Abstract
Genomic imprinting is a developmentally important mechanism that involves both differential DNA methylation and allelic histone modifications. Through detailed comparative characterization, a large imprinted domain mapping to chromosome 7q21 in humans and proximal chromosome 6 in mice was redefined. This domain is organized around a maternally methylated CpG island comprising the promoters of the adjacent PEG10 and SGCE imprinted genes. Examination of Dnmt3l−/+ conceptuses shows that imprinted expression for all genes of the cluster depends upon the germline methylation at this putative “imprinting control region” (ICR). Similarly as for other ICRs, we find its DNA-methylated allele to be associated with trimethylation of lysine 9 on histone H3 (H3K9me3) and trimethylation of lysine 20 on histone H4 (H4K20me3), whereas the transcriptionally active paternal allele is enriched in H3K4me2 and H3K9 acetylation. Our study reveals a novel placenta-specific transcript, TFPI2, which is expressed from the maternal allele in both humans and mice. Deficiency for the histone methyltransferase EHMT2 (also known as G9A) or for the Polycomb group protein EED, involved in repressive H3K9me2 and H3K27me3 respectively, leads to biallelic expression of Tfpi2 in the extra-embryonic lineages, whereas the other genes in the cluster maintain correct imprinting. Apart from the putative ICR, however, no other promoter regions within the domain exhibited allele-specific repressive histone modifications. This unexpected general lack of repressive histone modifications suggests that this domain may utilize a different silencing mechanism as compared to other imprinted domains.
Genomic imprinting is a mechanism of transcriptional regulation that results in gene expression from only one allele accord-ing to its parental origin. The monoallelic expression of imprinted genes results from epigenetic modifications that differentially mark the parental alleles during gametogenesis (Verona et al. 2003). The two parental copies of an imprinted gene often show differences in DNA methylation, histone tail modifications, nucleosome density, and replication timing (Ragha et al. 2006). The exact nature of the epigenetic “germ line mark,” and the way it can effect the expression in cis of genes many kilobases away, remains to be determined. However, coregulation by single cis-acting imprinting control regions (ICRs) has been described for several regions. ICRs are required for establishing and maintaining imprinting throughout the domain and may provide the principle explanation for clustering, one of the most characteristic features of imprinted genes (Delaval and Feil 2004; Lewis and Reik 2006). In each case, ICRs have been shown to correspond to differentially methylated regions (DMRs) that acquire allele-specific DNA methylation in one of the parental germlines due to the action of the de novo DNA methyltransferase DNMT3L–DNMT3A complex (Bourc’his et al. 2001; Kaneda et al. 2004).
There are several ways in which differential DNA methylation can lead to the silencing of one parental allele. The most simple is through direct silencing of a promoter, where the nontranscribed allele is heavily methylated. The second mechanism involves the ability of the methylation-sensitive transcription factor CTCF to form an insulator on the unmethylated allele. Here, downstream promoters are unable to interact with upstream enhancers through the formation of higher order chromatin structures, such as loops. This mechanism is known to control the expression of the reciprocally imprinted Igf2 and H19 genes (Murrell et al. 2004; Kurukuti et al. 2006). A third mechanism involves DNA methylation–dependent repression of a long noncoding RNA (ncRNA) transcript on one of the parental alleles only. Consequently, the ncRNA transcript is expressed from the unmethylated allele of the ICR only, and is then thought to recruit Polycomb proteins such as EZH2 and EED to the locus. Enrichment of repressive histone modifications to the region consequently silences the surrounding genes. This mechanism is thought to be involved in the imprinting of both the Kcnq1 and Igf2r domains (Sleutels et al. 2002; Mancini-DiNardo et al. 2006).
The proximal region of mouse chromosome 6 (mChr 6) contains two imprinted domains, which are regulated independently, since their respective phenotypes can be separated by a T77H reciprocal translocation breakpoint mapping to mChr6 3.2 (Beechey 2000). The centromeric (proximal) domain contains the Peg10 gene and appears responsible for the early embryonic lethality observed when two copies of this region are maternally inherited. The telomeric (distal) domain includes Mest and is responsible for growth restriction when both alleles are maternally derived (http://www.mgu.har.mrc.ac.uk/research/imprinting/maps/new/imprin-viewmaps6.html).
The orthologous mouse and human PEG10 clusters contain both ubiquitous and placental-specific imprinted genes, which are thought to be regulated by a single, centrally located DMR (Ono et al. 2003). We report here a comprehensive assessment of the allelic expression, DNA methylation, and histone modifications in this large chromosomal interval. Unlike other known imprinted clusters, there is no evidence for a long ncRNA arising from this DMR, suggesting an alternative imprinting mechanism. The Peg10 domain had previously been defined by an ∼1.6-Mb genomic region between Calcr and Asb4 (Ono et al. 2003), which we confirm in this study following further investigation of an additional 15 flanking genes using a systematic expressed single nucleotide polymorphism (SNP) approach in fetal tissues. The orthologous human imprinted interval, however, was reduced in size to between CALCR and PPP1R9A since the telomeric genes between and including PON3 and DLX5 were found to be biallelically expressed. We show, for the first time, that TFPI2, mapping to this interval, is expressed predominantly from the maternal allele in both mice and humans and is transcribed almost exclusively in extra-embryonic tissues. By performing studies on knockout mice and cells deficient for EHMT2 (also known as G9A) and EED, we find that repressive histone methylation is required to silence the paternal allele of this newly placental-specific imprinted gene.
Results
Assessing the allelic expression of genes within the proximal mChr6/hChr7q22 domains
To more precisely characterize the boundaries of the imprinted cluster on proximal mChr6, we investigated allele-specific expression of 15 additional genes (Shfm1, Pdk4, 170034M03Rik, AL987662, AK038694, Acn9, Asns, Tac1, Slc25a13, Dync1i1, Bet1, AK052535, Gng11, Gngt1, and Tfpi2) using RT-PCR amplification across transcribed SNPs. We utilized reciprocal crosses between C57BL/6 (B6) × Mus musculus molosinus (JF1) and Mus musculus castaneus (CAST) at embryonic days E12.5 and E14.5 in various embryonic tissues, yolk sac, and placenta. We confirm the ubiquitous imprinting previously described for the paternally expressed Peg10 and Sgce transcripts, and the maternally expressed Asb4 gene (Piras et al. 2000; Mizuno et al. 2002). Extra embryonic-specific preferential maternal expression was observed for Ppp1r9a, Pon2, and Pon3 (Fig. 1A; see Fig. 6, below) (Ono et al. 2003; Nakabayashi et al. 2004). In addition, we found that the gene encoding Tissue Factor Pathway Inhibitor 2 (Tfpi2) is expressed from the maternal allele in mouse placenta and yolk sac and is weakly expressed from both alleles in embryonic tissues (Fig. 1A; Fig. 6, below). Tfpi2 is a serine protease that plays an important role in extracellular matrix digestion and tissue remodeling (Miyagi et al. 1996; Udagawa et al. 1998). All remaining transcripts for the genes described above were biallelically expressed (data not shown).
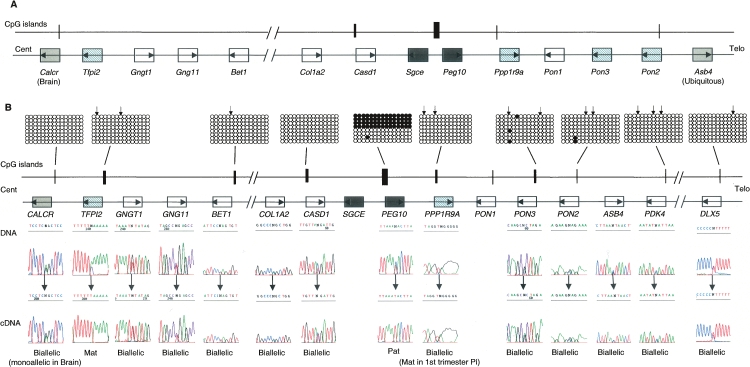
Figure 1.
