Abstract
To identify human intronic sequences associated with 5′ splice site recognition, we performed a systematic search for motifs enriched in introns downstream of both constitutive and alternative cassette exons. Significant enrichment was observed for U-rich motifs within 100 nucleotides downstream of 5′ splice sites of both classes of exons, with the highest enrichment between positions +6 and +30. Exons adjacent to U-rich intronic motifs contain lower frequencies of exonic splicing enhancers and higher frequencies of exonic splicing silencers, compared with exons not followed by U-rich intronic motifs. These findings motivated us to explore the possibility of a widespread role for U-rich motifs in promoting exon inclusion. Since cytotoxic granule-associated RNA binding protein (TIA1) and TIA1-like 1 (TIAL1; also known as TIAR) were previously shown in vitro to bind to U-rich motifs downstream of 5′ splice sites, and to facilitate 5′ splice site recognition in vitro and in vivo, we investigated whether these factors function more generally in the regulation of splicing of exons followed by U-rich intronic motifs. Simultaneous knockdown of TIA1 and TIAL1 resulted in increased skipping of 36/41 (88%) of alternatively spliced exons associated with U-rich motifs, but did not affect 32/33 (97%) alternatively spliced exons that are not associated with U-rich motifs. The increase in exon skipping correlated with the proximity of the first U-rich motif and the overall “U-richness” of the adjacent intronic region. The majority of the alternative splicing events regulated by TIA1/TIAL1 are conserved in mouse, and the corresponding genes are associated with diverse cellular functions. Based on our results, we estimate that ∼15% of alternative cassette exons are regulated by TIA1/TIAL1 via U-rich intronic elements.
Splicing is the process that ensures the production of functional mRNA from precursor (pre)-mRNA in eukaryotic organisms. It entails the accurate, covalent joining of exon sequences and removal of intron sequences by the spliceosome, a multisubunit complex consisting of five small nuclear RNAs (snRNAs) and a multitude of protein factors (Kramer 1996). Splicing relies on the identification of short and loosely conserved sequences, namely, the 5′ and 3′ splice sites, and the intronic branch site and polypyrimidine tract upstream of the 3′ splice site (Kramer 1996). The core splicing signals are necessary but insufficient to promote accurate splicing, as numerous sequences of similar functional potential as bona fide splice sites, termed pseudo splice sites, are present in pre-mRNAs. To ensure proper splicing, additional sequences located in exons and introns function to promote (enhancers) or prevent (silencers) splice site recognition. Together with the core splicing signals summarized above, enhancer and silencer elements comprise a major component of what has been termed the splicing code (for reviews, see Cartegni et al. 2002; Matlin et al. 2005; Blencowe 2006). In addition to their critical roles in the recognition and regulation of splice site selection in a cell type–independent manner, specific enhancers and silencers are also important elements for the regulation of alternative splicing (AS) in a cell/tissue, differentiation/developmental stage, or condition-specific manner.
During the past several years, much interest has been directed toward the identification of splicing regulatory sequences (for review, see Chasin 2007). Initial approaches involved the analysis of disease alleles and experimentally directed mutations that affect splicing of minigene reporter transcripts (Pagani et al. 2000; Cartegni and Krainer 2002; Pagani et al. 2003; Cartegni et al. 2006). SELEX (systematic evolution of ligands by exponential enrichment) methodology was employed to identify sequences in random oligonucleotide pools that promote splicing when inserted into exon sequences (Liu et al. 1998; Cavaloc et al. 1999). More recently, statistical and computational approaches using large data sets of genomic and transcript sequences have been successfully applied to the identification of enhancers or silencers (Fairbrother et al. 2002; Zhang and Chasin 2004; Zhang et al. 2005; Stadler et al. 2006). Fewer studies, however, have focused on the identification of functional intronic splicing elements, although several very recent reports have employed comparative genomic sequence analyses to identify splicing elements flanking constitutive (Zhang et al. 2005; Voelker and Berglund 2007; Yeo et al. 2007) and alternative exons (Minovitsky et al. 2005; Voelker and Berglund 2007; Yeo et al. 2007).
Exonic and intronic regulatory elements are thought to be primarily recognized by splicing factors in a sequence-specific manner. These factors generally harbor conserved RNA binding domains such as RNA recognition motifs (RRMs) and additional domains that function to recruit other factors to the pre-mRNA or to bridge to other factors already bound to the pre-mRNA. Ultimately, binding of factors to enhancers and silencers regulates formation of the spliceosome (Jurica and Moore 2003). To date, numerous splicing factors have been identified and shown to bind to exonic regulatory elements, including the well studied family of serine-arginine-repeat (SR) proteins (Graveley 2000; Lin and Fu 2007). Splicing factors that bind to silencer sequences include members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family (Zhu et al. 2001; Caceres and Kornblihtt 2002; Martinez-Contreras et al. 2007). However, depending on the sequence context and location with respect to splice sites, SR proteins and hnRNP proteins can also function to repress or promote splice site recognition, respectively (e.g., Hui et al. 2005).
Other examples of splicing factors are the cytotoxic granule-associated RNA binding protein (TIA1) and TIA1-like 1 (TIAL1; also referred to as TIAR) (Del Gatto-Konczak et al. 2000). These proteins are closely related and each contains three RRMs and a glutamine (Q)-rich domain (Tian et al. 1991; Kawakami et al. 1992). Using in vitro SELEX, TIA1 and TIAL1 were shown to preferentially bind uridine-rich (U-rich) sequences (Dember et al. 1996). In addition to their roles in splicing, TIA1 and TIAL1 function in translation repression by aggregating with mRNAs to form stress granules (Lopez de Silanes et al. 2005). A signature U-rich motif was identified in the 3′ untranslated region of gene targets repressed by TIA1 (Lopez de Silanes et al. 2005). In the context of splicing, it was shown in vitro that TIA1 binds to U-rich sequences downstream of 5′ splice sites and that it interacts with the U1 snRNP-specific U1-C protein through its Q-rich domain to recruit U1snRNP to 5′ splice sites (Forch et al. 2002). The interaction between TIA1 and U1 snRNP mutually stabilizes the association of these two factors with the 5′ splice site region (Del Gatto-Konczak et al. 2000; Gesnel et al. 2007). Consistent with important roles for TIA1 or TIAL1 proteins in promoting splicing via binding U-rich sequences, several in vitro studies employing different pre-mRNA substrates have shown that single or multiple mutations in U-rich sequences proximal to 5′-splice sites can affect the inclusion levels of the adjacent, upstream exons (Le Guiner et al. 2001; Shukla et al. 2004; Zuccato et al. 2004). Not surprisingly, given the many similarities between TIA1 and TIAL1, it has also been shown previously that these proteins have redundant activities in splicing (Le Guiner et al. 2003; Izquierdo et al. 2005; Shukla et al. 2005).
In order to systematically identify putative intronic splicing elements, we performed a search for motifs that are enriched in intronic sequences downstream of 5′ splice sites. U-rich motifs of 5–10 nucleotides (nt) were among the most highly enriched sequences and displayed peaks within positions +6 to +30 downstream of 5′ splice sites of both constitutive and alternative exons. Both exon classes with flanking regions that contain U-rich sequences have lower frequencies of exonic splicing enhancers (ESEs) and higher frequencies of exonic splicing silencers (ESSs), compared with exons that are located upstream of introns that lack U-rich sequences. Simultaneous knockdown of TIA1 and TIAL1 proteins in HeLa cells resulted in increased exon skipping of alternatively spliced exons located upstream of U-rich introns. The extent of increase in exon skipping correlated with the overall “U-richness” of the adjacent intronic sequence and proximity of the first U-rich motif to the exon–intron boundary. Based on our findings, we estimate that ∼15% of cassette-type alternative exons (i.e., exons that can be skipped or included in mRNA) are associated with adjacent U-rich intronic sequences and regulated by TIA1/TIAL1 proteins. These regulated AS events are often conserved between human and mouse, and the corresponding genes are associated with diverse cellular functions.
