Abstract
Agouti signaling protein (ASIP) functions to regulate pigmentation in mice, while its role in many other animals and in humans has not been fully determined. In this study, we identify a 190-kb tandem duplication encompassing the ovine ASIP and AHCY coding regions and the ITCH promoter region as the genetic cause of white coat color of dominant white/tan (AWt) agouti sheep. The duplication 5′ breakpoint is located upstream of the ASIP coding sequence. Ubiquitous expression of a second copy of the ASIP coding sequence regulated by a duplicated copy of the nearby ITCH promoter causes the white sheep phenotype. A single copy ASIP gene with a silenced ASIP promoter occurs in recessive black sheep. In contrast, a single copy functional wild-type (A+) ASIP is responsible for the ancient Barbary sheep coat color phenotype. The gene duplication was facilitated by homologous recombination between two non-LTR SINE sequences flanking the duplicated segment. This is the first sheep trait attributable to gene duplication and shows nonallelic homologous recombination and gene conversion events at the ovine ASIP locus could have an important role in the evolution of sheep pigmentation.
The genomic organization of the agouti signaling protein gene (ASIP) is generally highly conserved in mammalian species, including the mouse (Bultman et al. 1992), human (Kwon et al. 1994), horse (Rieder et al. 2001), pig (Leeb et al. 2000), cow (Girardot et al. 2005), and dog (Kerns et al. 2004). In the mouse genome, various insertions and deletions affecting ASIP and nearby loci have resulted in deregulated ASIP expression causing yellow pigmentation, adult onset obesity, diabetes, tumor growth, and embryonic lethality (Wolff et al. 1999). Other mice mutations in both the coding and regulatory regions affect ASIP expression and function and subsequent coat pigmentation patterns (Bennett and Lamoreux 2003; Eppig et al. 2005). ASIP alleles causing coat color variation have also been characterized in domestic dogs (Kerns et al. 2004), cats (Eizirik et al. 2003), pigs (Drogemuller et al. 2006), horses (Rieder et al. 2001), rats (Kuramoto et al. 2001), and foxes (Vage et al. 1997). Although ASIP transcripts are present in various human tissues, including liver, kidney, heart, and adipose tissue, the role of ASIP in humans is not clear (Voisey and van Daal 2002) but may include both pigmentation (Kanetsky et al. 2002; Bonilla et al. 2005; Voisey et al. 2006) and energy homeostasis (Voisey et al. 2002).
In mammalian species, coat color is an important form of camouflage and can be an integral part of social communication and recognition (Sponenberg 1997). The standard wild-type sheep coat color is generally dark-bodied with a pale belly, similar to other mammalian wild-type coat color patterns (Sponenberg 1997). However, this wild-type coat color pattern is much rarer in domestic sheep, where coat color is an important breed characteristic and production trait. In domestic breeds, unlike in their wild ancestors, the lack of natural selection allows coat color variants to arise and segregate. As a result of artificial selection for white fibers, the white coat phenotype has reached a high frequency in certain breeds and shows autosomal dominant inheritance. In mice the dark dorsal and pale ventral wild-type coat color pattern has been shown to be caused by the spatial expression of different transcripts from a single ASIP gene (Vrieling et al. 1994), presumably controlled by different regulatory elements.
The contribution of ASIP to coat color patterns of domestic sheep has been investigated via classical genetics since the early 1920s. The dominant white or tan (AWt) ASIP allele is responsible for the phaeomelanic (yellow/red) phenotype in modern sheep breeds, while the most recessive allele, non-agouti (Aa) results in eumelanic (black/brown) phenotypes (Brooker and Dolling 1969; Adalsteinsson 1970). Another ASIP allele, badgerface (Ab), is characterized by a pale dorsal phaeomelanic and darker ventral eumelanic pattern; it is recessive to AWt and dominant to Aa (Brooker and Dolling 1969). Independent loci controlling color directly and structural features of the hair/wool coat also modify the phaeomelanic colors such that wool areas are typically white (Sponenberg 1997). Despite strong selection for white wool color, the frequency of the recessive non-agouti (also known as self-color black) Aa allele in the Australian Merino flock was estimated at 0.03 (Hayman and Cooper 1965). While strong comparative and classical genetic evidence for ASIP involvement in sheep dominant white and recessively colored coat phenotypes exist, the molecular genetic cause has remained unresolved (Parsons et al. 1997; Parsons et al. 1999a, b).