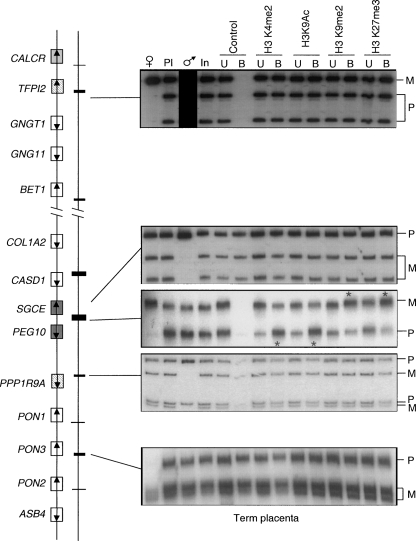
Genomic comparison between proximal mouse chromosome 6 and human chromosome 7q21 imprinted domains. (A) Schematic representation of mouse Peg10 domain, showing the relative organization of genes and CpG islands. (B) Schematic representation of the orthologous human domain. The methylation status of all human promoter CpG islands was examined in liver-, muscle-, lymphocyte-, and placenta-derived DNA. Methylation patterns were assessed first by restriction digestion of bisulfite PCR products. The positions of the restriction sites used are shown (↓). Patterns obtained by sequencing bisulfite PCR products are examples obtained from placenta-derived DNA. Similar patterns were obtained in all tissues analyzed. Each circle represents a single CpG dinucleotide on the strand, a methylated cytosine (●), or an unmethylated cytosine (◯). Only the first 15 CpG dinucleotides from each CpG island are shown. The only evidence for differential methylation was at the known DMR located between the promoters for PEG10 and SGCE; all other promoter CpG islands were hypomethylated. The imprinted expression for all genes in the cluster was analyzed in first trimester fetal tissues and term placenta. DNA sequence traces for heterozygous term placenta samples are shown for all genes, and the resulting RT-PCR sequences. Open boxes depict genes that are biallelic in all tissues throughout gestation, whereas dark gray boxes represent the ubiquitously imprinted, paternally expressed genes. Light gray boxes represent genes that are ubiquitously maternally expressed, while hatched boxes are the placental-specific maternally expressed genes. Arrows show the direction of transcription for each gene.
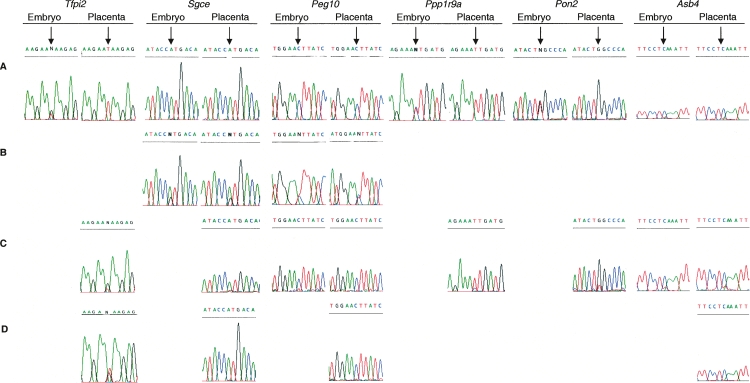
Figure 6.
Allele-specific expression of imprinted genes within the proximal mouse chromosome 6 domain in wild types and mice carrying chromatin remodeling targeted gene deletions. Sequence traces obtained from RT-PCR products are shown for the following: (A) wild-type embryo and placenta from C57BL/6J × M. mus castaneus; (B) the reactivation of the repressed allele for Peg10 and Sgce in Dnmt3l−/+ (C57BL/6J × M. mus castaneus) embryo and trophoblast giant cells from placenta; (C) allele-specific expression of genes in Ehmt2−/− (C57BL/6J × JF1) embryo and placenta, only Tfpi2 imprinting is affected; and (D) maintained imprinting in Eed−/− (JF1 × CD1) TS cells for all genes except Tfpi2. Ppp1r9a and Pon2 display biallelic expression in wild-type TS cells, so their imprinting status could not be assessed in the Eed−/− TS cell line.
To assess if imprinting is conserved within the human orthologous region on hChr7, we assessed the allelic expression for CALCR, TFPI2, GNGT1, GNG11, BET1, COL1A2, CASD1, SGCE, PEG10, PPP1R9A, PON2, PON3, DLX5, PDK4, and ASB4 (Fig. 1B). For the genes in which we identified transcribed SNPs, imprinting analysis was carried out in first trimester fetal tissues and term placenta. No SNPs were identified for SGCE. As expected, human PEG10 was paternally expressed in all human tissues analyzed and at all gestational ages (Ono et al. 2001; Smallwood et al. 2003). Paternal expression of SGCE was confirmed by semiquantitative RT-PCR on mUPD7 and pUPD7 cell lines (Supplemental Fig. 1). We have previously described the maternal expression of PPP1R9A in first trimester placenta and fetal tissues (Nakabayashi et al. 2004); however, we now demonstrate that in term placenta imprinting is relaxed, with biallelic expression being detectable in all samples analyzed. Biallelic expression in first trimester, term placentas, and all fetal tissues was observed for GNGT1, GNG11, BET1 COL1A2, CASD1, PON2, PON3, PDK4, and DLX5 (Supplemental Table 1). We found that the expression of TFPI2 was limited to the placenta (Supplemental Fig. 1), and maternal expression was observed in four of eight first trimester placenta samples (8- to 14-wk gestation). The remaining four showed biallelically expression. This is suggestive of either polymorphic imprinting (between individuals) or cell-type–specific imprinting in a tissue containing multiple cell types (where different tissue biopsies vary in the proportion of different cell types). Interestingly, this gene also showed a gestational age-related relaxation of imprinting with only one out of 19 term placenta samples showing maternal expression. Imprinting of Calcr has been shown to be restricted to mouse brain (Hoshiya et al. 2003). To confirm tissue-specific imprinting in the embryo, we assessed allelic expression of CALCR in various fetal tissues. CALCR was biallelically expressed in all tissues except brain, where four out of five samples were monoallelic. Parental origin of expression could not be assigned since maternal DNA samples from heterozygous sample sets were either unavailable or uninformative (data not shown).
The involvement of differential DNA methylation in establishing imprinting within the Peg10/PEG10 domain
It has previously been proposed that imprinting of this cluster is controlled by the maternally methylated germline DMR encompassing the promoter CpG island associated with the Peg10 and Sgce gene promoters (Ono et al. 2003). We analyzed the DNA methylation status of all human promoter-associated CpG islands throughout the domain in embryonic and placenta, using bisulfite sequencing and combined bisulfite restriction analysis (COBRA). We can confirm that the only DMR within this domain is located within the previously described PEG10/SGCE promoter CpG island, while all remaining CpG islands were unmethylated on both parental alleles in placenta and embryonic-derived DNAs (Fig. 1B).