Results
Motif searching in intronic sequences adjacent to 5′ splice sites
We employed a computational approach to identify putative splicing regulatory elements in intronic sequences downstream of 5′ splice sites. Two different populations of 100-nt intronic sequences were analyzed. One population (“Const”) was obtained from the Hollywood database (Holste et al. 2006) and consists of 109,225 intronic sequences adjacent to constitutively spliced exons identified using alignments of human genomic, cDNA and EST sequences (Holste et al. 2006). The other population (“Alt”) consists of 3872 intron sequences adjacent to recognized cassette-type alternative exons, which were also identified using alignments of human cDNA and EST sequences (Pan et al. 2006). For comparison and control purposes, 100 nt of “deep” intronic sequences (500 nt downstream of the exon–intron boundary) from both sets (Const and Alt) of genes were retrieved.
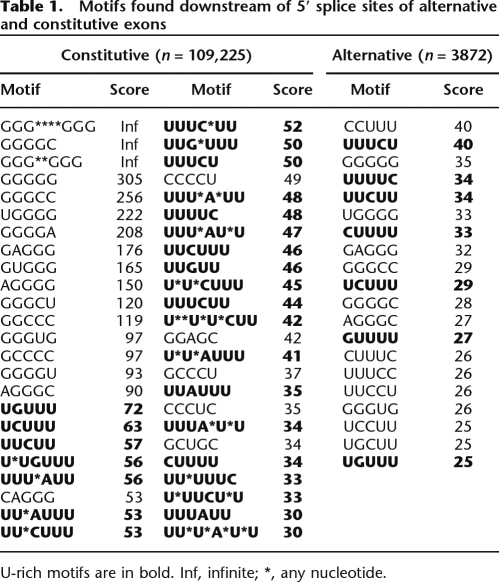
To identify motifs that are enriched in the Const and Alt populations of intronic sequences, SeedSearcher (Barash et al. 2001; Fagnani et al. 2007) was employed. This algorithm searches for enrichment of motifs of variable length and composition in test sequences relative to control sequences. In order to exclude the 5′ splice site consensus sequence, searches were conducted at each position starting from nucleotide +7 in the Const and Alt (test) populations and were compared to the searches in the deep intronic (control) sequences. The set of motifs that are the most significantly enriched in the Const and Alt intronic sequences are reported in Table 1. Most of these motifs can be classified as G-rich motifs or U-rich motifs (Table 1, shown in bold; see below).
Table 1.
Motifs found downstream of 5′ splice sites of alternative and constitutive exons
U-rich motifs are in bold. Inf, infinite; *, any nucleotide.
A recent genome-wide search for intronic motifs governing splicing decisions resulted in the identification of G-rich motifs as part of dichotomous signals present in exon flanking sequences (Zhang et al. 2005). U-rich sequences, however, have not been computationally analyzed or experimentally investigated on a large scale and therefore form the main focus of this article. Previous studies have shown that mutations introduced in specific U-rich sequences downstream of individual 5′ splice sites can result in the skipping of the preceding exons (Le Guiner et al. 2001; Shukla et al. 2004; Zuccato et al. 2004), and in vitro studies have shown that U-rich sequences downstream of two different exons can promote 5′ splice site selection (see Introduction) (Forch et al. 2000). However, it was not determined whether U-rich intronic sequences downstream of 5′ splice sites play a more widespread role in 5′ splice site recognition and splicing regulation. Taken together with our findings summarized above revealing enrichment of intronic U-rich motifs proximal to 5′ splice sites, it is possible that these motifs could play a more widespread role in splicing regulation.
Analysis of distribution of U-rich motifs in introns
In order to identify the distributions of U-rich motifs downstream of the two populations of exons, the intronic sequences were searched from the +1 position using a series of motifs with the following number of U nucleotides, over specified lengths: 3/3, 3/4, 3/5, 4/4, 4/5, 4/6, 5/5, 5/6, 5/7, 6/6, 6/7, 7/7. The results show that all of these U-rich motifs are significantly enriched between nucleotides +6 and +30 in introns downstream of constitutive exons (Fig. 1A; Supplemental Fig. 1). Some of the U-rich motifs, specifically those with at least one flexible base (e.g., 4/5, 4/6), are also enriched between nucleotides +6 and +30 in introns downstream of alternative exons (Fig. 1B; Supplemental Fig. 1).
Figure 1.
Positional bias of intronic U-rich motifs. Distribution of U-rich (4/5 U’s) motifs in the first 100 nucleotides of intronic sequences downstream of constitutively spliced (CS) (A) and alternatively spliced (AS) (B) exons. U-rich motifs are enriched between nucleotides +6 and +30 of introns downstream of CS and AS exons. The difference in the nature of the plots between CS and AS events (i.e., more smooth for CS events) is due to different data set sizes (109,225 vs. 3872). U-rich motifs are not enriched in the corresponding intronic region downstream of pseudo exons (Supplemental Fig. 1). Other U-rich motifs (3/3, 3/4, 4/4, 5/5, 5/6, 6/6, 6/7, 7/7 Us) were also assessed and showed similar distributions in introns adjacent to constitutive exons, whereas only motifs that have at least one positionally flexible nucleotide showed similar distributions in introns downstream of alternative exons (Supplemental Fig. 1).
To assess the possible functional significance of the U-rich motifs, we analyzed their occurrences downstream of pseudo exons. Pseudo exons are intragenic sequences that are flanked by sequences closely matching consensus splice sites, and are of a length that is comparable to true exons. However, unlike true exons, pseudo exons lack sufficient enhancer elements to promote splicing, and they are also enriched in silencer elements that likely further prevent their inclusion in spliced transcripts (Zhang and Chasin 2004). Therefore, if the U-rich intronic motifs generally act to promote splicing, we would expect to find that they are not enriched downstream of pseudo 5′ splice sites. Consistent with this prediction, none of the U-rich motifs were enriched between +6 and +30 downstream 2309 pseudo exons (Supplemental Fig. 1). These results suggest that U-rich motifs located between nucleotides +6 to +30 downstream of authentic exons may function as intronic splicing enhancer (ISE) elements.
U-richness of intron sequences downstream of weakly defined exons
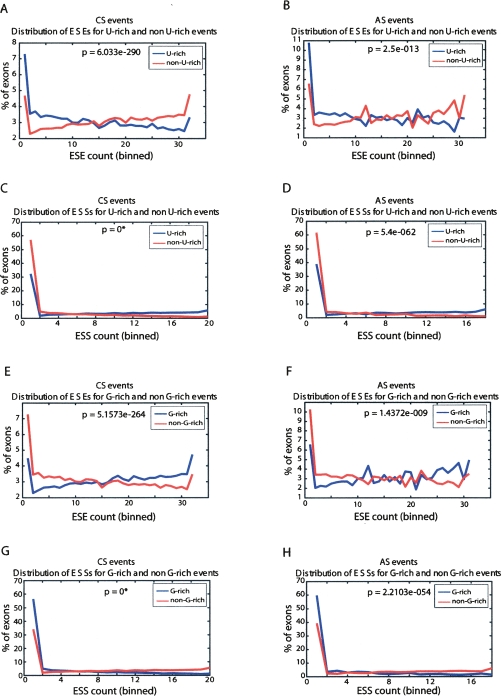
The possibility that U-rich motifs function as ISEs was further explored by investigating their relationship to the strength of splicing signals in the upstream exons. We considered that the enrichment of U-rich motifs in intronic sequences downstream of constitutive and alternative exons could function generally to facilitate the recognition and regulation of weakly defined exons. If this were the case, it would be expected that the frequency of U-rich motifs in the intronic regions downstream of exons is related to the strength of splicing signals that contribute to the recognition of the upstream exons. Accordingly, we first applied a statistical scoring method (for details, see Methods) to ask whether the relative frequencies of any of the U-rich motifs (as defined above) in the downstream introns represent a feature that “discriminates” exons based on their overall content of ESE or ESS motifs or based on the strength of the splice sites flanking the exon sequences. For these analyses, we used sets of ESE and ESS motifs from experimentally validated computational searches performed by Zhang and Chasin (2004). Our analyses revealed that the presence of relatively short U-rich motifs (e.g., 3/4 U’s) in the downstream intron sequences can discriminate both constitutive and alternative exons based on their relative ESE and ESS content (Supplemental Fig. 2). However, in every comparison made, overall U-content (as tested at different percentage cut-offs) was found to represent the best discriminating feature. Notably, the feature of >28%–30% Us in flanking intronic sequences was found to best discriminate adjacent exons scored on the basis of both ESE and ESS frequencies (Supplemental Fig. 2).