To identify the molecular genetic cause of coat color variation in domestic sheep, we have used sequence analysis of genomic DNA, BAC clones, and RT-PCR products from Romanov, Texel, Merino, and ancient Barbary sheep to characterize the genomic structure of the ASIP locus. We report a tandem duplication of a 190-kb portion of the ovine genome is responsible for the dominant white coat color (Aw/t) allele of domestic sheep. An asymmetric competitive PCR assay was developed and used to show that the multiple copy ASIP alleles consistently segregate with the dominant white coat color in Merino sheep. RT-PCR experiments were used to demonstrate alternative splicing of ASIP transcripts and show expression in multiple tissues of white sheep and lack of expression of the single copy alleles in black sheep. 5′RACE experiments determined that unlike the silent single copy ASIP of recessive black Merino sheep, a single copy Barbary sheep ASIP with a functional promoter is responsible for the wild-type pale belly phenotype.
Results
The ovine ASIP gene intron and exon structure
By sequencing the 5379-base-pair (bp) ovine ASIP gene from Merino sheep, (GenBank accession no. EU420022), we determined the genomic organization of the ovine ASIP coding exons 2, 3, and 4 and introns 2 and 3 to be similar to that reported for the bovine, human, and mouse genes (Fig. 1). Each coding exon is flanked by a consensus splice donor and acceptor site. BLASTN of the ASIP sequence against GenBank databases showed greatest identity to the bovine ASIP sequence (95.2%). The ovine gene encodes a putative 133 amino acid protein, which is 98%, 76%, and 74% identical to the bovine (133 amino acid), mouse (131 amino acid), and human (132 amino acid) proteins, respectively.
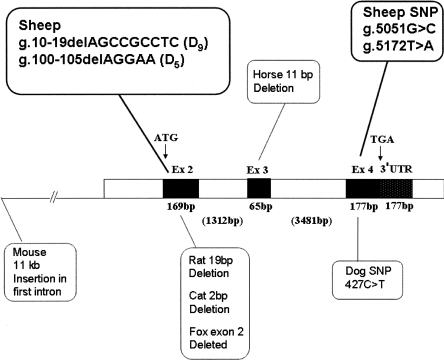
Figure 1.
Schematic of the structure of the ovine ASIP gene showing the sizes of the three coding exons (black) and the intervening intron sequences (white). The exon–intron organization of the ovine gene is similar to that reported for the bovine, human, and mouse genes. Coding exons 2, 3, and 4 are separated by 1312- and 3481-bp intron sequences, respectively. The nucleotide positions of various described recessive black “non-agouti” mutations for the mouse, rat, horse, cat, fox, and dog are shown. The positions of four mutations identified from sheep in this study are shown.
Mutation screening
To identify possible recessive mutations causing pigmentation, we sequenced the entire ASIP coding and noncoding sequence of Romanov, Texel, and Merino sheep and identified four coding sequence mutations compared with the published ovine ASIP sequence (Fig. 1; Parsons et al. 1999b). A previously reported 5-bp deletion, g.100-105delAGGAA (denoted D5), was present in exon 2 (Smit et al. 2002). A novel 9-bp deletion, g.10-19delAGCCGCCTC (denoted D9), was also found to be present in exon 2 10 bp from the ATG start codon, and two SNPs were found in exon 4 (g.5051G>C and g.5172T>A). Both the D5 deletion and the g.5172T>A SNP in exon 4 would be predicted to independently cause functional changes to the agouti protein. The D5 deletion would result in a frame shift followed by a premature stop codon 63 amino acids downstream of the start site, truncating the agouti protein before the functionally important cysteine signaling domain (amino acids 91–130). The g.5172T>A SNP would predict a change of cysteine (amino acid 123) to serine, which would disrupt the highly conserved signaling region of the protein. The D9 deletion would result in the loss of a tripeptide (SRL), which may affect the function of the ASIP transport leader sequence but not the remainder of the protein, while the synonymous g.5051G>C SNP in exon 4 would not be expected to disrupt ASIP function.