The DMR that overlaps the transcriptional start sites for Peg10 and Sgce is likely to be the ICR for the domain. Germline-derived differential methylation in this region is maintained throughout the genome-wide epigenetic reprogramming that occurs during pre- and post-implantation development (Ono et al. 2003). To assess if DNMT3L is involved in the establishment of DNA methylation at the Peg10/Sgce DMR in the female germline, we investigated E8.5 mice that had inherited a deletion of the Dnmt3l gene from homozygous mutant mothers (Bourc’his et al. 2001). Indeed, lack of maternally derived methylation was observed at the Peg10/Sgce DMR in the Dnmt3l−/+ embryos (Supplemental Fig. 2). This led to an increased expression of Peg10 and Sgce in the embryo itself and in trophoblast giant cells (TGC) of the placenta (Fig. 2). This correlates with reactivation of the normally silenced maternal allele for both genes (Fig. 6, below). Conversely, in Dnmt3l−/+ TGCs and the visceral yolk sac, the maternally expressed genes all showed a strong reduction in expression (Fig. 2), with the residual expression being from both parental alleles (data not shown). This remaining expression is likely to be endogenous, since the placental-specific genes exhibited incomplete silencing of the paternal allele in wild-type placenta (Ono et al. 2003). As the Dnmt3L gene is expressed exclusively in the gametes (Bourc’his et al. 2001), the observed loss of imprinting (LOI) results from a failure to establish methylation at this locus in the growing oocytes of Dnmt3l−/− females.
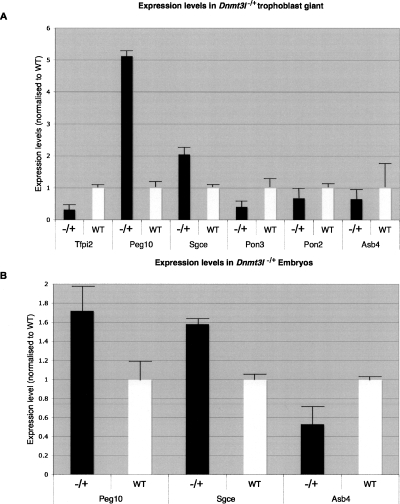
Figure 2.
Comparison of proximal mChr6 gene expression in wild-type and Dnmt3l−/+ embryo and trophoblast giant cells. (A) Expression of genes in trophoblast giant cells isolated from both Dnmt3l−/+ and wild-type placenta. The paternally expressed genes show an increase in expression, while the maternally expressed genes all show a decrease. (B) Increased expression in E8.5 Dnmt3l−/+ embryos compared with wild type for the paternally expressed genes Peg10 and Sgce. The expression of Asb4 exhibits a decrease in expression in Dnmt3l−/+ embryos. All expression values are presented as a ratio to the housekeeping gene Gapdh and are normalized to wild-type values.
The timing of imprinted expression within the Peg10 domain in early development
To ascertain the timing of imprinting in early embryonic stages, we studied early passage undifferentiated ES and TS cell lines from both Mus musculus domesticus (CD1) × JF1 and B6 × CAST hybrids. Previous work on other large imprinted clusters has suggested that ES and TS cells both mimic the epigenetic profiles of the inner cell mass and trophectoderm, respectively (Umlauf et al. 2004; Lewis et al. 2006). Similar to other tissues studied, we find that the ubiquitously imprinted Peg10 and Sgce genes show paternal expression in both ES and TS cells, with maternal repression already present, presumably due to the DNA methylation of their promoters on the maternal allele inherited from the oocyte. The placentally imprinted genes Ppp1r9a, Pon2, and Tfpi2 show different imprinting profiles. Ppp1r9a and Pon2 are biallelically expressed in both ES and TS cells, suggesting that the placental-specific paternal repression on the telomeric side of the cluster arises during differentiation of the trophectoderm lineages. A different result was obtained for Tfpi2 and Asb4, which are expressed from the maternal allele only in both ES and TS cells (Fig. 3), suggesting that these acquire their imprinted expression during preimplantation development. As the expression of Pon3 showed only basal levels in these cell lines, even at high PCR cycling, it was not possible to reliably assay the allelic origin of expression for this gene.
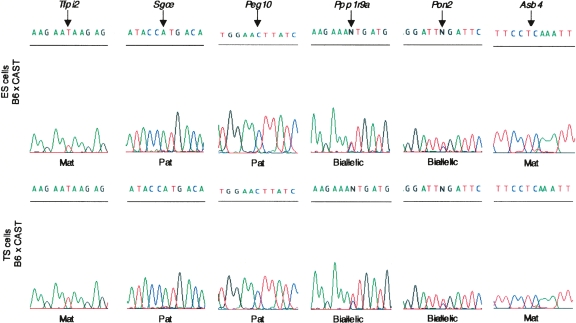
Figure 3.
Allele-specific expression in murine ES and TS cells. Sequence analysis of RT-PCR was used to investigate the allele-specific expression of genes within the Peg10 domain in undifferentiated C57BL/6J (B6) × M. mus castaneus (CAST) ES and TS cells. Both Peg10 and Sgce were paternally expressed (Pat) in both ES and TS cells, whereas Tfpi2 and Asb4 were maternally expressed (Mat) in ES and TS cells. Ppp1r9a and Pon2 show biallelic expression in both cell types. Pon3 expression could not be detected in either cell type.
Histone modifications and allelic repression in the Peg10/PEG10 domains
We next examined allelic histone modifications throughout the mouse and human Peg10/PEG10 domains to assess whether the evolutionary discordance in imprinting could be attributed to the absence of allelic histone modifications, as observed for the Kcnq1/KCNQ1 domain (Monk et al. 2006). Our analysis focused on various modifications on histone H3, including dimethylation at lysine 4 (H3K4me2) and acetylation of lysine 9 (H3K9ac) as markers of permissive chromatin, and dimethylation lysine 9 (H3K9me2), trimethylation lysine 9 (H3K9me3), and trimethylation of lysine 27 (H3K27me3) as markers of repressive chromatin. In addition, we also analyzed the repressive chromatin mark, trimethylation of lysine 20 of histone H4 (H4K20me3).
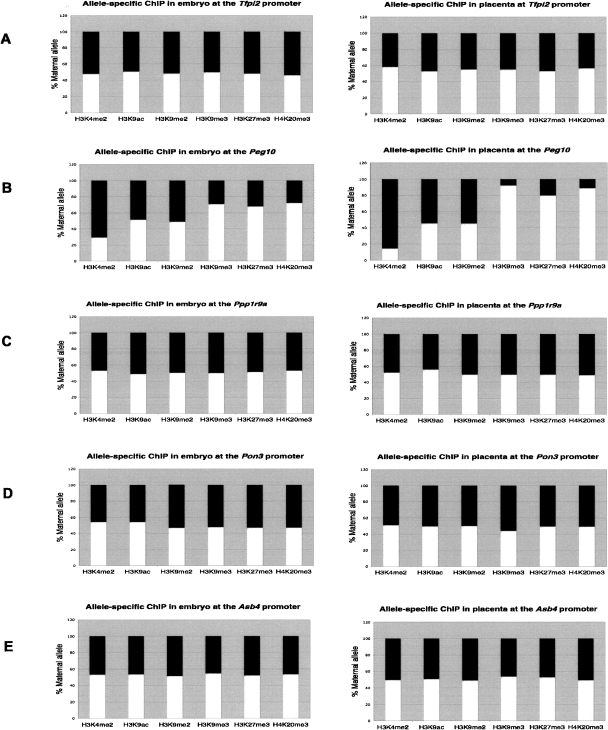
We carried out chromatin immunoprecipitation (ChIP) on unfixed chromatin from E14.5 placenta and embryos for both B6 × JF1 and B6 × CAST crosses. We ascertained allelic enrichment using polymorphisms mapping within 1 kb of the DMR. The active modification H3K4me2 precipitated on the paternal allele in both placenta and embryo, while inconsistent paternal precipitation for the permissive modification H3K9ac was observed. This suggested that the region of allelic acetylation was restricted to a much smaller interval, overlapping the transcriptional start sites for Peg10 and Sgce, which is ∼2 kb away from the polymorphism used. The same region showed precipitation of the repressive markers H3K9me3, H3K27me3, and H4K20me3 on the maternal allele, although no allelic enrichment was observed for H3K9me2. The data for this region are consistent with paternal transcription from the Peg10 and Sgce promoters. Using the same approach, allelic precipitation of active or repressive modification was not observed at any other imprinted gene promoters throughout the domain in either placenta or embryo (Fig. 4).
