genes and morphological features shared among chordates without the increases in complexity and redundancy that occurred along the vertebrate lineages. They are therefore attractive, simple, model organisms for studying chordate developmental mechanisms and evolution (Nishida 1987; Corbo et al. 1997; Harafuji et al. 2002; Davidson and Levine 2003; Satoh et al. 2003; Johnson et al. 2004; Meinertzhagen et al. 2004; Brown et al. 2007).
In the work presented here, we describe the construction of the genetic map for C. savignyi. We integrate this map with the existing genome sequence assembly, extending the contiguity of the assembly to the scale of chromosomes. We then use the extended assembly to compare the chromosomes of C. savignyi and C. intestinalis, to address questions about the persistence of genome architecture through evolution. The genetic map, extended assembly, and shareable markers produced by this work will facilitate forward genetics and comparative genomics in C. savignyi, an emerging model system.
The ∼170-Mb genomes of C. savignyi and C. intestinalis had both been sequenced to draft-level coverage (12.7× and 8×, respectively) (Dehal et al. 2002; Vinson et al. 2005). In C. intestinalis, 498 of the 4713 original WGS assembly fragments, representing 59% of the genome, were subsequently joined into larger composite fragments using BAC end sequencing and then mapped to the 14 chromosomes by fluorescent in situ hybridization (Shoguchi et al. 2006). Initial construction of the C. savignyi genome sequence, which was complicated by an extreme level of polymorphism (Small et al. 2007a), resulted in a double assembly that contained both haplomes of the single diploid individual that was sequenced (Vinson et al. 2005). Contiguity of the genome was significantly improved later by the merging of overlapping haplotypic fragments, and the parsing of a single reference sequence from the two alleles (Small et al. 2007b). Currently, the C. savignyi reference genome consists of 374 assembly fragments (“reftigs”) with an N50 size of 1.8 Mb.
The small number of reftigs, and their large sizes, provided the opportunity to construct a genetic map without first generating a physical map, which is usually required as a bridge between a sequence assembly and the mapping of assembly fragments to chromosomes. Furthermore, candidate markers for the map could be identified by computational screening instead of experimental isolation because the whole genome sequence was available. In light of the extreme degree of polymorphism in the C. savignyi population, informative markers were virtually guaranteed, but their amplification by PCR could be hampered by polymorphisms residing in primer sites. We therefore chose to develop markers whose primer binding sites would be in genomic regions that would have a low likelihood of being polymorphic, but which would amplify sequence that does bear sufficient neutral variation. Neighboring exons that would contain the primer sites, with the intervening intron providing polymorphisms as part of the amplicon, were the markers we settled on developing.
The extreme level of polymorphism meant that parents were sufficiently heterozygous for using a one-generation mapping cross. The choice of a one-generation mapping strategy meant that phase inference on the genotypes had to be incorporated into calculation of the map, which turned out to be straightforward, given the extreme level of polymorphism and the relatively even, close, spacing of markers that was achievable due to the availability of a sequence. Construction of the map enabled consolidation of the genome assembly into chromosomes containing ordered, and often oriented, reftigs.
The whole-chromosome reconstructions for both Ciona genomes afforded the opportunity to investigate the extent to which rearrangement events have broken up colinearity and synteny since the two species’ last common ancestor. (We use the term “synteny” to describe only retention of orthologous sequence on an ancestral chromosome, regardless of order or orientation, and the term “colinearity” to describe the conservation of order among orthologous sequences.) Comparisons in both flies and worms, over evolutionary distances comparable to those between the two Cionas, have shown that synteny has been highly conserved but colinearity has been almost completely lost (Richards et al. 2005; Drosophila 12 Genomes Consortium 2007; Hillier et al. 2007). In contrast, comparisons within classes of vertebrates (such as fish and mammals), whose members are not as diverged as the two Cionas, have shown that synteny has been largely abolished by interchromosomal rearrangements (Mouse Genome Sequencing Consortium 2002; Bourque et al. 2004, 2005; Gibbs et al. 2004; Jaillon et al. 2004; Zhao et al. 2004; Woods et al. 2005; Kasahara et al. 2007). Interchromosomal rearrangements are common even between mouse and rat, two species whose neutral evolutionary distance is less than one-tenth of that between the two Cionas (Gibbs et al. 2004).
To estimate the extent of synteny and colinearity between the two Cionas, we made use of previously generated pairwise sequence alignments of the two genomes (Brudno et al. 2003, 2007). By remapping these alignments to the C. savignyi chromosomes that resulted from the genetic mapping effort, we were able to investigate chromosomal evolution since the last common ancestor of the two species. Synteny and colinearity patterns in Ciona are much more similar to those found in the clades of Caenorhabditis and Drosophila, and quite distinct from vertebrates, which are much more closely related to Ciona than the other invertebrates. An intriguing hypothesis concerning the evolution of large-scale genome structure presents itself when these phylogenetic patterns are considered in light of the neutral theory (Kimura 1983). Specifically, the high efficacy of purifying natural selection, which is caused by the large effective population size of these invertebrate species, may be the root cause of the paucity of interchromosomal changes in comparison to an otherwise extreme divergence in genome structure.
Results
One-hundred-eighty-two highly informative and portable markers
A central feature of our approach was to target specific assembly fragments (“reftigs”) (Small et al. 2007a) for mapping. To ensure reliable amplification of marker loci in such a polymorphic species, we used PCR-based markers in which the primers bind to adjacent exons and amplify the intervening intronic sequence. To further enhance the likelihood of amplification, we restricted the primer binding sites to those with limited potential for synonymous polymorphism. This approach reduced amplification failure of candidate loci to 5%, a 10-fold reduction over primers targeted to untranslated sequences.
Before a pair of PCR primers could be used for mapping, it had to be characterized and validated. Marker characterization consisted of testing for amplification and determining which of eight enzymes with a 4-bp recognition sequence revealed an unambiguous banding pattern for each genotype. For each targeted region (one, two, or three per reftig, depending on reftig size) we selected multiple primer pairs in close physical proximity, but not amplifying the same intron. These nearby markers were expected to be genetically redundant, and thus co-segregation of alleles between them was used to verify correct targeting. The primer pair and enzyme combination producing the clearest banding pattern for each of the up to four segregating genotypes was then used as the marker for final mapping. Starting with almost 800 primer pairs and examining nearly 2000 combinations of amplicons digested with different enzymes, we identified 182 markers to 178 distinct target regions. One hundred forty-two markers were informative for recombination in both parents. The remaining markers were informative for recombination in just one parent. For two target regions, two different markers were used, each informative for a different parent. These markers were used to construct the full genetic map.
To test the portability of the characterized markers, a sample of 12 markers was tested on individuals from a population different from that of the cross. All 12 amplified robustly, and the segregating alleles were different from those of the mapping progeny (not shown). This demonstrates that the markers developed here for genetic mapping can be ported to other populations.