Based on the above observation that overall U-richness of the 100 nt of downstream intronic sequences appears to be the most significant feature that can discriminate upstream exons on the basis of their ESE and ESS content, we next plotted the frequencies of ESEs and ESSs within exons with and without U-rich flanking intronic regions (where >28% U-content is defined as U-rich, and <28% U-content is defined as non-U-rich). These plots reveal that constitutive (Fig. 2A) and alternative (Fig. 2B) exons adjacent to U-rich intronic sequences (blue line) are significantly more often associated with reduced frequencies of ESEs than exons that are adjacent to intronic sequences that are not U-rich (red line). Conversely, both classes of exons adjacent to U-rich intronic sequences (blue lines) display significantly increased frequencies of ESS motifs, compared with exons adjacent to introns that are not U-rich (red lines) (Fig. 2C,D). We note that using more conservative cut-offs for U-richness (e.g., >40% U-content defined as U-rich and <20% U-content defined as non-U-rich) results in an even more pronounced discrimination of upstream exons on the basis of their ESE and ESS frequencies (data not shown). When applying the same procedure as described above in relation to splice sites, a clear difference in the strength of the 5′ splice or 3′ splice sites as defined by a scoring matrix of Shapiro and Senapathy (1987) was not observed between exons that are adjacent or not adjacent to downstream U-rich intronic sequences. Similar results were obtained when applying a more sensitive splice site scoring method (Yeo and Burge 2004; data not shown).
Figure 2.
Intron sequences with elevated U-content are associated with exons having reduced frequencies of ESEs and increased frequencies of ESSs. Distribution of constitutively spliced (CS) (A) and alternatively spliced (AS) (B) exons with different frequencies of ESEs that are associated with either “U-rich” downstream intronic sequences (blue lines) or non-U-rich downstream intronic sequences (red lines). Panels C and D show the corresponding plots when comparing numbers of exons with different frequencies of ESSs. A single cut-off for U-richness of 28% (>28% U-content defined as U-rich, and <28% U-content defined as non-U-rich) was used. ESE and ESS counts (X-axis) are binned into equiprobable bins over the entire corresponding event set (CS/AS), so that higher counts bins have a higher X-value. Panels E–H repeat this analysis for ESE (E,F) and ESS (G,H) elements for G-rich and non-G-rich intronic flanks. A single threshold of 22%–25% was used to separate events that contain G-rich or non-G-rich flanking intron sequences. (*)P-value too small to compute.
Interestingly, different results from those described above were obtained when investigating the relationship between exon-associated splicing signals and downstream intronic regions that are enriched in G-rich motifs. For example, in contrast to the U-rich intronic sequences, we observe that both constitutive and alternative exons followed by introns with G-rich sequences contain a higher frequency of ESEs and a lower frequency of ESSs (Fig. 2E–H). These results suggest that G-rich sequences downstream of 5′ splice sites play a distinct role in 5′ splice site recognition as compared to U-rich sequences.
Together, the results described above indicate that U-rich motifs in intronic sequences downstream of exons may specifically function to compensate for reduced frequencies of ESEs, or increased frequencies of ESSs, in promoting constitutive splicing. Similarly, the presence of these motifs appears to relate to the ESE and ESS content of alternative exons, and it is possible that this relationship exists to facilitate regulation of the levels of splicing of these exons.
Effects of TIA1 and TIAL1 knockdowns on the splicing of exons adjacent to U-rich introns
TIA1 and TIAL1 were previously shown to bind preferentially to U-rich sequences using an in vitro SELEX approach (Dember et al. 1996). In addition, cross-linking experiments using HeLa nuclear extracts showed that TIA1 cross-linked to U-rich sequences downstream of a 5′ splice site in pre-mRNA reporter transcripts from the Drosophila msl-2 gene (Forch et al. 2000). Independently, a recent study showed that depletion of TIA1 and/or TIAL1 in HeLa cells resulted in increased skipping of exon 6 of the FAS gene, which is followed by U-rich intronic sequences (Izquierdo et al. 2005). Based on these observations and our findings described above, we next asked whether TIA1 and TIAL1 generally function to promote the recognition and regulation of splicing of exons followed by U-rich sequences. To test this, we used short interfering RNAs (siRNAs) to deplete TIA1 and TIAL1 proteins in HeLa cells, and RT-PCR assays were performed to determine the effects of depletion of these proteins on the in vivo splicing of a series of constitutive and alternative exons followed by U-rich or non-U-rich intronic sequences.
Individual pools of siRNA duplexes specific for TIA1 and TIAL1, as well as a pool of nontargeting (control) siRNA duplexes, were transfected into HeLa cells. Western blotting of the lysates showed that the levels of TIA1 and TIAL1 were reduced to ∼25% of the levels of the endogenous proteins, compared with the control siRNA-transfected cells (Fig. 3A). A total of 92 splicing events were initially selected for RT-PCR analysis from the Const and Alt populations (Supplemental Table 1). These events were initially selected using the following criteria: (1) the first 30 bases downstream intronic sequences for each set of Const and Alt sets have a wide range of percentage of U-content (see below and Methods), (2) the corresponding genes display expression in HeLa cells as determined from previous microarray and RT-PCR data (Pan et al. 2006; data not shown), and (3) the selected events exclude those that are predicted (Pan et al. 2006) to introduce premature termination codons (PTCs) that elicit nonsense mediated mRNA decay (NMD). This last selection criterion was included to maximize the potential of detecting splicing level changes that might otherwise be masked by effects of NMD. Semi-quantitative radioactive RT-PCR assays using primers specific for exons adjacent to each test constitutive and alternative exon were performed in duplicate on samples from two independent knockdown experiments (total of four repetitions). Representative RT-PCR experiments analyzing alternative exons are shown in Figure 3, B and C and quantification of the products from the four repetitions with standard deviations, is shown in Figure 3D.
Figure 3.
Alternative exons associated with U-rich downstream intronic sequences are differentially spliced following TIA1/TIAL1 depletion. (A) Immunoblot analysis of HeLa cells transfected with pools of siRNAs targeted against TIA1 and TIAL1 transcripts (lane 1), or transfected with a pool of control, nontargeting siRNAs (lanes 2–4). The upper panel was probed with an anti-TIA1 antibody, the middle panel with an anti-TIAL1 antibody, and the bottom panel with an anti-GAPDH antibody. The anti-TIA1/TIAL1 antibodies each detect a protein doublet, presumably corresponding to variants of these proteins. Lanes 3 and 4 are dilutions corresponding to 30% and 10%, respectively, of the sample loaded in lane 2. (B) RT-PCR analysis of alternative spliced exons associated with U-rich intronic sequences in HeLa cells transfected with either a pool of control nontargeting siRNAs (−) or pools of siRNAs specific to TIA1/TIAL1 (+). (C) RT-PCR analysis of alternative spliced exons associated with non-U-rich introns in HeLa cells transfected with either a pool of control, nontargeting siRNAs (−) or a pool of siRNAs specific to TIA1/TIAL1 (+). (D) Densitometric analyses of the resulting PCR bands were carried out using ImageQuant software. The increase in exon skipping upon TIA1/TIAL1 depletion was calculated as the ratio of the band intensities of exon-included (−)/exon-excluded (−) over the band intensities of exon-included (+)/exon-excluded (+). A ratio equal to 1, as shown by the horizontal line, indicates no change in the level of exon skipping upon TIA1/TIAL1 depletion. The means and standard errors of the ratios from two independent RT-PCR analyses of two independent siRNA transfection experiments were calculated for each alternatively spliced cassette exon and plotted using GraphPad Prism 4. Light gray bars correspond to U-rich alternatively spliced cassettes; dark gray bars correspond to non-U-rich AS cassettes. T/T siRNA indicates TIA1/TIAL1 siRNA; N-T siRNA, nontargeting (control) siRNA.