Figure 4.
Allelic histone modifications at different promoter sites in mouse embryo and placenta. Allele-specific chromatin composition was quantitated using polymorphisms at specific promoter region, in both embryo and placenta. ChIP was performed using antibodies recognizing the specific modification H3K4me2, H3K9ac, H3K9me2, H3K9me3, H3K27me3, and H4K20me3. Polymorphisms and primer locations are given in Supplemental Table 5. The ratio of the allele-specific histone modification at a specific promoter region was expressed as a percentage of maternal (white bars) or paternal (black bars) allele in the total (maternal + paternal, or 100%) immunoprecipitation. ChIP was performed at the Tfpi2 promoter (A), Peg10 DMR (B), Ppp1r9a promoter (C), Pon3 promoter (D), and the Asb4 promoter (E).
ChIP was also used next to assess histone modifications in human term placenta. Similar to the mouse, the only region with evidence of differential histone modifications was the DMR (Fig. 5). Antisera directed against H3K4me2 and H3K9ac precipitated on the paternal allele preferentially, and H3K9me2, H3K27me3, and H4K20me3 were precipitated on the repressed maternal allele. No allelic precipitation was observed 3.8 kb from the PEG10/SGCE DMR, suggesting that the interval with allele-specific differential allelic histone modification is small. Because we had previously observed gestational-age related relaxation of imprinting for PPP1R9A and TFPI2, we also preformed ChIP on first trimester (12–14 wk) human placenta, the same time interval in which these two genes are imprinted. As in the mouse, there was no detectable allelic enrichment for any modification of H3 or H4 within the promoters of these two genes (data not shown).
Figure 5.
Allele-specific histone methylation and acetylation in human term placenta. DNA extracted from antibody bound (B) and unbound (U) fractions were PCR amplified and followed by single strand confirmation polymorphism (SSCP) to differentiate parental origin. Polymorphism and primer locations are given in Supplemental Table 5. For each bound fraction, we determined the ratio between parental alleles, after correction against allelic ratio of the input chromatin. Asterisks indicate the lanes where, after correlation against the allelic ratio in the input chromatin, the allelic ratios were higher than 2. The only region of allelic enrichment was within the PEG10/SGCE DMR. The paternal relative enrichment for H3K4me2 was 2.3, and 2.1 for H3K9ac. The maternal relative enrichment for H3K9me2 was 3.3; for H3K27me3, 2.7. No allelic enrichment was observed within any other promoter region. Open boxes depict genes that are biallelic in all tissues throughout gestation, whereas dark gray boxes represent the ubiquitously imprinted, paternally expressed genes. Light gray boxes represent genes that are ubiquitously maternally expressed, while hatched boxes are the placental-specific maternally expressed genes. Arrows show the direction of transcription for each gene.
To confirm that methylation of H3K9 or H3K27 was not involved in allelic repression for genes mapping within the Peg10 domain, we assessed allelic expression in material from Ehmt2 and Eed null mice (Fig. 6). EHMT2 has recently been identified as a mammalian histone methyltransferase that is involved in methylation of lysine 9 of histone H3 (Tachibana et al. 2001). Imprinted expression was assessed in embryos and placentas derived from reciprocal Ehmt2−/− B6 × JF1 crosses at E9.5. Imprinted expression was maintained for Peg10 and Sgce, indicating that even though the histone modification was enriched on the transcriptionally repressed maternal allele, the EHMT2 contribution toward H3K9 methylation is dispensable for imprinted expression. Further studies are required to investigate the involvement of the remaining lysine 9 methyltransferases. The imprinting of Ppp1r9a, Pon2, and Asb4 was also sustained in the Ehmt2−/− placenta. However, EHMT2 is required for appropriate placental-specific imprinting of Tfpi2, since LOI, corresponding to an increase in total expression, was observed in the Ehmt2−/− placenta (Fig. 6; Supplemental Fig. 3).
We used a similar approach for assessing the involvement of H3K27me3 in the maintenance of imprinting of genes within this domain. The EED/EZH2 Polycomb complex has been shown to have methyltransferase activity specifically for lysine 27 of histone H3 (Cao and Zhang 2004). Using Eed−/− trophectoderm stem (TS) cell lines derived from CD1 × JF1 inter-subspecies crosses (Mager et al. 2003), imprinted expression was faithfully maintained for all imprinted genes, except Tfpi2, for which LOI and an increase in total expression were observed in the null TS cells, indicating the requirement for EED. The lack of allelic enrichment for H3K9me and H3K27me3 within the Tfpi2 promoter may be explained by the fact that expression of this transcript is limited to the syncytiotrophoblast in placenta (Udagawa et al. 1998), and therefore, any allelic enrichment of histone modifications in this single cell lineage will be difficult to ascertain in whole placenta ChIP experiments. To address this question, we performed ChIP on TS cells that showed maternal expression. This homogenous cell population did not show allelic histone enrichment at either Tfpi2 promoter (Supplemental Fig. 4).
Discussion
In this study, we have redefined the limits and characterized both the expression and epigenetic status of genes in the proximal mChr 6/hChr7q21 imprinting cluster. The proximal mChr6 domain in the mouse exhibits a maternal lethal phenotype in mice carrying partial uniparental duplications produced by the reciprocal translocation T77H (T77H MatDp.distal 6), presumably due to the absence of PEG10 in the placenta (Ono et al. 2006). To screen for additional imprinted genes in the mouse interval, we investigated the allelic expression status of an additional 15 genes found within or immediately adjacent to the proximal mChr6 cluster. All previously uncharacterized genes showed biallelic expression in mouse embryos and in the placenta, with the exception ofTfpi2, which showed imprinted, maternal expression in both the placenta and yolk sac. The only imprinted genes found in human fetal tissues were SGCE, PEG10, PPP1R9A, and TFPI2. Interestingly, PON2 and PON3 are not imprinted in human placenta, reminiscent of the limited placenta-specific imprinting recently described within the hChr11 domain (Monk et al. 2006). Human CALCR showed monoallelic expression at a high RT-PCR cycle number in fetal brain, although parent of origin could not be established due to absent parental samples.
Several previously reported imprinted human genes in this interval (Okita et al. 2003; Seoighe et al. 2006) were found to be biallelically expressed in our study. The earlier studies, however, used human monochromosomal somatic cell hybrids to obtain preliminary evidence of imprinted expression of COLIA2, GNGT1, DLX5, and PDK4. Of these, DLX5 was inferred to be monoallelically expressed and regulated by MECP2 and was therefore proposed to be a key candidate gene for Rett syndrome (Horike et al. 2005). Our data showing consistent biallelic expression of DLX5 in human brain strongly dispute a causative role in Rett syndrome, an argument also put forward recently by others (Schule et al. 2007).