Genetic map construction
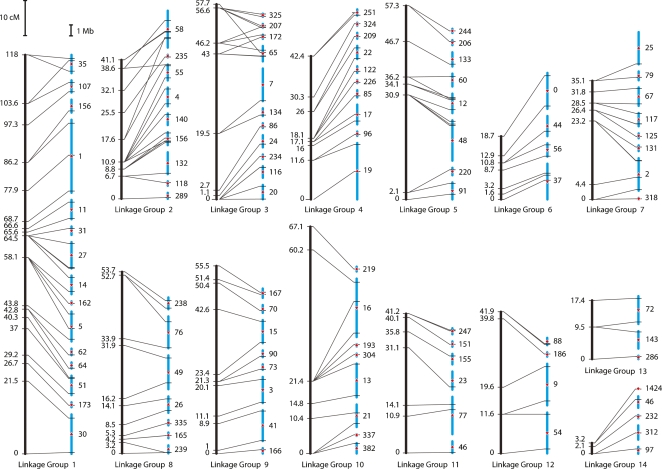
From the segregation data obtained by assaying the 182 markers in each of the 48 cross progeny, we constructed a genetic map. Map construction was accomplished with methodology that was specifically developed for this project because of the unusual nature of the cross and the markers, and proceeded in five phases: (1) assessment of linkage between pairs of markers, (2) formation of linkage groups, (3) parental haplotype inference within each linkage group, (4) ordering of markers within linkage groups, and (5) multipoint estimation of intermarker distances. The segregation data allowed us to resolve 110 of the 182 markers into distinct genetic loci clustering into 14 linkage groups (Fig. 1). The longest group was 118 cM, the shortest, 3.2 cM, with other groups falling between 17 and 61 cM. Collectively, the map spans 650 cM.
Figure 1.
Integrated genetic map and extended sequence assembly. Fourteen linkage groups in C. savignyi were identified by genetically mapping 182 markers, corresponding to 110 distinct genetic loci. Vertical black bars represent the genetic linkage map. All bars are drawn to scale; the length of the bars is proportional to the number of centimorgans (cM). Map distances at each mapped genetic locus are indicated to the left of each bar. Blue bars on the right of each pair represent reftigs. Tick marks indicate the physical location of the markers among the 104 mapped reftigs. Transverse lines link the location of each marker on the genetic and physical maps. Numbers to the right of each assembly fragment correspond to reftig identifiers in Version 2.1 of the assembly (Small et al. 2007a). When two or more markers on the same reftig allowed the orientation of the fragment to be resolved, the orientation is indicated with a red arrow pointing up or down for the positive or negative orientation, respectively. Red diamonds indicate unknown orientation. The gap size between reftigs is arbitrary since the true physical distance between mapped reftigs is unknown.
Weak linkage was detected between linkage groups 6 and 14, but only for alleles inherited from one parent. The comparative analysis performed between C. savignyi and C. intestinalis, discussed in greater detail below, suggests that this linkage is spurious. We, therefore, kept linkage groups 6 and 14 unlinked and separated.
Integrated genetic map
Targeting markers to specific reftigs enabled reconstruction of chromosomes corresponding to each of the linkage groups. For each linkage group, we ordered reftigs to be consistent with ordering of the markers determined by the genetic map. This let us place 104 of the largest reftigs, cumulatively representing 83% (146.8 Mb) of the reference sequence, onto chromosomes. In addition, we could orient 36 of the largest reftigs that contained multiple genetically resolved markers. Of the 56 reftigs >1 Mb, 54 have now been mapped. In only nine instances were we unable to confidently order small subgroups of reftigs (e.g., Fig. 1, chromosome 10 reftigs 304 and 193).
Our approach appears to have resulted in complete coverage of the genome. In fact, in an effort to extend the map and place additional reftigs, 11 of the 182 markers were actually developed after a preliminary map had been built. All of the additional markers were found to be linked to the previously identified linkage groups. A complete list of the reference sequence reftigs, and their inferred order and orientation, is provided in Supplemental Table 1.
Tight integration of the genetic map and sequence data enabled us to estimate additional genome parameters. The newly assembled 650-cM map accounts for 83% of the sequenced genome; therefore, given certain simplifying assumptions, we estimate the full genetic map length to be at most 783 cM. From 60 pairs of loci located on the same reftig, the average rate of recombination is 200 kb/cM, and recombination events appear to be roughly Poisson distributed. Finally, the regular spacing achieved by targeting markers to fragments puts any point on the genome within an average of 4.4 cM (880 kb) from the nearest established marker.
Disagreement between the genetic map and sequence assembly
For a small number of marker loci, the location on the genetic map and their presumed physical location were incongruent. Disagreements in ordering are easily identified on the integrated genetic map (Fig. 1) by intersecting transverse lines connecting corresponding loci on the linkage groups and chromosomes. Such differences could be due to problems with markers, such as amplification of an unintended target locus, a misassembly in the reference sequence, or an actual polymorphism between the mapping population and the sequenced individual. In the cases where ordering disagreed, we confirmed that the correct locus was amplified by subcloning. However, without additional sequencing data from the sequenced individual it is not possible to distinguish between a misassembly and a true polymorphism.
In just two cases, markers thought to reside on the same reftig mapped to different linkage groups. Of the five markers on reftig 156, one clustered with linkage group 1 while the other four clustered with linkage group 2. Of the two markers on reftig 46, one clustered with linkage group 11 and the other clustered with linkage group 14. We explored various possible causes of these inconsistencies by subcloning and sequencing amplicons, and by sensitive alignment of the reftigs to the C. intestinalis genome with BLAST (Altschul et al. 1990). We found that reftig 46 is highly repetitive, and that reftig 156 aligns well to C. intestinalis chromosome 9 (96% of BLAST alignments). Although reftig 156 is shown in both chromosomes 1 and 2 in Figure 1, for reference, we assigned it to chromosome 2 (Supplemental Table 2) because of the conserved synteny between C. savignyi chromosome 2 and C. intestinalis chromosome 9 (described in greater detail below). Similarly, reftig 46 is shown in both chromosomes 11 and 14 (Fig. 1), but has been assigned to chromosome 14 (Supplemental Table 2).
Extensive retention of synteny between C. savignyi and C. intestinalis
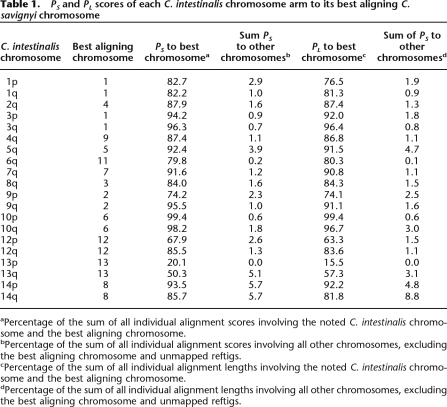
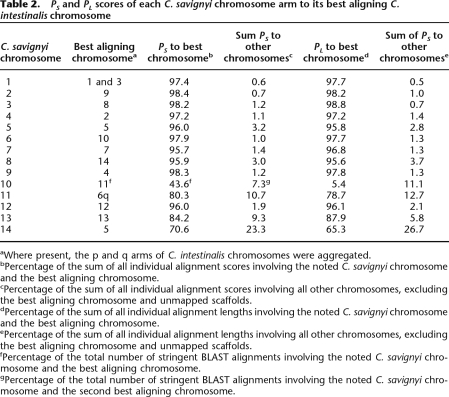
The construction of chromosomes in C. savignyi made it possible to compare conservation of synteny between it and C. intestinalis. Sensitive nucleotide-level alignments between these two species were previously generated using a combination of global and local (glocal) alignment (Brudno et al. 2007). To enrich for high-quality and putatively orthologous alignments, only reciprocal best alignments were used in our analyses. Construction of the C. savignyi chromosomes allowed remapping of these alignments from unordered reftigs to chromosomes. We identified syntenic chromosome pairs between the species by measuring the extent of alignment between the pairs. The extent of alignment was expressed as the percent score (PS) or percent length (PL). These were defined as the percentage of the sum of individual alignment scores or sum of individual alignment lengths, respectively, of all alignments associated with a chromosome in one species that corresponded to a particular chromosome in the second species. The scores and lengths of the alignments provided two different, but related, measures of the quality and extent of each alignment. Regardless of which metric was used, 11 C. savignyi chromosomes (2–9, 11–13) and 11 C. intestinalis chromosomes formed reciprocal best aligning chromosome pairs (Tables 1, 2), which we refer to as syntenic pairs.