Simultaneous knockdown of TIA1 and TIAL1 (TIA1/TIAL1) resulted in pronounced increases in exclusion (or “skipping”) levels of 23/27 (85%) of the alternatively spliced exons that are followed by U-rich intronic sequences (Fig. 3B,D), whereas the splicing levels of the other four exons in this group were affected to a minor extent or not at all. In contrast, the splicing levels of the 24 alternatively spliced exons that are not followed by U-rich intronic sequences were not significantly affected (Fig. 3C,D). Moreover, the splicing levels of constitutively spliced exons in HeLa cells were not significantly affected by depletion of TIA1/TIAL1 proteins, regardless of the U-richness of their downstream intronic sequences (Supplemental Fig. 3).
In order to further test the generality of the effects of TIA1/TIAL1 proteins on the splicing of alternative exons followed by U-rich sequences, we next tested whether TIA1/TIAL1 are also involved in the regulation of PTC-introducing AS events. For this analysis, we selected 36 additional cassette alternative exons, of which 10 are associated with U-rich (same criteria as above) downstream intronic sequences and are predicted to introduce a PTC upon inclusion, and eight are associated with U-rich intron sequences and are predicted to introduce a PTC upon exclusion. An additional set of exons associated with non-U-rich intronic sequences, eight of which are PTC upon inclusion AS events and 10 of which are PTC upon exclusion AS events, were also analyzed. In close agreement with the results described above, the U-rich and non-U-rich events that were not alternatively spliced in HeLa cells were not affected by the depletion of TIA1/TIAL1. Moreover, 13/14 (92.8%) of the PTC-introducing, alternatively spliced events associated with U-rich introns display TIA1/TIAL1-dependent changes in exon inclusion, whereas only one of nine (11%) of the alternative exons associated with non-U-rich introns displayed TIA1/TIAL1-dependent increases in exon skipping (Supplemental Fig. 4). When comparing all of the intron sequences of the AS events assayed above by RT-PCR assays, we observe that a U-rich threshold of 30% best discriminates the events that display inclusion level changes upon knockdown of TIA1/TIAL1 proteins. This is comparable to the computationally defined threshold for U-richness that best discriminates exons based on relative frequencies of ESE and ESS elements. Together, these observations suggest that TIA1/TIAL1 proteins may play a dominant role in regulating the inclusion levels of alternative exons flanked by U-rich motifs and that have relatively low ESE counts and/or relatively high ESS counts. Interestingly, the results also suggest that TIA1/TIAL1 may in some cases function to regulate gene expression by modulating the inclusion levels of exons that, when included or skipped, trigger transcript degradation by NMD.
In summary, the results described above indicate that reduced expression of TIA1/TIAL1 in HeLa cells predominantly affects the inclusion levels of alternatively spliced exons that are followed by U-rich sequences. Moreover, the results also indicate that the vast majority of such exons may be under the control of TIA1/TIAL1 proteins, and this thus implies that most genes with alternative exons flanked by U-rich regions are regulated by these proteins.
Variables modulating the effect of TIA1/TIAL1 knockdown
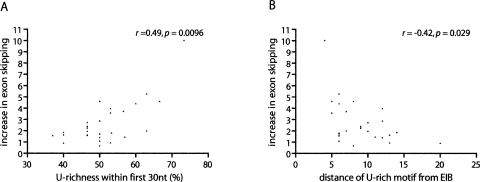
While knockdown of TIA1/TIAL1 resulted in reproducible increases in the levels of skipping of the vast majority of alternatively spliced exons followed by U-rich sequences (Fig. 3B), the extent of these effects were quite variable (the increased skipping levels ranged from ∼1.5- to 10-fold) (Fig. 3D). In order to assess which factors might determine the extent of the activity of TIA1/TIAL1 proteins, we investigated whether different parameters correlate with the magnitude of the change in exon skipping upon depletion of these proteins. These parameters included 5′ splice site strength, proximity of the first U-rich motif to the exon–intron boundary, proportion of U’s within the first 30 nt of the intron, and the presence of ESE and ESS motifs. Among these parameters, significant correlations (as measured using two-tailed Spearman’s correlation tests) were observed between the increase in level of exon skipping and the U-richness within the first 30 nt of the intron (r = 0.49, P = 0.0096) (Fig. 4A), as well as the proximity of the first U-rich motif from the exon–intron boundary (r = −0.42, P = 0.029) (Fig. 4B). These results suggest that both the proximity and overall U-richness are among the strongest determinants of TIA1/TIAL1-dependent regulation of AS. In contrast, although the frequencies of ESE and ESS motifs in the regulated exons are predictive of the presence of downstream U-rich motifs (see Fig. 2), neither the frequencies of these motifs nor the strength of the 5′ splice sites appeared to play dominant roles in establishing relative exon inclusion levels of TIA1/TIAL1-regulated exons.
Figure 4.
The increase in exon skipping correlates with the proportion of U’s and the proximity of the first U-rich motif to the exon–intron boundary. Spearman correlation tests showing significant correlations between the increase in exon skipping upon TIA1/TIAL1 silencing and proportion of U’s within the first 30 nt of the second intron of the splicing cassettes (A), and the proximity of the first U-rich motif to the exon–intron boundary (B). The correlations suggest that exons followed by extended TIA1/TIAL1 binding sites (longer stretches of U’s) and/or by binding sites that are located closer to the 5′ splice sites are more sensitive to the reduced levels of TIA1/TIAL1. EIB, exon–intron boundary; n = 27 alternative splicing cassettes.
Biological relevance of TIA/TIAL1-regulated AS
To assess the possible functional relevance of the AS events associated with U-rich downstream intronic sequences and regulated by TIA1/TIAL1, we first determined which of these events are conserved in mouse transcript sequences and have the potential to alter protein coding sequences. In previous studies, it was shown that only 10%–20% of cassette-type alternative exons are conserved between human and mouse transcripts (Modrek and Lee 2003; Nurtdinov et al. 2003; Sorek et al. 2004; Pan et al. 2005; Yeo et al. 2005). The remaining 80%–90% of cassette exons are either genome specific (i.e., they are only detected in transcripts from one species) or alternatively spliced in a species-specific manner (i.e., they involve conserved exons that are alternatively spliced in one species and constitutively spliced in the other species) (Modrek and Lee 2003; Nurtdinov et al. 2003; Sorek et al. 2004; Pan et al. 2005; Yeo et al. 2005). Consistent with the probable functional importance of many cassette-type AS events that are conserved between human and mouse, these events are significantly more often frame-preserving and regulated in a cell- or tissue-dependent manner than are species-specific cassette-type events (Pan et al. 2004; Resch et al. 2004; Sorek et al. 2004, Xing and Lee 2005).