The imprinted cluster in the mouse contains eight genes distributed along a 1.6-Mb interval. The orthologous human cluster contains only five imprinted genes spread over a larger region of ∼1.8 Mb. However, this interval is significantly reduced from a previously implied 3.2-Mb domain, since DLX5, ASB4, PON2, and PON3 are not imprinted as shown in this study. However, we cannot fully exclude the possibility that Tfpi2 belongs to an adjacent imprinted cluster controlled by an as yet uncharacterized maternally methylated ICR. This could explain why Tfpi2 is controlled by a different mechanism to the remaining genes within the Peg10 cluster. Targeted deletions of the Peg10 DMR are therefore needed to address its function as an ICR and to delineate the extent of the cluster it controls.
The novel imprinted gene TFPI2
We have directly demonstrated maternal expression of TFPI2 in placenta, in both humans and mouse. A recent bioinformatics screen predicted that this gene would be imprinted based on the unequal frequency of alleles in SNPs in expressed sequence tags (Seoighe et al. 2006). Expression of TFPI2 is almost exclusively found in the placenta, limited to the syncytiotrophoblast layer in humans. Tfpi2 is thought to be involved in the initial stages of placental development, when the fetal cytotrophoblasts adopt tumor-like properties and invade the endometrium (Udagawa et al. 1998). Tfpi2 itself is thought to monitor the highly regulated spatiotemporal expression of the serine proteases and matrix metalloproteinases, which are required to degrade the extracellular matrix, facilitating controlled placental invasion. It is plausible that Tfpi2 may be involved in the embryonic lethal phenotype observed in the uniparental proximal mChr6 (T77H MatDp.distal 6) mice. Overexpression of this gene could inhibit normal decidualization, intervillous remodeling, and appropriate placental invasion. Compared with the Peg10 model, however, this phenotype may be relatively subtle and therefore masked in the T77H MatDp.distal 6 mouse. A similar scenario was previously documented, in which the growth phenotypes associated with the lack of Cdkn1c and Phlda2 were masked by the effect of a lack of Ascl2 in PatDp.distal 7 mice (McLaughlin et al. 1996).
Epigenetic regulation within the mChr6/hChr7q22 cluster
We assessed the methylation status of all promoter CpG islands confirming the presence of a single DMR overlapping the promoters of the PEG10 and SGCE genes in both human and mouse. This conserved germline DMR was previously suggested as the ICR for the entire cluster of genes (Ono et al. 2003). The differential methylation normally observed at the Peg10/Sgce DMR was not present in the embryos and extra-embryonic material derived from Dnmt3l−/− mothers (Supplemental Fig. 2). These Dnmt3l−/− females failed to establish maternal methylation imprints during gametogenesis. The loss of methylation on the maternal allele results in the biallelic expression of the paternally expressed genes. Interestingly, the embryos of these females also demonstrate a reduction in transcription of the maternally expressed genes. Taken together, these findings indicate that all imprinted genes within the proximal mChr6 cluster require DNA methylation for establishment of their imprinted expression. These data are consistent with the previous findings that DNMT3L is required to repress paternally expressed genes on the maternal allele and for the expression of maternal specific genes in gene clusters regulated by a maternally methylated ICR (Bourc’his and Bestor 2006).
Our data suggest an imprinting mechanism independent of somatic DNA methylation (at least in the placenta), which is reliant on a second asymmetric epigenetic mechanism between parental chromosomes. Histone modifications, specifically methylation and acetylation of histone H3, have been shown to correlate with the imprinting of the Kcnq1 domain. Interestingly, there is a restricted interval of allelic histone modifications in the human KCNQ1 domain compared with that of the mouse, and this correlates with the limited conservation of imprinting of genes in human placenta (Lewis et al. 2004; Umlauf et al. 2004; Monk et al. 2006). By use of native ChIP on both mouse and human material, we observed only limited regions of allelic enrichment for the modified histones. Enrichment of H3K4me3 and H3K9ac was associated with the transcriptionally active, hypomethylated paternal allele of the Peg10/Sgce DMR, whereas the silent, methylated maternal allele was associated with enriched H3K9me, H3K27me3, and H4K20me3. No other regions throughout the cluster showed any enrichment of allelic histone modifications, either in placenta or embryo. The differential H3K4me2 within this germline DMR may be important in consolidating the DNA methylation imprint established in the developing oocyte. The absence of H3K4me was recently implicated in the induction of DNA methylation and presumably provides a signal for ICRs to become DNA methylated by the DNMT3L–DNMT3A complex (Ooi et al. 2007). DPPA3 (also known as STELLA/PGC7) then protects this maternal germline methylation against the global demethylation that occurs after fertilization (Nakamura et al. 2007).
To explore whether repressive histone methylation could be involved in the imprinting of this domain, we assessed allelic expression in the Eed and Ehmt2 null tissues. It has been reported that EED is required for maintained repression of the imprinted X chromosome and several imprinted genes (Wang et al. 2001; Mager et al. 2003). The EED/EZH2 complex has methyltransferase activity, specifically for H3K27 (Cao and Zhang 2004) and appears to be required for DNA methylation at certain promoters (Vire et al. 2006). EHMT2 also has histone methyltransferase activity but is directed toward euchromatic H3K9 (Tachibana et al. 2001). To date, only a few genes have been identified as targets of EHMT2, which has been proposed to be required for differential methylation of the somatic DMR and appropriate imprinting of Snprn gene in ES cells, although no in vivo evidence for such a role was obtained (Xin et al. 2003). We recently demonstrated that EHMT2 is involved in the in vivo maintenance of the placental imprinting at the Kcnq1 domain (Wagschal et al. 2008). DNA methylation of the Peg10/Sgce DMR is maintained in both the Eed and Ehmt2 mutants (Supplemental Fig. 4), and allelic expression was maintained for all genes within the cluster except Tfpi2. LOI of Tfpi2 was observed in both Eed mutant TS cells and Ehmt2 mutant placenta compared with wild type, which corresponded with increases in total expression. Tfpi2 mRNA levels were also shown to be greatly increased in a recent expression micro-array experiment using Ehmt2 mutant placenta (Wagschal et al. 2008). Together, these results suggest that the paternal allele silencing of Tfpi2 occurs through either a direct mechanism, requiring H3K9 and H3K27 corroboration through the combined actions of EHMT2 and EED, or indirectly through the involvement of a yet to be identified transcription factor. In the mouse, Tfpi2 has a rarely used, alternatively first exon. No allelic enrichment was observed at this alternative promoter either (data not shown). Interestingly the imprinting of Meg3 on mChr12 was shown to require EED (Mager et al. 2003), and there was also no subsequent enrichment of H3K27me in developing mouse embryos (Carr et al. 2007). This may suggest that the EED/EZH2 repressive complex is necessary for establishing the silent chromatin state early in development and that a second mechanism is then required for somatic maintenance. Another possibility for the lack of allelic enrichment in these experiments is because the native ChIP was performed on whole placenta and because expression of Tfpi2 could be limited to certain cell types within the placenta. In this scenario, the specific Tfpi2 expressing cell types may have allelic chromatin differences that are diluted out by the nonexpressing cell types. However, the lack of allelic enrichment in TS cells makes this explanation much less likely.