Table 1.
PS and PL scores of each C. intestinalis chromosome arm to its best aligning C. savignyi chromosome
aPercentage of the sum of all individual alignment scores involving the noted C. intestinalis chromosome and the best aligning chromosome.
bPercentage of the sum of all individual alignment scores involving all other chromosomes, excluding the best aligning chromosome and unmapped reftigs.
cPercentage of the sum of all individual alignment lengths involving the noted C. intestinalis chromosome and the best aligning chromosome.
dPercentage of the sum of all individual alignment lengths involving all other chromosomes, excluding the best aligning chromosome and unmapped reftigs.
Table 2.
PS and PL scores of each C. savignyi chromosome arm to its best aligning C. intestinalis chromosome
aWhere present, the p and q arms of C. intestinalis chromosomes were aggregated.
bPercentage of the sum of all individual alignment scores involving the noted C. savignyi chromosome and the best aligning chromosome.
cPercentage of the sum of all individual alignment scores involving all other chromosomes, excluding the best aligning chromosome and unmapped scaffolds.
dPercentage of the sum of all individual alignment lengths involving the noted C. savignyi chromosome and the best aligning chromosome.
ePercentage of the sum of all individual alignment lengths involving all other chromosomes, excluding the best aligning chromosome and unmapped scaffolds.
fPercentage of the total number of stringent BLAST alignments involving the noted C. savignyi chromosome and the best aligning chromosome.
gPercentage of the total number of stringent BLAST alignments involving the noted C. savignyi chromosome and the second best aligning chromosome.
Initially, we thought that C. savignyi chromosome 10 might represent an exception to the pattern of conserved synteny. It aligned best to chromosome 13, but this chromosome accounted for only 7.8% of the PS score of chromosome 10 to the C. intestinalis genome. Upon closer inspection of the C. intestinalis sequence, however, we realized that chromosome 11 and the scaffolds that comprise it—although they were assembled by Shoguchi and colleagues (Shoguchi et al. 2006)—were entirely absent from the publicly available version of the assembly. Since chromosome 11 was absent from the assembly, there were no alignments involving it. To determine if this missing chromosome might represent the syntenic match to C. savignyi chromosome 10, we obtained C. intestinalis chromosome 11 and used BLAST (Altschul et al. 1990) to generate alignments between C. savignyi chromosome 10 and a complete C. intestinalis genome. Of the resultant BLAST alignments, which were stringently filtered, 43.6% corresponded to the newly appended chromosome 11. C. intestinalis chromosome 1 was the second best hit, garnering just 7.3% of the BLAST alignments. A significant fraction of the alignments (21.6%) were to unmapped C. intestinalis reftigs. We conclude that C. savignyi chromosome 10 is syntenic with C. intestinalis chromosome 11.
Chromosome 14 was the only linkage group for which we were unable to identify a syntenic match. The PS score (70.6%) indicates that it aligns best with C. intestinalis chromosome 5. However, C. savignyi chromosome 5 and C. intestinalis chromosome 5 form a reciprocal best aligning pair. Further analysis revealed that the total extent of alignment of chromosome 14 to any C. intestinalis chromosome was limited. Using BLAST (Altschul et al. 1990) to conduct a more sensitive search, we attempted to identify a region of C. intestinalis orthologous to chromosome 14. Still, no regions of significant similarity were found. In fact, the distribution of BLAST alignments indicated that the reftigs comprising chromosome 14 are composed of highly repetitive regions.
Extensive intrachromosomal rearrangements between C. savignyi and C. intestinalis
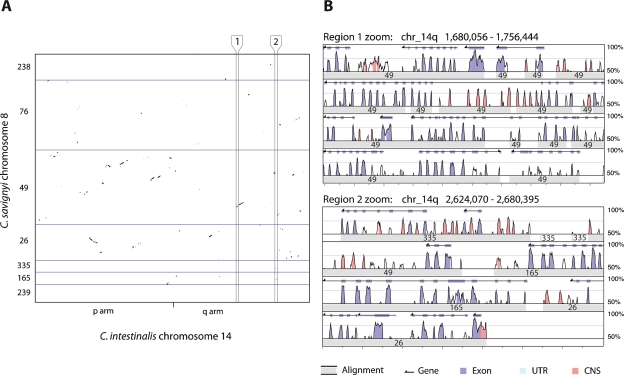
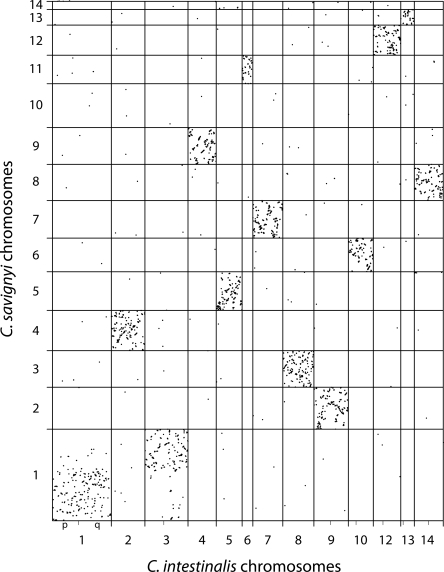
Next, we examined the retention of colinearity between the syntenic pairs of chromosomes. The remapping of alignments from reftigs in C. savignyi to chromosomes allowed us to plot the physical distribution of alignments on the syntenic pairs. Colinearity between the two species appears to have been almost completely abolished, with only small blocks (<<1 Mb) remaining (Fig. 2). Rearrangements have been extensive, as the blocks also do not appear to be arranged along either main diagonal. Rearrangements also appear to have occurred frequently between the different arms of the chromosomes in the seven instances where both chromosome arms have been assembled in C. intestinalis. When the distribution of alignments between all pairs of chromosomes is viewed simultaneously, the extent of the retained synteny and loss of colinearity is truly remarkable (Fig. 3; Supplemental Fig. 1).
Figure 2.