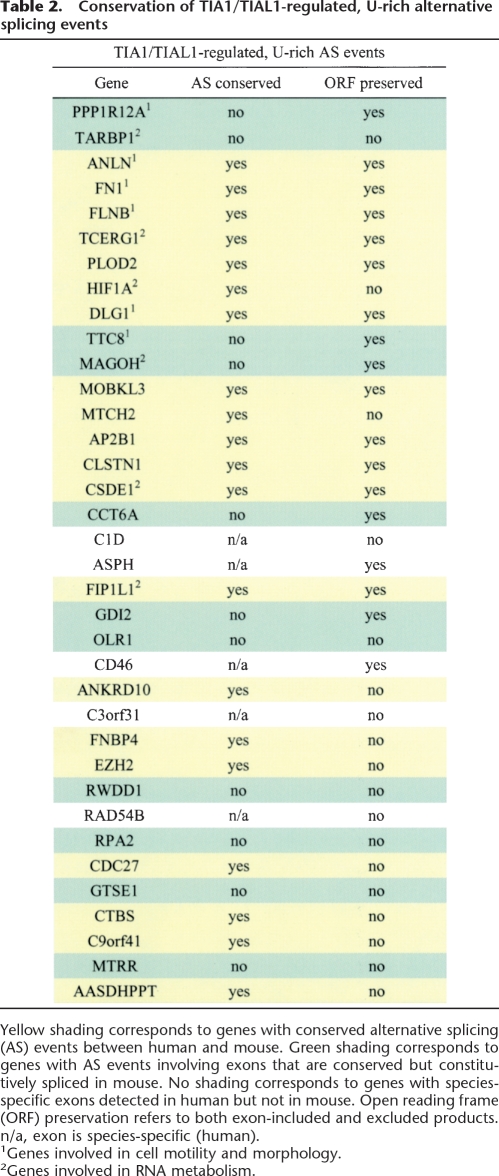
We initially assessed the conservation of the 36 human AS events shown above to be regulated by TIA1/TIAL1, by searching mouse EST and cDNA databases for the presence of the orthologous, exon-included, and exon-excluded transcripts. Conservation of both isoforms was detected for 20/36 (55.5%) of these cassette AS events (Table 2, yellow shading). This represents a significant enrichment for conserved AS events over the proportion (17.7%) observed in the total data set (n = 3872). Eleven of the 36 (30.5%) cassette-type AS events show conservation for only one of the isoforms, suggesting that these events may represent cases of conserved exons that are spliced in a species-specific manner (Table 2, green shading). The remaining five cassette-type alternative exons were only detected in human transcripts, indicating that these may represent genome-specific exons (Table 2). We next assessed whether, overall, AS events flanked by U-rich downstream intronic sequences (scored as described above) are more conserved than the AS events not flanked by U-rich intron sequences. A large collection of orthologous human and mouse sequences were mined for AS events using alignments of EST and cDNA sequences (see Methods). Regions with sufficient coverage in both species were used to determine which cassette exons are conserved in both species and which are not conserved. Consistent with the results described above involving the experimentally characterized TIA1/TIAL1-dependent AS events, in the larger data set, exons that are flanked by U-rich regions are also enriched in the group of conserved AS events between human and mouse. These results thus provide evidence that TIA1/TIAL1 proteins and U-rich ISEs often mediate the regulation of AS events that are conserved between human and mouse.
Table 2.
Conservation of TIA1/TIAL1-regulated, U-rich alternative splicing events
Yellow shading corresponds to genes with conserved alternative splicing (AS) events between human and mouse. Green shading corresponds to genes with AS events involving exons that are conserved but constitutively spliced in mouse. No shading corresponds to genes with species-specific exons detected in human but not in mouse. Open reading frame (ORF) preservation refers to both exon-included and excluded products. n/a, exon is species-specific (human).
1Genes involved in cell motility and morphology.
2Genes involved in RNA metabolism.
Finally, we sought to determine whether TIA1/TIAL1 regulate AS of functionally related sets of genes. Previous studies have shown that specific sets of coregulated AS events often involve genes that belong to the same functional processes and pathways (Ule et al. 2005; Fagnani et al. 2007; Ip et al. 2007). AS events with U-rich flanking intronic sequences, with known or predicted regulation by TIA1/TIAL1 proteins, were analyzed for enrichment of specific Gene Ontology (GO) terms. The AS events were classified as U-rich using increasing cut-offs for U-richness (i.e., >30%, >35%, >40%). GOstat (Beissbarth and Speed 2004) was employed to assess whether there is enrichment of specific GO terms in the U-rich groups relative to the entire set of AS events in the starting data set. However, no specific GO terms were scored as being significantly enriched among the set of genes with U-rich intronic sequences adjacent to alternative exons. It appears therefore that TIA1/TIAL1 regulate the AS of genes associated with diverse functions.
Discussion
A major challenge of splicing research is to elucidate the “splicing code,” specifically, the full set of motifs and other sequence features that are responsible for the accurate recognition of splice sites and regulation of AS. To date, the majority of published studies have focused on the identification and characterization of exonic sequence motifs, namely, ESEs and ESSs (for review, see Chasin 2007). While recent studies employing computational analyses of conserved intronic sequences flanking alternative exons have resulted in the identification or prediction of some new splicing regulatory motifs (Minovitsky et al. 2005; Voelker and Berglund 2007; Yeo et al. 2007), the extent of the function of these and other ISE and silencer motifs is not known. Moreover, how exonic and intronic motifs, together with the splice sites, combine to achieve a concerted splicing outcome is not well understood.
In the present study, we have performed a systematic analysis of human intronic sequences downstream of 5′ splice sites, with the goal of identifying sequence elements that have a widespread function in splicing regulation. Computational analyses revealed significant enrichment of U-rich motifs between nucleotides +6 to +30 downstream of both constitutive and alternative exons. No such enrichment was observed downstream of pseudo exons. The property of overall U-richness (i.e., the percentage U content) in the first 100 nt of downstream intronic sequence was found to display significant relationships with the frequencies of ESEs and ESSs in the preceding exons. Downstream U-rich intronic regions correlate with reduced frequencies of ESEs and with increased frequencies of ESSs in the upstream exons. These sequence relationships suggest an important and potentially widespread role for U-rich intronic regions in controlling the recognition and regulation of the upstream exons. Consistent with this prediction, siRNA-mediated depletion of the closely related proteins TIA1 and TIAL1, which were previously implicated in promoting the recognition of 5′ splice sites via binding U-rich sequences, resulted in a general reduction in inclusion levels, specifically of alternatively spliced exons upstream of U-rich intronic sequences. Our results thus reveal a widespread role for recognition of U-rich intronic sequences by TIA1/TIAL1 proteins in the regulation of AS.
Based on the proportion of cassette-type alternative exons that are flanked by U-rich intronic regions (with a conservative cut-off of >40% U content) and the high rate at which exons associated with such flanking U-rich regions display a change in inclusion level upon depletion of TIA1/TIAL1, our results predict that ∼15% of alternatively spliced exons are regulated by these proteins. In contrast, none of the constitutive exons flanked by U-rich intron sequences displayed a significant change in inclusion level following depletion of TIA1 and TIAL1 proteins. Among the possible reasons for this lack of response are that constitutive exons may be “buffered” from TIA1/TIAL1 depletion by having, on average, stronger splice sites, higher average ESE counts, and lower average ESS counts, relative to alternatively spliced exons (Zhang and Chasin 2004; Zheng et al. 2005). In particular, the ∼75% depletion level achieved for TIA1/TIAL1 proteins may not be sufficient to result in significant effects on the inclusion levels of constitutive exons compared with alternative exons. In this regard, it is also noteworthy that the proportion of U’s in the downstream 30 nt of intronic sequence was correlated with the magnitude of the increase in exon skipping level of the associated alternative exons upon TIA1/TIAL1 depletion. This observation, which is consistent with the results of previous SELEX experiments showing that TIA1 and TIAL1 preferentially bind U-rich sequences in vitro (Dember et al. 1996), thus supports a widespread and direct role for TIA1/TIAL1 proteins in promoting inclusion of alternative exons via binding of U-rich intronic sequences. Also supporting this mechanistic relationship was the observation that the effect of depletion of TIA1/TIAL1 on exon skipping levels was more pronounced when the U-rich regions were closer to the 5′-splice site. Previous studies have shown that TIA1 and related proteins function to promote 5′ splice site recognition by forming direct interactions with components of U1 snRNP (Forch et al. 2002). Thus, binding of TIA1 and possibly TIAL1 via U-rich sequences proximal to the 5′-splice site presumably generally favors interactions with U1 snRNP to facilitate exon inclusion. While it is not clear why the overall U-richness of flanking intronic sequences correlates with a stronger TIA1/TIAL1-dependent effect on splicing, it is possible that a higher number of potential binding sites increases the probability of a single TIA1/TIAL1 protein effecting splicing stimulation. However, it is also conceivable that TIA1 and/or TIAL1 could bind via multiple sites in a cooperative manner such that binding of a single, proximal protein effects a direct response. It also cannot be excluded that TIA1 and/or TIAL1 promote increased exon inclusion through additional, as yet unidentified contacts with the splicing machinery, and in this scenario, the binding of multiple TIA1/TIAL1 proteins to proximal U-rich sequences would similarly favor increased exon inclusion levels.