Recent theories on the evolution of imprinting have suggested that placental development in mammals has been a driving force for imprinted expression (Reik and Lewis 2005; Wagschal and Feil 2006). This is supported since our closest placenta-less relatives, the monotremes, do not appear to have imprinted genes (Hore et al. 2007). The ancestral mechanism of placental-specific imprinting is thought to be similar to imprinted X chromosome inactivation and reliant upon the acquisition of repressive histone modifications through the action of an ncRNA, as suggested by the mouse Kcnq1 and Igf2r clusters (Reik and Lewis 2005). It is interesting to note that imprinted expression of the PEG10 gene, and potentially the surrounding domain, arose as a consequence of retrotransposon integration (Suzuki et al. 2007). The mechanism controlling imprinted expression of this domain is centered on the PEG10 DMR, which arose with the transposition event. Since we were unable to detect the presence of antisense transcripts (data not shown), we suggest that this region is unlikely to act as the promoter for a ncRNA. Moreover, knockout of Peg10 does not affect the imprinting of other nearby transcripts (Ono et al. 2006). A potential model might involve CTCF binding to the unmethylated paternal allele of the DMR. Indeed, a genome-wide CTCF ChIP (Barski et al. 2007) reveals that the human PEG10 DMR contains two CTCF binding regions, one located between the transcription start sites for PEG10 and SGCE and the other within the first intron of PEG10. CTCF does not have an obvious binding sequence, but the high level of conservation between mammalian genomes in this region (data not shown) suggests that the CTCF sites will also be present in other species. CTCF binding is known to be required for insulator function, intrachromosomal interactions, and chromatin composition at the Igf2/H19 locus (Hark et al. 2000; Kurukuti et al. 2006; Han et al. 2008), and the PEG10 domain may also utilize one or all of these CTCF functions. Together these data indicate that each individual imprinted cluster can be subject to different mechanisms and evolutionary pressures.
Methods
Collection of human material
A total of 65 fetal tissue sets (8–18 wk) with corresponding maternal blood sample were obtained from termination of pregnancies at Queen Charlotte’s and Chelsea Hospital, London. Local ethical approval for obtaining fetal tissues was granted by the Research Ethics Committee of Hammersmith, Queens Charlotte’s and Chelsea, and Acton Hospitals’ Research Ethics Committee (2001/6028). An additional 31 human fetal brain samples were obtained from the MRC Tissue Bank. A set of 240 term placenta trio samples consisting of multiple-site placental samples with corresponding maternal and paternal blood samples were collected from consecutive consenting pregnancies at Queen Charlotte’s and Chelsea Hospital (local ethics approval 2001/6029). All samples were washed in sterile PBS and snap-frozen in liquid nitrogen and stored at –80°C.
Cell lines and mouse crosses
Wild-type mouse embryos and placentas were produced by crossing C57BL/6 females with either JF1 or M. musculus castaneus (CAST) male mice. Reciprocal crosses between heterozygous C57BL/6 females and JF1 males carrying a gene-trap allele produced Ehmt2 null mice (Wagschal et al. 2008). Material from three targeted embryos from three separate litters were analyzed. Animal husbandry and breeding were licensed by Direction Departementale des Services Veterinaries. Mice derived from reciprocal C57BL/6 × CAST crosses were a kind gift from Dr. G. Kelsey (Babraham Institute, Cambridge, UK). For expression studies on undifferentiated ES cells and TS cells, RNA was kindly provided by Dr. W. Reik (Babraham Institute, Cambridge, UK). RNA and DNA from Dnmt3l−/+ mice was isolated and extracted as previously described (Bourc’his et al. 2001). Eed null and wild-type TS cells were derived and cultured as previously described (Kalantry et al. 2006).
Analysis of allelic expression
Total RNA from embryonic and placental material was isolated using TRIzol (Sigma) according to manufacturer’s instructions. The RNA was treated with DNase1 (Invitrogen) for 30 min at 37°C followed by phenol:chloroform (1:1) extraction and isopropanol precipitation. The RNA was transcribed into cDNA using MMLV reverse transcriptase (Promega) and amplified using primers specific to each gene of interest (for primer sequences, see Supplemental Table 2). All polymorphisms were identified by interrogating SNP databases or genomic sequencing or were previously published. All PCR products were sequenced in both forward and reverse orientation by using an ABI Prism 3100 DNA sequencer (Applied Biosystems).
Real-time expression
All Q-PCR were run in triplicate on 96-well plates using an ABI Prism 7700 sequence detector (Applied Biosystems) following the manufacturer’s protocol. All primers were optimized using SYBR Green and melt curve analysis to ensure that amplicons were free of primer dimer products. Thermal cycle parameters included Taq polymerase activation for 10 min at 95°C for one cycle, repetitive denaturation for 15 sec at 95°C, and annealing for 1 min at 60°C for 40 cycles. All resulting triplicate cycle threshold (Ct) values had to be with 1 Ct of each other. The quantitative values for each triplicate were determined as a ratio with the level of Gapdh, which was measured in the same sample and then averaged to provide relative expression values (for primer sequences, see Supplemental Table 3).
Methylation analysis
Genomic DNA was isolated by standard proteinase K digestion and phenol:chloroform extraction. The DNA was subsequently modified using the sodium bisulfite conversion kit EZ DNA Methylation-Gold (Zymo Research). PCR primers were designed to all promoter CpG islands that are present on the March 2006 UCSC sequence browser (http://genome.ucsc.edu/). PCR amplification, cloning, and sequencing were performed as previously described (for primer sequences, see Supplemental Table 4; Monk et al. 2006).
Chromatin immunoprecipitation
Six term and four first trimester human placenta samples were used for ChIP. Approximately 300 mg of material was used for each ChIP assay. Three mouse livers or placentas were used for mouse ChIP experiments. We carried out ChIP as previously described (Umlauf et al. 2004; Monk et al. 2006). Briefly, we purified nuclei on a sucrose gradient and incubated them with MNase to obtain chromatin fragments of 1–6 nucleosomes in length. We incubated 20 μg of chromatin with 10 μg of antibody overnight at 4°C and then incubated it for 4 h with Protein A–Sepharose beads. For ChIP we used the following antisera directed against H3K4me2 (07-030), H3K9me2 (07-441), H3K9me3 (060,904,589), H3K9ac (07-352), H3K27me3 (07-449), and H4K20me3 (07-463) (Upstate Biotechnology). Human ChIP used an affinity-purified antibody to H3K27me3 (Donated by Y. Zhang, University of North Carolina, Chapel Hill). We eluted the chromatin–antibody complexes from the beads and purified the DNA by phenol:chloroform extraction and ethanol precipitation. All DNA from ChIP was assayed by either PCR-SSCP or PCR and enzymatic digestion using HOT-STOP PCR. Where possible, all polymorphisms mapped within 2 kb of the transcription start site or within the first exon (for primer sequences, see Supplemental Table 5). BLAT searches using the mouse UCSC sequence browser (http://genome.ucsc.edu/) with mouse build Feb. 2006 and Illumina ChIP data sets revealed that all mouse PCR amplified regions showing enrichment for H3K4 and H3K27 in mouse ES cells (Mikkelsen et al. 2007). Only ChIP sample sets that showed enrichment for additional ICRs were used in the analysis. Relative band intensities of the maternal and paternal bands were determined using ImageMaster VDS software (Amersham Biosciences).
Acknowledgments
We thank J. Frost and S. Abu-Amero for technical assistance and critical reading of the manuscript. We also thank T. Magnuson and S. Chamberlain for growing the Eed TS cell lines. This work was supported by grants from The Wellcome Trust, WellBeing of Women, the Medical Research Council, The March of Dimes, The Agence de Recherche Contre le Cancer (ARC) Programme ARECA, The Agence Nationale de Recherche (ANR, Programme Blanc), the Genome Canada/Ontario Genomic Institute, and the Hospital for Sick Children (SickKids) Foundation. D.M. was supported by an EMBO short-term fellowship; A.W., an ARC Ph.D. fellowship; and L.P.K., a Graduate Scholarship from the Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.077115.108.