Conservation of synteny and loss of colinearity between C. savignyi and C. intestinalis. (A) Plot of alignments between C. intestinalis chromosome 14 and its C. savignyi ortholog, chromosome 8, reveals the extent of intrachromosomal rearrangement since these species diverged from their last common ancestor. Ordered C. savignyi reftig identifiers are shown along the Y-axis, with blue lines indicating boundaries between reftigs. The minor tick on the X-axis represents the boundary between the short (p) and long (q) chromosome arms of C. intestinalis. The aspect ratio of the plot is one. Alignments within the regions bounded by vertical black lines and labeled 1 and 2, are enlarged in panel B. (B) Enlargement of alignments within regions 1 and 2 reveals the quality of the alignments shown in panel A. Region 1 shows a set of colinear alignments, while region 2 shows a series of alignments that are linear in C. intestinalis, but widely separated in C. savignyi. This view of the alignments between the two species was adapted from the VISTA browser (http://genome.lbl.gov/vista). Each region is shown as a set of four rows of alignment plots. Within each set, each subsequent row is a continuation of the previous row. The X-axis of each plot indicates the position in the C. intestinalis genome; major tick marks are spaced at 4-kb intervals. Wide gray bars in each row represent the location of an aligning C. savignyi segment; the superimposed number indicates the C. savignyi reftig identifier. The height of the peaks is the percent identity between the sequences in 100-bp sliding windows. The area under the line is colored according to the key if the identity between the two species is >70%. Gene predictions for the C. intestinalis genome are shown at the top of each plot. (UTR) Untranslated region; (CNS) conserved noncoding sequence.
Figure 3.
Retention of synteny between C. savignyi and C. intestinalis chromosomes. Alignments (Brudno et al. 2007) between all C. savignyi and all C. intestinalis chromosomes were plotted by concatenating the chromosomes in each species and adjusting alignment coordinates accordingly. Unmapped reftigs and scaffolds and their alignments from both genomes (17% of C. savignyi and 46% of C. intestinalis) have been excluded. Where at least some portion of both the short (p) and long (q) chromosome arms of each of the C. intestinalis have been reconstructed, the boundary between arms is indicated by a minor tick mark on the X-axis. Alignments appear as dots because of their length relative to the displayed scale. The aspect ratio of the plot is one; the greater length of the Y-axis reflects the greater number of mapped fragments in C. savignyi.
A recent fusion occurred between ancestral chromosomes 1 and 3 in C. savignyi
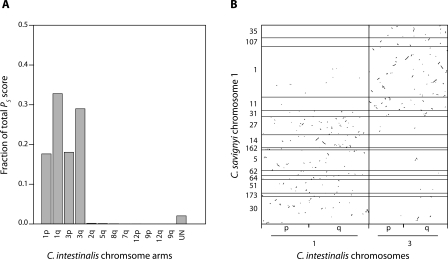
C. savignyi chromosome 1 is syntenic with both chromosomes 1 and 3 in C. intestinalis. Alignments to these two chromosomes jointly represent 97.7% of the PS score involving C. savignyi chromosome 1 (Fig. 4A). The C. intestinalis chromosomes also reciprocally align best to chromosome 1 (>82% of PS score). The physical distribution of alignments (Fig. 4B) reveals two distinct regions on chromosome 1, each aligning almost exclusively to just one of the C. intestinalis chromosomes. Thus, synteny appears to be retained in the two regions, but within each, the colinear blocks have been extensively rearranged. Only a small number of alignments between chromosome 1 and the pair of chromosomes in C. intestinalis occur between nonsyntenic regions.
Figure 4.
Conserved synteny between C. savignyi chromosome 1 and both chromosomes 1 and 3 of C. intestinalis. (A) Fraction of total alignment score between C. savignyi chromosome 1 and the various chromosome arms of C. intestinalis. The majority of alignments involve chromosomes 1 and 3. Alignments to unmapped C. intestinalis scaffolds were aggregated and labeled “UN.” (B) Distribution of alignments between C. savignyi chromosomes 1 and C. intestinalis chromosomes 1 and 3. Alignments cluster in specific subregions. Black horizontal lines indicate boundaries between the ordered reftigs of chromosome 1. Minor ticks indicate the boundary between the short (p) and long (q) arms of the C. intestinalis chromosomes. The aspect ratio of the plot is one.
The boundary between the two regions appears to be located either on reftig 31 or in the gap between it and reftig 27. Strong linkage in both parents between markers on either side of the boundary make it highly unlikely that the observation is due to an error in linkage or a segregating fusion/fission polymorphism. Instead, the conservation of synteny between the two regions on chromosome 1 and the two chromosomes in C. intestinalis suggests either a fusion between ancestral chromosomes in C. savignyi, or a fission of an ancestral chromosome in C. intestinalis. We reasoned that if there had been a fission event in C. intestinalis, either ancient or recent, there would not be two distinct regions of synteny in C. savignyi. For an ancient fission event, intrachromosomal rearrangements in C. savignyi after the event would have distributed blocks of colinearity to the two chromosomes in C. intestinalis along the entire length of chromosome 1, eliminating the distinct syntenic regions. Likewise, if a recent fission event in C. intestinalis occurred, rearrangements in the period prior to the event would have shuffled colinear blocks in C. savignyi along the entire length of chromosome 1, and again distinct regions of synteny should not be observed. Thus, a fusion event must have occurred between ancestral chromosomes 1 and 3 in the C. savignyi lineage. Had the fusion been ancient, the distinct regions of synteny on chromosome 1 would have degraded over time. Since only a few rearrangements appear to have occurred between the distinct regions in C. savignyi, the fusion is likely to be relatively recent.
Assigning unmapped reftigs in both species to chromosomes
The near absence of interchromosomal rearrangements makes it possible to use the interspecies comparison to infer to which chromosome unmapped reftigs or scaffolds belong. For example, an unmapped reftig in C. savignyi that aligns primarily to a chromosome in C. intestinalis can be putatively assigned to the chromosome that is syntenic to the known C. intestinalis chromosome. We assigned reftigs or scaffolds only if >75% of the fragment’s cumulative glocal alignment score corresponded to a single chromosome in the other species. Using this procedure, we assigned 123 C. savignyi reftigs to chromosomes (Supplemental Table 3), amounting to an additional 11% (18.8 Mb) of the sequenced genome. Applying the converse procedure to unmapped scaffolds in C. intestinalis, we mapped 155 C. intestinalis scaffolds to chromosomes (Supplemental Table 4), also amounting to an additional 11% (18.6 Mb).
Discussion
Traditionally, the construction of a genetic map precedes the generation of a whole-genome sequence, and involves the laborious creation of a physical map as an intermediate. For C. savignyi, the availability of a high-quality genome sequence greatly facilitated construction of a genetic map, and obviated the need for a physical map. The sequence enabled us to generate reliable markers despite the extreme level of variation in this species; because of the variation, these markers were highly informative. The strategy we employed for generating the map may serve as a model for building genetic maps for other organisms, many of which will also have their genomes sequenced before a genetic map is built, and many of which will also be highly polymorphic.
CAINS markers
Development of CAINS markers was a key part of the strategy to leverage the genome sequence for genetic map construction. In general, the most useful genetic marker assays capture lots of polymorphism while allowing reliable amplification, and they are easily shared and detected by different laboratories working on different mapping populations; the final set of marker assays should be widely dispersed to maximize the fraction of the genome that is tightly linked to a marker. The set of CAINS makers we developed meet all of these criteria, and should additional marker development be required in the future, it will be straightforward to add to the initial CAINS set because of the large number of potential CAINS sites in the genome, and because the map position of most future marker candidates is predictable from our current work.