The widespread functional importance of regulation of AS by TIA1/TIAL1 via binding U-rich sequences is supported by the observation that the majority of the alternative exons flanked by U-rich sequences that have been experimentally validated are conserved in mouse. Moreover, the proportion of conserved alternative exons adjacent to U-rich intronic sequences is higher than the proportion of conserved exons adjacent to intronic sequences that are not U-rich. Previous studies have identified a role for TIA1 and TIAL1 in the regulation of exon 6 of the FAS gene; this event functions to regulate the level of the pro-apoptotic FAS isoform leading to regulation of the apoptotic process (Izquierdo et al. 2005). In addition, TIA1 regulates the inclusion level of exon 6 of the PPP1R12A (MYPT1) gene involved in smooth muscle phenotypic variations (Shukla et al. 2005). Our data greatly expands the number of AS events and associated gene functions that are predicted to be regulated by TIA1/TIAL1. While the experimentally defined and computationally predicted sets of these events are not significantly enriched in specific GO terms, many of the genes in our data sets with validated or predicted TIA1/TIAL1-dependent AS events function in cell morphology, cell migration, and RNA metabolism, in addition to a range of other cellular functions (see Table 2). Given that both TIA1/TIAL1 are widely expressed (Beck et al. 1996), our results, taken together with the previous studies on these proteins, indicate that TIA1/TIAL1 participate in the regulation of diverse cellular functions via binding to U-rich elements adjacent to target alternative exons. Interestingly, both proteins show higher expression levels in brain and testis (Beck et al. 1996), organs that are known to have relatively complex AS patterns. It is interesting to note that no overlap was observed between gene targets reported to be repressed by TIA1 at the translation level (Lopez de Silanes et al. 2005) and genes regulated by TIA1/TIAL1 at the AS level. This suggests that TIA1/TIAL1 regulates distinct subsets of genes at the splicing and translation levels.
The results of the present study reveal that presence of U-rich sequences downstream of alternative exons is highly predictive of regulation by TIA1/TIAL1 proteins, and therefore advance our understanding of the sequence code responsible for the regulation of AS. In this regard, the results complement and extend a recent, pioneering study linking clusters of YCAY motifs in exonic and intronic regions to regulation of altenative splicing by the RNA binding neural-specific splicing regulator Nova (Ule et al. 2006). The information from such studies should facilitate not only the prediction of splicing factor regulatory activities but also the consequences of disease mutations. For example, disease genes such as the cystic fibrosis transmembrane-conductance regulator (CFTR) (Kerem et al. 1989; Riordan et al. 1989; Rommens et al. 1989); filamin B (FLNB), which is disrupted in a number of skeletal disorders (Krakow et al. 2004); procollagen lysine hydroxylase 2 (PLOD2), one of the genes responsible for Bruck syndrome (van der Slot et al. 2003; Ha-Vinh et al. 2004); and tetratricopeptide repeat protein 8 (TTC8), which is disrupted in Bardet-Biedl syndrome (Ansley et al. 2003) contain exons with downstream functional U-rich regions (Zuccato et al. 2004; this study). An additional gene that contains an exon followed by U-rich intronic sequences (this study), oxidized low density lipoprotein receptor 1 (OLR1), is associated with increased risk of Alzheimer’s disease and myocardial infarction (Luedecking-Zimmer et al. 2002; Lambert et al. 2003). Many disease mutation screening protocols assess at least 50 nt of flanking intronic sequence of individual exons, although mutations in these regions are often not investigated further if they do not directly alter splice site consensus sequences. Our results warrant the careful investigation of the possible consequences of mutations in intronic regions that target U-rich sequences proximal and downstream of alternatively spliced exons.
Methods
Sequence data sets
The 109,225 human constitutive exons and adjacent 100 nt of downstream intronic sequences in the Const data set were obtained from the Hollywood database (Holste et al. 2006), and the 3872 human alternative exons and adjacent 100 nt of downstream intronic sequences in the Alt data set were mined as previously described and documented (Pan et al. 2006). The data set of 100-nt-long “deep”-intron sequences (located ∼500 nt from each upstream exon) was obtained from the corresponding genes in both the Const and Alt data sets. The data set of 2309 pseudo exons and flanking intronic sequences was obtained from Zhang and Chasin (2004) (http://www.columbia.edu/cu/biology/faculty/chasin/xz3/pseudos5.doc).
Motif searching
The intronic sequences from the Const and Alt data sets were aligned at +1 and scanned for motifs using a modified version of the SeedSearcher algorithm (Barash et al. 2001). SeedSearcher identifies sequence motifs that discriminate a set of query sequences from a set of control sequences. In the present study, the Const and Alt query sequences were compared against the set of corresponding deep-intron sequences as the control set. SeedSearcher was configured to search for motifs comprising short subsequences (or “seeds”) of 5–12 nt with various degrees of sequence flexibility (see Results). Each motif was scored by assigning a hypergeometric P-value representing the probability of observing at least the same number of sequences from the query group with the motif, given its abundance in the positive and negative groups (Barash et al. 2001). To perform the searches, the large Const set of 109,225 intronic sequences was randomly partitioned into 27 subsets, each of similar size to the Alt set. Each of the 27 subsets was searched for motifs as described above. Motifs with a Bonferroni-corrected hypergeometric P-value above 0 (−log10 scaled) were reported and then searched against all the other subsets of the data, as well as in the entire group of 109,225 intronic sequences. This threshold was set to be permissive to avoid missing motifs that might appear even more significant when searching against the complete data set. The top 20 highest scoring SeedSearcher motifs in the “Alt” set, and corresponding scores (Bonferroni-corrected hypergeometric P-values) are shown in Table 1. Scores are −log10 scaled and range from 25 to more than 40.
Computational analysis of the distribution of U-rich-motifs across introns
The distribution of U-rich motifs of variable length and composition (3/3, 3/4, 3/5, 4/4, 4/5, 4/6, 5/5, 5/6, 5/7, 6/6, 6/7, 7/7) was determined by counting motifs independently at every nucleotide position in the +1 aligned intronic sequences of each data set (Const, Alt, and Pseudo). The graphic representation of the distribution of U-rich motifs for each sequence population was generated using Matlab.
Statistical analysis
Jensen Shannon divergence (Lin 1991) was employed to score the differences of ESE and ESS frequencies in exons upstream of U-rich or G-rich and non-U-rich or non-G-rich intronic regions. The Jensen Shannon divergence was computed using U-rich or G-rich motifs (e.g., 3/4, 4/5,. . .) and U-rich or G-rich thresholds (e.g., number of non-overlapping U-rich motifs) in the first 100 nt to define the two populations, U-rich or G-rich and non-U-rich or non-G-rich (see Supplemental Fig. 2; data not shown). The P-values reported in Figure 2 are based on Kolmogorov-Smirnov test.
Two-tailed Spearman correlation tests were performed to assess for correlations between the effect of TIA1/TIAL1 knockdown on the splicing of U-rich AS cassettes and different variables (see Fig. 4). These tests were carried out using the GraphPad Prism 4 statistical package.
A binomial tail probability was used to compute an enrichment of U-rich AS events in the AS events conserved between human and mouse in the Alt set. For this, the number of U-rich events (%U > 40) that are also found to be conserved in the Alt set was compared against the total number of conserved events in U-rich (>40%) and non-U-rich (<20%) events in the Alt set.
Transfection experiments
HeLa cells were grown in DMEM medium supplemented with 10% fetal calf serum and 1% streptomycin-penicillin antibiotics. The cells were grown in 35-mm tissue culture dishes. Two pools of four siRNAs, each specific to TIA1 and TIAL1 (50 nM each gene, 100 nM total), and nontargeting control were obtained from Dharmacon Inc. and transfected into 50%–70% confluent HeLa cells using 4 μL of DharmaFECT 1 reagent. Forty-eight hours after the first transfection, a second round of transfection was performed using identical conditions. Cells were harvested 48 h after the second transfection.