References
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Wang Z., Wei G., Chepelev I., Zhao K., Wei G., Chepelev I., Zhao K., Chepelev I., Zhao K., Zhao K. High-resolution profiling of histone modifications in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beechey C.V. Peg1/Mest locates distal to the currently defined imprinting region on mouse proximal chromosome 6 and identifies a new imprinting region affecting growth. Cytogenet. Cell Genet. 2000;90:309–314. doi: 10.1159/000056794. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T., Bestor T. Origins of extreme sexual dimorphism in genomic imprinting. Cytogenet. Genome Res. 2006;113:36–40. doi: 10.1159/000090813. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H., Xu G.L., Lin C.S., Bollman B., Bestor T.H., Lin C.S., Bollman B., Bestor T.H., Bollman B., Bestor T.H., Bestor T.H. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Cao R., Zhang Y., Zhang Y. SUZ12 is required for both the histone methyltransferases activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Carr M.S., Yevtodiyenko A., Schmidt C.L., Schmidt J.V., Yevtodiyenko A., Schmidt C.L., Schmidt J.V., Schmidt C.L., Schmidt J.V., Schmidt J.V. Alelle-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics. 2007;89:280–290. doi: 10.1016/j.ygeno.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K., Feil R., Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Han L., Lee D.H., Szabo P.E., Lee D.H., Szabo P.E., Szabo P.E. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol. Cell. Biol. 2008;28:1124–1135. doi: 10.1128/MCB.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Leverose J.M., Tilghman S.M., Schoenherr C.J., Katz D.J., Ingram R.S., Leverose J.M., Tilghman S.M., Katz D.J., Ingram R.S., Leverose J.M., Tilghman S.M., Ingram R.S., Leverose J.M., Tilghman S.M., Leverose J.M., Tilghman S.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hore T.A., Rapkins R.W., Graves J.A., Rapkins R.W., Graves J.A., Graves J.A. Construction and evolution of imprinted loci in mammals. Trends Genet. 2007;23:440–448. doi: 10.1016/j.tig.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cheng J.F., Kohwi-Shigematsu T., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaird imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Hoshiya H., Meguro M., Kashiwagi A., Okita C., Oshimura M., Meguro M., Kashiwagi A., Okita C., Oshimura M., Kashiwagi A., Okita C., Oshimura M., Okita C., Oshimura M., Oshimura M. Calcr, a brain-specific imprinted mouse calcitonin receptor gene in the imprinted cluster of the proximal region of chromosome 6. J. Med. Genet. 2003;48:208–211. doi: 10.1007/s10038-003-0006-6. [DOI] [PubMed] [Google Scholar]
- Kalantry S., Mills K.C., Yee D., Otte A.P., Panning B., Magnuson T., Mills K.C., Yee D., Otte A.P., Panning B., Magnuson T., Yee D., Otte A.P., Panning B., Magnuson T., Otte A.P., Panning B., Magnuson T., Panning B., Magnuson T., Magnuson T. The polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat. Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Sado T., Tsujimoto N., Li E., Sasaki H., Tsujimoto N., Li E., Sasaki H., Li E., Sasaki H., Sasaki H. Essential role for de novo DNA methylation Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Pugacheva E., Murrell A., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Murrell A., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Zhao Z., Labonenkov V., Reik W., Ohlsson R., Labonenkov V., Reik W., Ohlsson R., Reik W., Ohlsson R., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin confirmation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Reik W., Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–90. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- Lewis A., Mitsuya K., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Mitsuya K., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Umlauf D., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Smith P., Dean W., Walter J., Higgins M., Feil R., Reik W., Dean W., Walter J., Higgins M., Feil R., Reik W., Walter J., Higgins M., Feil R., Reik W., Higgins M., Feil R., Reik W., Feil R., Reik W., Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lewis A., Green K., Dawson C., Redrup L., Huynh K.D., Lee J.T., Hemberger M., Reik W., Green K., Dawson C., Redrup L., Huynh K.D., Lee J.T., Hemberger M., Reik W., Dawson C., Redrup L., Huynh K.D., Lee J.T., Hemberger M., Reik W., Redrup L., Huynh K.D., Lee J.T., Hemberger M., Reik W., Huynh K.D., Lee J.T., Hemberger M., Reik W., Lee J.T., Hemberger M., Reik W., Hemberger M., Reik W., Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- Mager J., Montgomery N.D., de Villena F.P., Magnuson T., Montgomery N.D., de Villena F.P., Magnuson T., de Villena F.P., Magnuson T., Magnuson T. Genomic imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo D., Steele S.J., Levorse J.M., Ingham R.S., Tilghman S.M., Steele S.J., Levorse J.M., Ingham R.S., Tilghman S.M., Levorse J.M., Ingham R.S., Tilghman S.M., Ingham R.S., Tilghman S.M., Tilghman S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighbouring genes. Genes & Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin K.J., Szao P., Haegal H., Mann J.R., Szao P., Haegal H., Mann J.R., Haegal H., Mann J.R., Mann J.R. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placenta spongiotrophoblasts. Development. 1996;122:265–270. doi: 10.1242/dev.122.1.265. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Alvarez P., Brockman W., Kim T.K., Koche R.P., Brockman W., Kim T.K., Koche R.P., Kim T.K., Koche R.P., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi Y., Yasumitsu H., Mizushima H., Koshikawa N., Matsuda Y., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Yasumitsu H., Mizushima H., Koshikawa N., Matsuda Y., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Mizushima H., Koshikawa N., Matsuda Y., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Koshikawa N., Matsuda Y., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Matsuda Y., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Itoh H., Hori T.A., Aoki I., Misugi K., Miyazaki K., Hori T.A., Aoki I., Misugi K., Miyazaki K., Aoki I., Misugi K., Miyazaki K., Misugi K., Miyazaki K., Miyazaki K. Cloning of the cDNA encoding mouse PP5/TFPI2 and mapping of the gene to chromosome 6. DNA Cell Biol. 1996;15:947–954. doi: 10.1089/dna.1996.15.947. [DOI] [PubMed] [Google Scholar]
- Mizuno Y., Sotomaru Y., Katsuzawa Y., Kono T., Merguro M., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Sotomaru Y., Katsuzawa Y., Kono T., Merguro M., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Katsuzawa Y., Kono T., Merguro M., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Kono T., Merguro M., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Merguro M., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Oshimura M., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Kawai J., Tomaru Y., Kiyosawa H., Nikaido I., Tomaru Y., Kiyosawa H., Nikaido I., Kiyosawa H., Nikaido I., Nikaido I., et al. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 2002;290:1499–1505. doi: 10.1006/bbrc.2002.6370. [DOI] [PubMed] [Google Scholar]
- Monk D., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Kelsey G., Stanier P., Feil R., Moore G.E., Stanier P., Feil R., Moore G.E., Feil R., Moore G.E., Moore G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A., Heeson S., Reik W., Heeson S., Reik W., Reik W. Interactions between differentially methylated regions partitions the imprinted Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889– 893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Makino S., Minagawa S., Smith A.C., Bamford J.S., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Makino S., Minagawa S., Smith A.C., Bamford J.S., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Minagawa S., Smith A.C., Bamford J.S., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Smith A.C., Bamford J.S., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Bamford J.S., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Stanier P., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Preece M.A., Parker-Katiraee L., Paton T., Oshimura M., Parker-Katiraee L., Paton T., Oshimura M., Paton T., Oshimura M., Oshimura M., et al. Genomic imprinting of PPP1R9A encoding neurabin 1 in skeletal muscle and extra-embryonic tissues. J. Med. Genet. 2004;79:965–972. doi: 10.1136/jmg.2003.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Ikawa M., Yoneda Y., Okabe M., Yoneda Y., Okabe M., Okabe M., et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Okita C., Meguro M., Hoshiya H., Haruta M., Sakamoto Y.K., Oshimura M., Meguro M., Hoshiya H., Haruta M., Sakamoto Y.K., Oshimura M., Hoshiya H., Haruta M., Sakamoto Y.K., Oshimura M., Haruta M., Sakamoto Y.K., Oshimura M., Sakamoto Y.K., Oshimura M., Oshimura M. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics. 2003;81:556–559. doi: 10.1016/s0888-7543(03)00052-1. [DOI] [PubMed] [Google Scholar]