Relative to SSLP markers, CAINS markers do require more work in their initial characterization. Each CAINS marker must be individually amplified, restriction-digested, and examined to determine if multiple alleles are segregating. However, SSLP markers were determined not to be an option in this species due to a prohibitively high rate of amplification failure. Other markers, such as AFLP, are useful for mapping individual loci (Veeman et al. 2008), but cannot be targeted a priori to specific regions in the genome.
The C. savignyi genetic map
Targeting marker development to specific regions on the reftigs facilitated construction of an apparently complete, high-quality map. Conventional mapping strategies generally use randomly distributed markers, but chance and biases in the distribution of these markers often result in maps with substantial gaps in coverage. For example, prior to the work of Jacob and colleagues (Jacob et al. 1995) there were 550 loci on the 1500-cM map of the rat, but because of the uneven distribution of these markers there were still multiple linkage groups on at least five chromosomes. Jacob and colleagues finally achieved full coverage of the rat genetic map with a concerted effort using SSLP makers at a density of 0.29 Loci per Centimorgan (L/cM) (Jacob et al. 1995). In zebrafish, an early map constructed of RAPD makers to a density of 0.16 L/cM contained four more linkage groups than chromosomes (Postlethwait et al. 1994). The marker density of the C. savignyi map is 0.14 L/cM lower than the efforts mentioned, but the number of linkage groups (14) is consistent with the number of chromosomes observed in this species (Colombera et al. 1988), suggesting completeness.
Additional evidence also suggests that the map is complete. The remaining unmapped reftigs appear to be unbiased in their distribution among the chromosomes. The additional 11% of the genome mapped by comparison with C. intestinalis showed roughly even distribution among the existing chromosomes. Therefore, the 24 Mb of remaining unmapped sequence might also be expected to distribute evenly among the remaining 14 chromosomes. In this scenario, just 1.7 Mb (8.5 cM) per chromosome remains unmapped. Furthermore, the remaining sequence is likely to simply intercalate into the gaps between mapped assembly fragments. While coverage is not exceedingly dense, the relatively even spacing of markers, on average, put any point in the genome within 4.4 cM of the nearest marker.
While we made every reasonable effort to resolve disagreements between the genetic map and sequence assembly, a small number of disagreements remain. These observed disagreements are not unexpected, and are common when genetic maps and sequence assemblies are integrated. Without additional sequence data, it is impossible to determine whether differences are due to misassemblies in the reference sequence or are the result of polymorphisms between the mapping population and the sequenced individual. However, given the extreme frequency of nucleotide level and insertion/deletion polymorphism found in this organism, it would not be surprising to detect rarer types of polymorphisms such as intrachromosomal rearrangements.
The C. savignyi sequence assembly as the basis for genetic map construction
Of the 374 reftigs of the C. savignyi reference sequence, we definitively ordered 104, and putatively placed an additional 123, on the genetic map. Cumulatively, 227 reftigs representing 94% (167 Mb) of the sequenced genome have been placed onto chromosomes. A brief review of the way in which genomic resources for C. savignyi were generated over the past few years may be instructive in this context. The C. savignyi genome was sequenced to almost 13× coverage, due to an increase in the number of high-quality base pairs that could be obtained from Sanger sequence traces while the sequencing effort was ongoing. The high coverage, though obtained serendipitously, turned out to be a key investment that facilitated downstream efforts. First, the sequence was assembled such that the two haplomes assembled separately, at an average depth of >6× coverage. In effect, the “double assembly” was like a single assembly at draft level coverage, and as a result the contiguity and size of assembly fragments was already of high quality (Vinson et al. 2005). Then, our group generated a single reference sequence from the double assembly, which resulted in 374 “reftigs” of an N50 length of 1.8 Mb (Small et al. 2007a). In the course of doing so, we also obtained insights into the dynamics of genetic variation in large populations that would have been impossible to obtain with a standard assembly strategy (Small et al. 2007b). The small number of reftigs, and their large average length, allowed the direct construction of a genetic map without a physical map intermediate. In our view, had the initial sequence data been only slightly less deep, our strategy would have not worked at all, or required (at every stage) far more effort. Both the reference sequence as well as the genetic map would have likely been of considerably lower comprehensiveness and quality had the initial investment in very deep sequencing not been made.
Retention of synteny since the last common ancestor of C. savignyi and C. intestinalis
Ordering of the reference genome sequence according to the genetic map enabled the comparison of larger genome structure between the two Ciona species. For 12 of 14 C. savignyi chromosomes, a single syntenic chromosome was identified in C. intestinalis. Of the two remaining chromosomes in C. savignyi, further analysis revealed that chromosome 1 contained two distinct regions of synteny. The region of chromosome 1 between 0 cM and 64.5 cM is syntenic to C. intestinalis chromosome 1, while the remainder is syntenic to C. intestinalis chromosome 3. This observation is best explained by a relatively recent chromosomal fusion in C. savignyi. Only recent fusion in C. savignyi would result in distinct blocks of syntenic, but highly rearranged, sequence on a single C. savignyi chromosome. This change in karyotype is remarkable considering the otherwise stable set of chromosomes between these species. The few rearrangements that did occur between the two distinct regions of chromosome 1 suggest that the fusion is old enough not to be a segregating polymorphism, though we cannot rule out this possibility with the available evidence.
The only C. savignyi chromosome lacking a syntenic match is 14, which appears to be highly repetitive and might simply represent a small degenerate chromosome unique to C. savignyi and similar to chromosome 4 in D. melanogaster (Sun et al. 2000). Interestingly, we also found that the alleles of linkage group 14 markers that were derived from one parent were weakly linked to chromosome 6. How it is possible that the markers show linkage in only one parent is a matter of speculation. One explanation is that the mapping population contains a segregating karyotype polymorphism in which one parent harbors a fusion of chromosomes 6 and 14.
The Cionas as evidence of the deleteriousness of interchromosomal rearrangements in vertebrates
Comparisons among flies and worms have shown a plethora of intrachromosomal rearrangements among a dearth of interchromosomal rearrangements within each clade (Richards et al. 2005; Drosophila 12 Genomes Consortium 2007; Hillier et al. 2007). Our comparisons indicate that the Cionas, despite a larger number of chromosomes and a statistically greater chance of exhibiting interchromosomal changes, exhibit the same extreme bias toward intrachromosomal events. In contrast, among the many vertebrates that have been compared (human, mouse, rat, dog, chicken, and certain fishes), both types of rearrangement are common. This difference between the vertebrates and invertebrates could be due to inherent biological differences, or the relatively small population sizes of vertebrates, which prevents the purging of mildly deleterious mutations by selection.
The Cionas have a very large effective population size (Ne ∼ 1.5 × 106) (Small et al. 2007b). In light of Kimura’s Neutral Theory (Kimura 1983), which states that selection is more efficient in larger populations, the paucity of interchromosomal rearrangements provides strong support that such rearrangements are deleterious in these species, as might be expected since such rearrangements can produce aneuploid genotypes. Flies and worms show a similar pattern of genome rearrangement, and also have relatively large populations sizes (Fay et al. 2002; Cutter et al. 2006; Hillier et al. 2007). In most vertebrates, however, the balance of rearrangements is much less skewed toward intrachromosomal events (Mouse Genome Sequencing Consortium 2002; Bourque et al. 2004, 2005; Gibbs et al. 2004; Zhao et al. 2004; Jaillon et al. 2004; Woods et al. 2005; Kasahara et al. 2007), and vertebrates have population sizes one to four orders of magnitude smaller than the invertebrates discussed here (Frankham 1995; Chimpanzee Sequencing and Analysis Consortium 2005; Tenesa et al. 2007). Thus, one parsimonious explanation for this fundamental difference between invertebrates and vertebrates might be that vertebrates as a clade have generally had population sizes in which fixation of a greater number of potentially deleterious alleles is favored by the inability of selection to overcome the effects of genetic drift.