Transcript and Western blot analyses
Total RNA was isolated from one-third of the cells from each transfection experiment using RNAesy columns (Qiagen). First strand cDNA was synthesized from 0.5 μg of DNase I–treated RNA with Superscript reverse transcriptase (Invitrogen) according to supplier’s recommendations. Radioactive PCR amplification (20 cycles beginning at 30 sec at 94°C, 30 sec at 60°C, and 30 sec at 72°C with a decrease in annealing temperature of 0.5°C per cycle, followed by five cycles with 30 sec at 94°C, 30 sec at 50°C and 30 sec at 72°C) of each splicing cassette was performed using 0.5 μCi/μL of [α-32P]dCTP (GE Healthcare). Oligonucleotide primer pairs for each splicing event analyzed are available upon request. The PCR reaction products were separated by nondenaturing 5% PAGE. Gels were dried and product bands were quantified using PhosphorImager analysis and Image Quant software (Molecular Dynamics).
Total protein was extracted from two thirds of the collected siRNA-transfected cells using RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, protease inhibitor cocktail; Roche). Twenty micrograms of total protein extract were separated by SDS-PAGE on 4%–12% gels (Invitrogen) and transferred to Hybond-C super membranes (Amersham Biosciences). Immunoblot detection of TIA1, TIAL1, and GAPDH was performed using goat polyclonal antibodies and rabbit polyclonal antibody, respectively (Santa Cruz Biotechnology), as per the manufacturer’s instructions. Immunoreactive protein was detected using enhanced chemiluminescence of horseradish peroxidase-conjugated anti-goat or anti-rabbit (Bio-Rad) secondary antibodies with Hyperfilm ECL film (Amersham Biosciences).
Acknowledgments
We thank Sandy Pan, Jeff McDonald, and Bhooma Thiruvahindrapuram for providing sequence data and Arneet Saltzman for providing RT-PCR primers and expression data. We also thank Dr. Andrew Paterson for advice on statistical procedures. The work was supported by a grant from the Canadian Cystic Fibrosis Foundation (CCFF) to L.C.T. and J.Z. Research in B.J.B.’s laboratory was supported by grants from the Canadian Institutes of Health Research and National Cancer Institute of Canada, and in part by a grant from Genome Canada funded through the Ontario Genomics Institute. I.A. was the recipient of CCFF Studentship and The Hospital for Sick Children Research Training Awards.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073155.107.
References
- Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., Ross A.J., Eichers E.R., Teslovich T.M., Eichers E.R., Teslovich T.M., Teslovich T.M., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Barash Y., Bejerano G., Friedman N., Bejerano G., Friedman N., Friedman N. A simple hyper-geometric approach for discovering putative transcription factor binding sites. Lect. Notes Comp. Sci. 2001;2149:278–293. [Google Scholar]
- Beck A.R., Medley Q.G., O’Brien S., Anderson P., Streuli M., Medley Q.G., O’Brien S., Anderson P., Streuli M., O’Brien S., Anderson P., Streuli M., Anderson P., Streuli M., Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA1 and TIAL1. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P., Speed T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Caceres J.F., Kornblihtt A.R., Kornblihtt A.R. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Chew S.L., Krainer A.R., Chew S.L., Krainer A.R., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R., Calarco J.A., de Stanchina E., Krainer A.R., de Stanchina E., Krainer A.R., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y., Bourgeois C.F., Kister L., Stevenin J., Bourgeois C.F., Kister L., Stevenin J., Kister L., Stevenin J., Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. Adv. Exp. Med. Biol. Vol. 623. 2007. Searching for splicing motifs; pp. 85–106. [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Gesnel M.C., Stevenin J., Breathnach R., Stevenin J., Breathnach R., Breathnach R. The RNA-binding protein TIA1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember L.M., Kim N.D., Liu K.Q., Anderson P., Kim N.D., Liu K.Q., Anderson P., Liu K.Q., Anderson P., Anderson P. Individual RNA recognition motifs of TIA1 and TIAL1 have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Fagnani M., Barash Y., Ip J.Y., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Barash Y., Ip J.Y., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Ip J.Y., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Shai O., Lee L., Rozenhek A., Mohammad N., Lee L., Rozenhek A., Mohammad N., Rozenhek A., Mohammad N., Mohammad N., et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B., Yeh R.F., Sharp P.A., Burge C.B., Sharp P.A., Burge C.B., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J., Granneman S., Seraphin B., Anderson P., Valcarcel J., Seraphin B., Anderson P., Valcarcel J., Anderson P., Valcarcel J., Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Forch P., Puig O., Martinez C., Seraphin B., Valcarcel J., Puig O., Martinez C., Seraphin B., Valcarcel J., Martinez C., Seraphin B., Valcarcel J., Seraphin B., Valcarcel J., Valcarcel J. The splicing regulator TIA1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesnel M.C., Theoleyre S., Del Gatto-Konczak F., Breathnach R., Theoleyre S., Del Gatto-Konczak F., Breathnach R., Del Gatto-Konczak F., Breathnach R., Breathnach R. Cooperative binding of TIA1 and U1 snRNP in K-SAM exon splicing activation. Biochem. Biophys. Res. Commun. 2007;358:1065–1070. doi: 10.1016/j.bbrc.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha-Vinh R., Alanay Y., Bank R.A., Campos-Xavier A.B., Zankl A., Superti-Furga A., Bonafe L., Alanay Y., Bank R.A., Campos-Xavier A.B., Zankl A., Superti-Furga A., Bonafe L., Bank R.A., Campos-Xavier A.B., Zankl A., Superti-Furga A., Bonafe L., Campos-Xavier A.B., Zankl A., Superti-Furga A., Bonafe L., Zankl A., Superti-Furga A., Bonafe L., Superti-Furga A., Bonafe L., Bonafe L. Phenotypic and molecular characterization of Bruck syndrome (osteogenesis imperfecta with contractures of the large joints) caused by a recessive mutation in PLOD2. Am. J. Med. Genet. A. 2004;131:115–120. doi: 10.1002/ajmg.a.30231. [DOI] [PubMed] [Google Scholar]
- Holste D., Huo G., Tung V., Burge C.B., Huo G., Tung V., Burge C.B., Tung V., Burge C.B., Burge C.B. HOLLYWOOD: A comparative relational database of alternative splicing. Nucleic Acids Res. 2006;34:D56–D62. doi: 10.1093/nar/gkj048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Hung L.H., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Hung L.H., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A., Neumuller N., Reither G., Haas S.A., Bindereif A., Reither G., Haas S.A., Bindereif A., Haas S.A., Bindereif A., Bindereif A. Intronic CA-repeat and CA-rich elements: A new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip J.Y., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Topp J.D., Blencowe B.J., Lynch K.W., Blencowe B.J., Lynch K.W., Lynch K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo J.M., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J., Castelo R., Guigo R., Bilbao D., Valcarcel J., Guigo R., Bilbao D., Valcarcel J., Bilbao D., Valcarcel J., Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kawakami A., Tian Q., Duan X., Streuli M., Schlossman S.F., Anderson P., Tian Q., Duan X., Streuli M., Schlossman S.F., Anderson P., Duan X., Streuli M., Schlossman S.F., Anderson P., Streuli M., Schlossman S.F., Anderson P., Schlossman S.F., Anderson P., Anderson P. Identification and functional characterization of a TIA1-related nucleolysin. Proc. Natl. Acad. Sci. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C., Chakravarti A., Buchwald M., Tsui L.C., Buchwald M., Tsui L.C., Tsui L.C. Identification of the cystic fibrosis gene: Genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Krakow D., Robertson S.P., King L.M., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Robertson S.P., King L.M., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., King L.M., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Acuna D., Shapiro S.S., Takafuta T., Shapiro S.S., Takafuta T., Takafuta T., et al. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat. Genet. 