- Ono R., Kobayashi S., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F., Kobayashi S., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F., Kohda T., Kaneko-Ishino T., Ishino F., Kaneko-Ishino T., Ishino F., Ishino F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- Ono R., Shiura H., Aburatani H., Kohda T., Kaneko-Ishino T., Ishino F., Shiura H., Aburatani H., Kohda T., Kaneko-Ishino T., Ishino F., Aburatani H., Kohda T., Kaneko-Ishino T., Ishino F., Kohda T., Kaneko-Ishino T., Ishino F., Kaneko-Ishino T., Ishino F., Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003;13:1696–1705. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Suzuki-Migishima R., Ogonuki N., Miki H., Ogonuki N., Miki H., Miki H., et al. Deletion of Peg10, an imprinted gene acquired from a retransposon, causes early embryonic lethality. Nat. Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Tempst P., Lin S.P., Allis C.D., Lin S.P., Allis C.D., Allis C.D., et al. DNMT3L connects unmethyated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras G., Kharroubi A., Kozlov S., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Kharroubi A., Kozlov S., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Kozlov S., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L., Gilbert D.J., Jenkins N.A., Stewart C.L., Jenkins N.A., Stewart C.L., Stewart C.L. Zac1 (Lot1), a potential tumour suppressor gene, and the gene for ε-sarcoglycan are maternally imprinted genes: Identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 2000;20:3308–3315. doi: 10.1128/mcb.20.9.3308-3315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragha K., Latos P.A., Spahn L., Latos P.A., Spahn L., Spahn L. The imprinted mouse Igf2r/Air cluster—a model maternal imprinting system. Cytogenet. Genome Res. 2006;113:165–177. doi: 10.1159/000090829. [DOI] [PubMed] [Google Scholar]
- Reik W., Lewis A., Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- Schule B., Li H.H., Fisch-Kohl C., Purmann C., Francke U., Li H.H., Fisch-Kohl C., Purmann C., Francke U., Fisch-Kohl C., Purmann C., Francke U., Purmann C., Francke U., Francke U. DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am. J. Hum. Genet. 2007;81:492–506. doi: 10.1086/520063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C., Nembaware V., Scheffler K., Nembaware V., Scheffler K., Scheffler K. Maxium likelihood inference of imprinting and allele-specific expression from EST data. Bioinformatics. 2006;22:3032–3039. doi: 10.1093/bioinformatics/btl521. [DOI] [PubMed] [Google Scholar]
- Sleutels F., Zwart R., Barlow D.P., Zwart R., Barlow D.P., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smallwood A., Papageorghiou A., Nicholaides K., Alley M.K., Jim A., Nargund G., Ojha K., Campbell S., Banerjee S., Papageorghiou A., Nicholaides K., Alley M.K., Jim A., Nargund G., Ojha K., Campbell S., Banerjee S., Nicholaides K., Alley M.K., Jim A., Nargund G., Ojha K., Campbell S., Banerjee S., Alley M.K., Jim A., Nargund G., Ojha K., Campbell S., Banerjee S., Jim A., Nargund G., Ojha K., Campbell S., Banerjee S., Nargund G., Ojha K., Campbell S., Banerjee S., Ojha K., Campbell S., Banerjee S., Campbell S., Banerjee S., Banerjee S. Temporal regulation of the expression of sycytin (HERV-W), maternally imprinted PEG10, and SGCE in human placenta. Biol. Reprod. 2003;69:286–293. doi: 10.1095/biolreprod.102.013078. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Ono R., Narita T., Pask A.J., Shaw G., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Ono R., Narita T., Pask A.J., Shaw G., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Narita T., Pask A.J., Shaw G., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Pask A.J., Shaw G., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Shaw G., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Kohda T., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Marshall Graves J.A., Kohara Y., Alsop A.E., Kohara Y., Alsop A.E., Alsop A.E., et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Fukushima T., Shinkai Y., Sugimoto K., Fukushima T., Shinkai Y., Fukushima T., Shinkai Y., Shinkai Y. Set domain-containing protein G9a, is a novel lysine-preferring mammalian histone methylatransferase with hyperactivity and specific selectivity to lysine 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Udagawa K., Miyagi Y., Hirahara F., Miyagi E., Nagashima Y., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Miyagi Y., Hirahara F., Miyagi E., Nagashima Y., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Hirahara F., Miyagi E., Nagashima Y., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Miyagi E., Nagashima Y., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Nagashima Y., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Minaguchi H., Misugi K., Yasmitsu H., Miyazaki K., Misugi K., Yasmitsu H., Miyazaki K., Yasmitsu H., Miyazaki K., Miyazaki K. Specific expression of PP5/TFPI2 mRNA by syncytiotrophoblasts in human placenta as revealed by in situ hybridisation. Placenta. 1998;19:217–223. doi: 10.1016/s0143-4004(98)90011-x. [DOI] [PubMed] [Google Scholar]
- Umlauf D., Goto Y., Cao R., Cerqueria F., Wagschal A., Zhang Y., Feil R., Goto Y., Cao R., Cerqueria F., Wagschal A., Zhang Y., Feil R., Cao R., Cerqueria F., Wagschal A., Zhang Y., Feil R., Cerqueria F., Wagschal A., Zhang Y., Feil R., Wagschal A., Zhang Y., Feil R., Zhang Y., Feil R., Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Verona R.I., Mann M.R., Bartolomei M.S., Mann M.R., Bartolomei M.S., Bartolomei M.S. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Morey L., Van Eynde A., Bernard D., Vanderwinder J.M., Van Eynde A., Bernard D., Vanderwinder J.M., Bernard D., Vanderwinder J.M., Vanderwinder J.M., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wagschal A., Feil R., Feil R. Genomic imprinting in the placenta. Cytogenet. Genome Res. 2006;113:90–98. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- Wagschal A., Sutherland H., Woodfine K., Henckel A., Chebli K., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Sutherland H., Woodfine K., Henckel A., Chebli K., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Woodfine K., Henckel A., Chebli K., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Henckel A., Chebli K., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Chebli K., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Schulz R., Oakey R.J., Bickmore W.A., Feil R., Oakey R.J., Bickmore W.A., Feil R., Bickmore W.A., Feil R., Feil R. G9a histone methyltransferases contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 2008;28:1104–1113. doi: 10.1128/MCB.01111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mager J., Chen Y., Schneider E., Cross J.C., Nagy A., Magnuson T., Mager J., Chen Y., Schneider E., Cross J.C., Nagy A., Magnuson T., Chen Y., Schneider E., Cross J.C., Nagy A., Magnuson T., Schneider E., Cross J.C., Nagy A., Magnuson T., Cross J.C., Nagy A., Magnuson T., Nagy A., Magnuson T., Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Xin Z., Tachibana M., Guggiara M., Heard E., Shinkai Y., Wagstaff J., Tachibana M., Guggiara M., Heard E., Shinkai Y., Wagstaff J., Guggiara M., Heard E., Shinkai Y., Wagstaff J., Heard E., Shinkai Y., Wagstaff J., Shinkai Y., Wagstaff J., Wagstaff J. Role of histone methyltransferases G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]