Methods
Mapping population
Linkage was evaluated in 48 F1 progeny resulting from a cross of two wild C. savignyi individuals obtained from Santa Barbara, California. Sperm and eggs were first isolated from both parents and then mixed. The larvae were allowed to settle onto plates and were reared in tanks supplied with a continuous flow of fresh seawater for ∼2 mo. Muscle tissue was excised from adults, and genomic DNA was extracted.
Genetic markers
Each marker assay consisted of a pair of PCR primers and a specific restriction enzyme used to reveal restriction fragment length polymorphisms among alleles. Like cleaved amplified polymorphic sequences (CAPS) (Konieczny and Ausubel 1993), each pair of primers was designed to amplify a specific segment of genomic DNA selected from the genome sequence. Primers for each candidate marker were always designed to bind to adjacent exons and amplify the intervening intronic sequence. We call these markers cleaved amplified intronic sequences (CAINS). As C. savignyi is diploid, there were up to four banding patterns for each locus, with each one representing a composite pattern of two cleaved allelic amplicons.
The exons used for generating candidate markers were annotated in version 1.0 of C. savignyi assembly using the Ensembl genome annotation pipeline. Only exons supported by alignment to either a C. savignyi EST or a homolog in another species were used. Some additional candidate markers were generated using the publicly available genome annotation from Ensembl (www.ensembl.org/Ciona_savignyi), which is based on version 2.1 of the C. savignyi assembly.
Synonymous substitution polymorphisms are common in coding regions of Ciona because of the generally high polymorphism rate; therefore, only primer binding sites with limited potential for mismatches, particularly in the 3′-end of the binding site, were selected. We used Primer3 (Rozen and Skaletsky 2000) to identify an initial set of high-quality primer binding sites independently from the upstream and downstream exon sequences. Upon these potential upstream and downstream binding sites, we then imposed a set of criteria to limit potential mismatches. The criteria were based on the degeneracy of the codons encoding the amino acid sequence at the binding site. The upstream binding site criteria were:
the 5′ end must begin on the first position of a codon;
the 3′ end must terminate on the second position of a codon that is fourfold degenerate or less;
the two codons preceding the 3′ terminal codon must be twofold degenerate or less.
The downstream binding site criteria were:
the 5′ end must begin on the second position of a codon;
the 3′ end must terminate on the first position of a codon;
the two 3′ terminal codons must be twofold degenerate or less.
Because of these criteria, binding site lengths were constrained to multiples of three. Empirically, we found amplifications improved with longer primers; therefore, only primers of 20, 23, or 26 nucleotides (nt) were used. After filtering binding sites identified by Primer3 by our criteria, all combinations of the remaining acceptable upstream and downstream primers were added to a database of candidate markers.
Marker selection and characterization
Prior to mapping, at least one polymorphic marker to each target region had to be identified. Target regions were selected on reftigs >650 kb in size. Previous small-scale mapping efforts (not shown) suggested that recombination events would be rare between loci <1.5 Mb apart in a mapping cross the size of the one used in this study (96 meioses). On reftigs <1.5 Mb, one target region was selected. On fragments 1.5–3 Mb, two widely spaced target regions were selected. On the small number of fragments >3 Mb, three well-spaced target regions were selected. Uncharacterized markers from each target region were drawn from our large database of candidate CAINS markers. From these markers, four with the best annealing temperature, length, and other characteristics were selected for evaluation and characterization.
Due to the large number of markers evaluated, we used a three-stage process to identify at least one marker from each target region that was informative for recombination, preferably for both parents. In the first stage, potentially polymorphic loci were identified. Amplification with the markers from each region was tested using genomic DNA pooled from all individuals in the cross. Amplicons were then digested with each of the eight restriction enzymes used. The resulting banding patterns were evaluated for brightness and number of bands. Complex banding patterns were indicative of the number of segregating alleles. In the second stage, we selected two robustly amplifying markers with complex patterns and proceeded with amplification and digestion from 12 individuals from the cross. This let us determine if multiple distinguishable alleles were segregating. We often repeated stage two in order to identify a marker and enzyme combination revealing four banding patterns among the mapping progeny, which was indicative of a fully informative marker locus.
All PCR amplifications were carried out in Applied Biosystems optical 384-well reaction plates on an Applied Biosystems 9700 PCR machine. Common PCR reaction buffer (10 mM Tris, 45 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs) and conventional Taq polymerase were used for all assays. The PCR cycling conditions were as follows: initial melting: 94°C for 30 sec; 35 cycles amplification: 94°C for 15 sec, 56°C for 15 sec, and 72°C for 1 min; final elongation: 72°C for 3 min.
All amplicons were digested with one of eight restriction enzymes (AccII, AluI, HaeIII, HhaI, MboI, MseI, MspI, RsaI), each representing a different 4-bp palindrominc recognition sites. All digests were carried out in 1× Buffer 2 from New England Biolabs. For consistency, we adjusted all digests to 10 mM MgCl2, accommodating for the quantity of PCR reaction mixture digested.
Digestion fragments were separated by agarose gel electrophoresis. The agarose used was a combination of 1.4% (w/v) Seakem LE (Cambrex Bio Science Rockland, Inc.) and 1.5% (w/v) Metaphor agarose (Cambrex Bio Science Rockland, Inc.) prepared with standard 1× Tris-Borate-EDTA (TBE) buffer. Electrophoresis was carried out in large-format horizontal gel boxes (Owl A6 Millipede, Thermo Fisher Scientific, Inc.). Four multichannel pipette-compatible combs were used to run 196 reactions per gel. The electrophoresis buffer used was 0.5× TBE, and gels were typically electrophoresed at 10 V/cm. Separated bands were visualized by post-staining with ethidium bromide in water (0.10 μg/mL).
Mapping
Assessment of linkage between pairs of markers
We initially considered all markers with four distinct marker phenotypes (banding patterns). Denoting the ordered parental genotypes as AB and CD, the F1 progeny have genotypes of either AC, AD, BC, or BD. If the relationship between marker genotype and phenotype (banding pattern) was known, establishment of linkage could be assessed by simply counting recombinants. In the absence of such information, we consider all possible ways to assign the banding patterns to genotypes. While at a given marker there are 24 possible ways to assign banding patterns to genotypes, our inability to distinguish the order of the two parents, or the order of the two haplotypes within a parent, results in 72 unique ways to assign the banding patterns for the two markers to genotypes. For each of these 72 possible assignments, we estimate the recombination fraction between the two markers (assuming no sex difference in recombination) and a LOD score (log10 likelihood ratio) comparing the hypothesis that the two markers are linked to the hypothesis that they are not linked (and so have recombination fraction = 1/2). The inferred assignment is that with the greatest LOD score (and so the greatest likelihood). A more detailed description of the methodology can be found in the Supplemental material.