2004;36:405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lambert J.C., Luedecking-Zimmer E., Merrot S., Hayes A., Thaker U., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Luedecking-Zimmer E., Merrot S., Hayes A., Thaker U., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Merrot S., Hayes A., Thaker U., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Hayes A., Thaker U., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Thaker U., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Desai P., Houzet A., Hermant X., Cottel D., Pritchard A., Houzet A., Hermant X., Cottel D., Pritchard A., Hermant X., Cottel D., Pritchard A., Cottel D., Pritchard A., Pritchard A., et al. Association of 3′-UTR polymorphisms of the oxidised LDL receptor 1 (OLR1) gene with Alzheimer’s disease. J. Med. Genet. 2003;40:424–430. doi: 10.1136/jmg.40.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guiner C., Lejeune F., Galiana D., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F., Lejeune F., Galiana D., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F., Galiana D., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F., Breathnach R., Stevenin J., Del Gatto-Konczak F., Stevenin J., Del Gatto-Konczak F., Del Gatto-Konczak F. TIA1 and TIAL1 activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- Le Guiner C., Gesnel M.C., Breathnach R., Gesnel M.C., Breathnach R., Breathnach R. TIA1 or TIAL1 is required for DT40 cell viability. J. Biol. Chem. 2003;278:10465–10476. doi: 10.1074/jbc.M212378200. [DOI] [PubMed] [Google Scholar]
- Lin J. Divergence measures based on the Shannon entropy. IEEE Trans. Inf. Theory. 1991;37:145–151. [Google Scholar]
- Lin S., Fu X.-D., Fu X.-D. SR proteins and related factors in alternative splicing. In: Blencowe B.J., Graveley B.R., Graveley B.R., et al., editors. Alternative splicing in the postgenomic era. Landes Bioscience; Georgetown, TX: 2007. pp. 108–123. [Google Scholar]
- Liu H.X., Zhang M., Krainer A.R., Zhang M., Krainer A.R., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I., Galban S., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Galban S., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Indig F.E., Falco G., Zhan M., Gorospe M., Falco G., Zhan M., Gorospe M., Zhan M., Gorospe M., Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedecking-Zimmer E., DeKosky S.T., Chen Q., Barmada M.M., Kamboh M.I., DeKosky S.T., Chen Q., Barmada M.M., Kamboh M.I., Chen Q., Barmada M.M., Kamboh M.I., Barmada M.M., Kamboh M.I., Kamboh M.I. Investigation of oxidized LDL-receptor 1 (OLR1) as the candidate gene for Alzheimer's disease on chromosome 12. Hum. Genet. 2002;111:443–451. doi: 10.1007/s00439-002-0802-7. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J., Revil T., Chabot B., Cloutier P., Shkreta L., Fisette J., Revil T., Chabot B., Shkreta L., Fisette J., Revil T., Chabot B., Fisette J., Revil T., Chabot B., Revil T., Chabot B., Chabot B. hnRNP proteins and splicing control. In: Blencowe B.J., Graveley B.R., Graveley B.R., et al., editors. Alternative splicing in the postgenomic era. Landes Bioscience; Georgetown, TX: 2007. [Google Scholar]
- Matlin A.J., Clark F., Smith C.W., Clark F., Smith C.W., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Minovitsky S., Gee S.L., Schokrpur S., Dubchak I., Conboy J.G., Gee S.L., Schokrpur S., Dubchak I., Conboy J.G., Schokrpur S., Dubchak I., Conboy J.G., Dubchak I., Conboy J.G., Conboy J.G. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B., Lee C.J., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Nurtdinov R.N., Artamonova I.I., Mironov A.A., Gelfand M.S., Artamonova I.I., Mironov A.A., Gelfand M.S., Mironov A.A., Gelfand M.S., Gelfand M.S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Niksic M., Giglio L., Faraguna D., Baralle F.E., Giglio L., Faraguna D., Baralle F.E., Faraguna D., Baralle F.E., Baralle F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- Pagani F., Buratti E., Stuani C., Baralle F.E., Buratti E., Stuani C., Baralle F.E., Stuani C., Baralle F.E., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Babak T., Siu H., Hughes T.R., Morris Q.D., Siu H., Hughes T.R., Morris Q.D., Hughes T.R., Morris Q.D., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Frey B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Blencowe B.J., Blencowe B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Maquat L.E., Frey B.J., Blencowe B.J., Frey B.J., Blencowe B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A., Xing Y., Alekseyenko A., Modrek B., Lee C., Xing Y., Alekseyenko A., Modrek B., Lee C., Alekseyenko A., Modrek B., Lee C., Modrek B., Lee C., Lee C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J.R., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., Zielenski J., Lok S., Plavsic N., Chou J.L., Lok S., Plavsic N., Chou J.L., Plavsic N., Chou J.L., Chou J.L., et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rommens J.M., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., Cole J.L., Kennedy D., Hidaka N., Kennedy D., Hidaka N., Hidaka N., et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Shapiro M.B., Senapathy P., Senapathy P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Dirksen W.P., Joyce K.M., Le Guiner-Blanvillain C., Breathnach R., Fisher S.A., Dirksen W.P., Joyce K.M., Le Guiner-Blanvillain C., Breathnach R., Fisher S.A., Joyce K.M., Le Guiner-Blanvillain C., Breathnach R., Fisher S.A., Le Guiner-Blanvillain C., Breathnach R., Fisher S.A., Breathnach R., Fisher S.A., Fisher S.A. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J. Biol. Chem. 2004;279:13668–13676. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- Shukla S., Del Gatto-Konczak F., Breathnach R., Fisher S.A., Del Gatto-Konczak F., Breathnach R., Fisher S.A., Breathnach R., Fisher S.A., Fisher S.A. Competition of PTB with TIA proteins for binding to a U-rich cis-element determines tissue-specific splicing of the myosin phosphatase targeting subunit 1. RNA. 2005;11:1725–1736. doi: 10.1261/rna.7176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Shamir R., Ast G., Shamir R., Ast G., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Stadler M.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B., Yeo G.W., Schneider A., Xiao X., Burge C.B., Schneider A., Xiao X., Burge C.B., Xiao X., Burge C.B., Burge C.B. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Streuli M., Saito H., Schlossman S.F., Anderson P., Streuli M., Saito H., Schlossman S.F., Anderson P., Saito H., Schlossman S.F., Anderson P., Schlossman S.F., Anderson P., Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Wang H., Clark T., Fraser C., Ruggiu M., Clark T., Fraser C., Ruggiu M., Fraser C., Ruggiu M., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B., Gaasterland T., Blencowe B.J., Darnell R.B., Blencowe B.J., Darnell R.B., Darnell R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- van der Slot A.J., Zuurmond A.M., Bardoel A.F., Wijmenga C., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Zuurmond A.M., Bardoel A.F., Wijmenga C., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Bardoel A.F., Wijmenga C., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Wijmenga C., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., Abraham D.J., Black C.M., Verzijl N., Black C.M., Verzijl N., Verzijl N., et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- Voelker R.B., Berglund J.A., Berglund J.A. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C.J., Lee C.J. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C.B., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B., Van Nostrand E., Holste D., Poggio T., Burge C.B., Holste D., Poggio T., Burge C.B., Poggio T., Burge C.B., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Nostrand E.L., Liang T.Y., Nostrand E.L., Liang T.Y., Liang T.Y. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Chasin L.A., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Leslie C.S., Chasin L.A., Leslie C.S., Chasin L.A., Chasin L.A. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C.L., Fu X.D., Gribskov M., Fu X.D., Gribskov M., Gribskov M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA. 2005;11:1777–1787. doi: 10.1261/rna.2660805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A.R., Mayeda A., Krainer A.R., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- Zuccato E., Buratti E., Stuani C., Baralle F.E., Pagani F., Buratti E., Stuani C., Baralle F.E., Pagani F., Stuani C., Baralle F.E., Pagani F., Baralle F.E., Pagani F., Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]