For each of the markers with just two or three distinct banding patterns, we considered linkage to each of the fully informative markers, and not to each other. The establishment of linkage required that we consider all possible partitions of the four marker genotypes to the two or three observed banding patterns. The establishment of linkage between a partially informative marker and a fully informative marker cannot be accomplished by simply counting recombination events, as for many individuals it will not be clear, for example, whether there was 0 or 1 recombination events. Estimation of the recombination fraction between two markers was thus accomplished by maximum likelihood via the EM algorithm (Dempster et al. 1977).
Formation of linkage groups
The pairwise linkage results (among all fully informative markers and between incompletely informative markers and the fully informative markers) were used to establish initial linkage groups. Two markers were placed in the same group if the estimated recombination fraction between them was not >0.25 and the LOD score for a test of linkage was at least 4.5. The transitive property (if A is linked to B and B is linked to C, then A is linked to C) was used to close the linkage groups.
Parental haplotype inference within each linkage group
We used the pairwise linkage information to infer the parental haplotypes within each linkage group, though recognizing that the order of the two parents and the order of the two haplotypes within each parent cannot be identified. (Thus, the haplotypes in one linkage group cannot immediately be attached to the haplotypes in another linkage group.) The haplotypes were formed starting with a pair of closely linked markers, and then working through the rest of the markers in the linkage group, considering one additional marker at a time.
Ordering of markers within linkage groups and multipoint estimation of intermarker distances
Taking the inferred parental haplotypes to be known, marker order was determined by considering all possible orders of markers or, for the larger linkage groups, all possible orders for a sliding window of markers. The chosen order was that with the maximum likelihood (that is, the marker order for which the observed data were most probable). Multipoint estimates of the recombination fractions between markers were also estimated by maximum likelihood, assuming no crossover interference. Likelihood calculations were performed via the Lander-Green algorithm (Lander and Green 1987). The estimated recombination fractions between adjacent markers were transformed to genetic distances using the Haldane map function (Haldane 1919). After the initial establishment of marker order, we used the locations of markers within reftigs to refine marker order, when possible.
Large gaps in the estimated linkage maps indicated the possibility that a linkage group should be split in two. In such cases, we calculated a LOD score comparing the hypothesis that the two linkage groups should remain merged with the hypothesis that they should be distinct. A linkage group was split into two if this LOD score was not large (<3). We similarly considered merging pairs of linkage groups; doing so required the consideration of the eight possible connections between the inferred haplotypes in one linkage group with those in a second linkage group.
Pairwise linkage calculations, the establishment of linkage groups, and the inference of parental haplotypes were accomplished with perl scripts written specific to the current data. The establishment of marker order and the multipoint estimation of intermarker distances were accomplished with R/qtl (Broman et al. 2003), an add-on package for the general statistical software, R (Ihaka and Gentleman 1996).
Synteny strength
The extent of alignment between two chromosomes was determined by calculating the percent score (PS) or percent length (PL). PS and PL were defined as the percentage of the sum of individual alignment scores or sum of individual alignment lengths, for PS and PL, respectively, of all alignments associated with a chromosome in one species that corresponded to a particular chromosome in the second species. To do this, we took advantage of a publicly available data set containing the whole-genome pairwise alignment of C. savignyi and C. intestinalis, constructed by Brudno and colleagues (Brudno et al. 2003, 2007) and available from http://pipeline.lbl.gov/data/Cioin2_cioSav2/. Each of the alignments in this data set had an associated length, score, and type. The type (M1, M2, and DM) indicated whether the aligned segment of either species was involved in other alignments (M1 or M2), or if the segments aligned singly and reciprocally to each other (DM, for dual-monotonic). To enrich for orthologous sequences, we used only the DM alignments. First, we remapped the alignments, which were based on the reftigs of version 2.0 of in the C. savignyi sequence (Small et al. 2007a), to the chromosomes we constructed. We then calculated PS and PL from the scores and lengths of each alignment. The total sum of scores or total sum of lengths for all alignments involving chromosome i was denoted Ti. The sum of the scores or lengths of all the alignments between an interspecies chromosome pair was denoted Sij. Thus, PS and PL were calculated as 100%*Sij/Ti for scores or lengths, respectively. Note that while Sij = Sji, the value of PS and PL depended on which species was being considered (because Sij/Ti ≠ Sji/Tj).
Acknowledgments
We thank Shawn Hoon and Balamurugan Kumarasamy for help with the genome annotation; Kerrin Small for prepublication access to the improved C. savignyi genome assembly; and Mehdi Yahyanejad and Phil Lacroute for discussions about methodology. This work was supported by NIH grants R24GM076171 to A.S. and HD038701 to W.C.S. M.M.H. was supported by the Stanford Genome Training Program (NIH/NHGRI).
Footnotes
[Supplemental material is available online at www.genome.org and at mendel.stanford.edu/sidowlab/ciona.html.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078576.108.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bourque G., Pevzner P.A., Tesler G., Pevzner P.A., Tesler G., Tesler G. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 2004;14:507–516. doi: 10.1101/gr.1975204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., Zdobnov E.M., Bork P., Pevzner P.A., Tesler G., Zdobnov E.M., Bork P., Pevzner P.A., Tesler G., Bork P., Pevzner P.A., Tesler G., Pevzner P.A., Tesler G., Tesler G. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res. 2005;15:98–110. doi: 10.1101/gr.3002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., Churchill G.A., Wu H., Sen S., Churchill G.A., Sen S., Churchill G.A., Churchill G.A. R/qtl: Qtl mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Brown C.D., Johnson D.S., Sidow A., Johnson D.S., Sidow A., Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Brudno M., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Do C.B., Couronne O., Dubchak I., Batzoglou S., Couronne O., Dubchak I., Batzoglou S., Dubchak I., Batzoglou S., Batzoglou S. Glocal alignment: Finding rearrangements during alignment. Bioinformatics. 2003;(Suppl. 1) 19:i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- Brudno M., Poliakov A., Minovitsky S., Ratnere I., Dubchak I., Poliakov A., Minovitsky S., Ratnere I., Dubchak I., Minovitsky S., Ratnere I., Dubchak I., Ratnere I., Dubchak I., Dubchak I. Multiple whole genome alignments and novel biomedical applications at the VISTA portal. Nucleic Acids Res. 2007;35:W669–W674. doi: 10.1093/nar/gkm279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Colombera D., Nishikawa T., Tagliaferri F., Nishikawa T., Tagliaferri F., Tagliaferri F. The chromosomes of 13 enterogonid ascidians from japan. Cytologia (Tokyo) 1988;53:45–51. [Google Scholar]
- Corbo J.C., Levine M., Zeller R.W., Levine M., Zeller R.W., Zeller R.W. Characterization of a notochord-specific enhancer from the brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Cutter A.D., Baird S.E., Charlesworth D., Baird S.E., Charlesworth D., Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–913. doi: 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B., Levine M., Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc. Natl. Acad. Sci. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Satou Y., Campbell R.K., Chapman J., Degnan B., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Campbell R.K., Chapman J., Degnan B., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Chapman J., Degnan B., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Degnan B., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Tomaso A.D., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Davidson B., Gregorio A.D., Gelpke M., Goodstein D.M., Gregorio A.D., Gelpke M., Goodstein D.M., Gelpke M., Goodstein D.M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H., Brinkmann H., Chourrout D., Philippe H., Chourrout D., Philippe H., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dempster A.P., Laird N.M., Rubin D.B., Laird N.M., Rubin D.B., Rubin D.B. Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Stat. Soc. B. 1977;39:1–38. [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Fay J.C., Wyckoff G.J., Wu C.-I., Wyckoff G.J., Wu C.-I., Wu C.-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1995;10:1500–1508. [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scott G., Steffen D., Worley K.C., Burch P.E., Steffen D., Worley K.C., Burch P.E., Worley K.C., Burch P.E., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. The combination of linkage values, and the calculation of distances between the loci of linked factors. J. Genet. 1919;8:299–309. [Google Scholar]
- Harafuji N., Keys D.N., Levine M., Keys D.N., Levine M., Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L.W., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H., Fulton L.A., Koboldt D.C., Waterston R.H., Koboldt D.C., Waterston R.H., Waterston R.H. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R., Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Jacob H.J., Brown D.M., Bunker R.K., Daly M.J., Dzau V.J., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Brown D.M., Bunker R.K., Daly M.J., Dzau V.J., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Bunker R.K., Daly M.J., Dzau V.J., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Daly M.J., Dzau V.J., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Dzau V.J., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Goodman A., Koike G., Kren V., Kurtz T., Lernmark A., Koike G., Kren V., Kurtz T., Lernmark A., Kren V., Kurtz T., Lernmark A., Kurtz T., Lernmark A., Lernmark A., et al. A genetic linkage map of the laboratory rat, Rattus norvegicus. Nat. Genet. 1995;9:63–69. doi: 10.1038/ng0195-63. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J.-M., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Aury J.-M., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Fischer C., Ozouf-Costaz C., Bernot A., Ozouf-Costaz C., Bernot A., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Johnson D.S., Davidson B., Brown C.D., Smith W.C., Sidow A., Davidson B., Brown C.D., Smith W.C., Sidow A., Brown C.D., Smith W.C., Sidow A., Smith W.C., Sidow A., Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Yamada T., Nagayasu Y., Doi K., Kasai Y., Nagayasu Y., Doi K., Kasai Y., Doi K., Kasai Y., Kasai Y., et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- Konieczny A., Ausubel F.M., Ausubel F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Green P., Green P. Construction of multilocus genetic linkage maps in humans. Proc. Natl. Acad. Sci. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen I.A., Lemaire P., Okamura Y., Lemaire P., Okamura Y., Okamura Y. The neurobiology of the ascidian tadpole larva: Recent developments in an ancient chordate. Annu. Rev. Neurosci. 2004;27:453–485. doi: 10.1146/annurev.neuro.27.070203.144255. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. iii. Up to the tissue restricted stage. Dev. Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Postlethwait J.H., Johnson S.L., Midson C.N., Talbot W.S., Gates M., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Johnson S.L., Midson C.N., Talbot W.S., Gates M., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Midson C.N., Talbot W.S., Gates M., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Talbot W.S., Gates M., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Gates M., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Ballinger E.W., Africa D., Andrews R., Carl T., Eisen J.S., Africa D., Andrews R., Carl T., Eisen J.S., Andrews R., Carl T., Eisen J.S., Carl T., Eisen J.S., Eisen J.S., et al. A genetic linkage map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Satoh N., Satou Y., Davidson B., Levine M., Satou Y., Davidson B., Levine M., Davidson B., Levine M., Levine M. Ciona intestinalis: An emerging model for whole-genome analyses. Trends Genet. 2003;19:376–381. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- Shoguchi E., Kawashima T., Satou Y., Hamaguchi M., Sin-I T., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Kawashima T., Satou Y., Hamaguchi M., Sin-I T., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Satou Y., Hamaguchi M., Sin-I T., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Hamaguchi M., Sin-I T., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Sin-I T., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Kohara Y., Putnam N., Rokhsar D.S., Satoh N., Putnam N., Rokhsar D.S., Satoh N., Rokhsar D.S., Satoh N., Satoh N. Chromosomal mapping of 170 bac clones in the ascidian Ciona intestinalis. Genome Res. 2006;16:297–303. doi: 10.1101/gr.4156606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K.S., Brudno M., Hill M.M., Sidow A., Brudno M., Hill M.M., Sidow A., Hill M.M., Sidow A., Sidow A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007a;8:R41. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K.S., Brudno M., Hill M.M., Sidow A., Brudno M., Hill M.M., Sidow A., Hill M.M., Sidow A., Sidow A. Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. 2007b;104:5698–5703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F.L., Cuaycong M.H., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Cuaycong M.H., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Craig C.A., Wallrath L.L., Locke J., Elgin S.C., Wallrath L.L., Locke J., Elgin S.C., Locke J., Elgin S.C., Elgin S.C. The fourth chromosome of Drosophila melanogaster: Interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A., Navarro P., Hayes B.J., Duffy D.L., Clarke G.M., Goddard M.E., Visscher P.M., Navarro P., Hayes B.J., Duffy D.L., Clarke G.M., Goddard M.E., Visscher P.M., Hayes B.J., Duffy D.L., Clarke G.M., Goddard M.E., Visscher P.M., Duffy D.L., Clarke G.M., Goddard M.E., Visscher P.M., Clarke G.M., Goddard M.E., Visscher P.M., Goddard M.E., Visscher P.M., Visscher P.M. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Nakatani Y., Hendrickson C., Ericson V., Lin C., Smith W.C., Nakatani Y., Hendrickson C., Ericson V., Lin C., Smith W.C., Hendrickson C., Ericson V., Lin C., Smith W.C., Ericson V., Lin C., Smith W.C., Lin C., Smith W.C., Smith W.C. Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J.P., Jaffe D.B., O’Neill K., Karlsson E.K., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Jaffe D.B., O’Neill K., Karlsson E.K., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., O’Neill K., Karlsson E.K., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Karlsson E.K., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Stange-Thomann N., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Anderson S., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Mesirov J.P., Satoh N., Satou Y., Nusbaum C., Satoh N., Satou Y., Nusbaum C., Satou Y., Nusbaum C., Nusbaum C., et al. Assembly of polymorphic genomes: Algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods I.G., Wilson C., Friedlander B., Chang P., Reyes D.K., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Wilson C., Friedlander B., Chang P., Reyes D.K., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Friedlander B., Chang P., Reyes D.K., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Chang P., Reyes D.K., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Reyes D.K., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Nix R., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Kelly P.D., Chu F., Postlethwait J.H., Talbot W.S., Chu F., Postlethwait J.H., Talbot W.S., Postlethwait J.H., Talbot W.S., Talbot W.S., et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Shetty J., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Shetty J., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Hou L., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Delcher A., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Zhu B., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Osoegawa K., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., de Jong P., Nierman W.C., Strausberg R.L., Fraser C.M., Nierman W.C., Strausberg R.L., Fraser C.M., Strausberg R.L., Fraser C.M., Fraser C.M., et al. Human, mouse, and rat genome large-scale rearrangements: Stability versus speciation. Genome Res. 2004;14:1851–1860. doi: 10.1101/gr.2663304. [DOI] [PMC free article] [PubMed] [Google Scholar]