Association analysis of the D9 and D5 deletions and A allele with recessive black Merino phenotypes
A coat color panel comprising 373 DNA samples from white (n = 183), self-color black (n = 142), and badgerface (n = 48) Australian Merino sheep was genotyped for the D5 and D9 indels and the g.5172T>A SNP. The self-color black Merinos displayed a typical agouti symmetrical pigmentation pattern of a dark body with a white blaze on the head and neck (Fig. 2A). A characteristic badgerface Merino with dark ventral and lighter dorsal symmetrical agouti pigmentation pattern is shown in Figure 2B. None of the white Merinos were homozygous for the D9, D5, or A alleles. Furthermore, 78 self-color black Merinos that were homozygous D5 were also homozygous T at the g.5172T>A SNP, and only 11% of the recessive black sheep were homozygous A. Approximately 16% of the white sheep and less than 2% of the black sheep had the D9 allele, and none of the animals had an allele with both of these deletions. During genotyping, we also deduced that 8% of the Merino sheep had all alleles—nondeleted alleles at the D9 and D5 positions (denoted N9 and N5) and the D9 and D5 alleles, confirming the previous report of another agouti-like locus (Smit et al. 2002). As none of the white Merinos were homozygous for the D9, D5, or A alleles, the presence of the functional N9, N5, and T haplotype thus seems necessary for a Merino sheep to be white. However, not all black animals (self-color and badgerface) in our panel could be explained by homozygosity of a recessive nonfunctional D5 or A allele. PCR products spanning ASIP exons 2, 3, and 4 in four self-color black Merino sheep homozygous for the N9, N5, and T alleles were amplified and sequenced directly. No other coding mutations that might predict a nonfunctional mature agouti protein were present.
Figure 2.
Illustration of sheep coat color patterns. Three coat colors in the Australian Merino; (A) the dominant white; (B) the non-agouti, also known as recessive self-color-black; and (C) the badgerface pattern. The dominant white coat color in Texel is displayed in D. Romanov sheep (E) appear to have a pattern similar to recessive self-color-black and are proposed as homozygous for the most recessive ASIP allele, Aa. (F) The Barbary sheep wild-type coat color. Hypothesized genotypes for ASIP are indicated.
ASIP expression in Merino sheep
Allele-specific expression of functional ASIP transcripts in white sheep skin
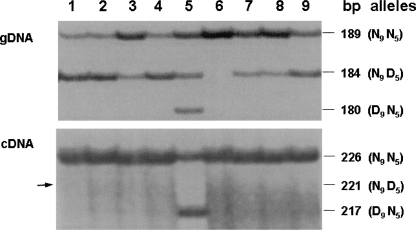
The expression of alleles with the protein altering D5 mutation and the D9 mutation were investigated in nine white Merino sheep. PCR and polyacrylamide electrophoresis were used to examine the expression of the variant alleles (Fig. 3). Seven white sheep with the N9N5 and N9D5 alleles and one sheep (Fig. 3, lane 5) with all three alleles (N9N5, N9D5, and D9N5) showed that the nondeleted N9N5 and deleted D9N5 alleles were preferentially expressed in Merino skin as visualized by the band intensities. The N9D5 allele was not amplified by competitive RT-PCR (Fig. 3, arrow, lanes 1–9) and was considered comparatively lowly expressed or not expressed at all.
Figure 3.
Allele-specific expression of ASIP transcripts in nine white Merino sheep. Autoradiographs of PCR products from gDNA and cDNA from the skin of nine white sheep are shown. The size difference between the 189 (226)- and 184 (221)-, and the 189 (226)- and 180 (217)-bp fragments is due to the corresponding D5 and D9 deletions in the latter fragments. Seven sheep had both the N9N5 and N9D5 alleles. In one sheep (lane 5) all three alleles N9N5, N9D5, and D9N5 were present. One sheep had only the nondeleted N9N5 alleles (lane 6). The nine white animals showed allele-specific expression of the functional N9N5 and D9N5 alleles. Expression of the nonfunctional N9D5 allele (arrow) was not detected. The different band intensities in the gDNA autoradiograph likely reflect the different allele copy numbers present in each animal.
ASIP expression in white sheep is controlled by an ITCH promoter
By use of gene-specific primers designed from ASIP ESTs (data not shown), RT-PCR analysis of skin and internal tissues from white and recessive black Merinos indicated that the dominant white phenotype of Merino sheep was caused by high levels of deregulated expression of the agouti gene from an itchy homolog E3 ubiquitin protein ligase (ITCH) gene promoter. The same alternatively spliced forms of ovine ASIP were expressed in liver, kidney, heart, spleen, and skin tissues tested from white Merinos but were not amplified from any tissues of a self-color black Merino (Fig. 4A–F). ASIP ubiquitous expression has also been reported in humans (Voisey et al. 2002) and cattle (Girardot et al. 2005) and represents a departure from what is seen in mice, where expression is generally confined to the skin (Bultman et al. 1992). RT-PCR (Fig. 4 A,B) and 3′ RACE (Fig. 4C) experiments also indicated all the ASIP transcripts started with two noncoding exons, designated It and It′, and the same transcripts were present in all the white Merino sheep tissues examined. The expression of ovine ITCH, however, was not affected with expression of the same order of magnitude in both the black and white sheep tissues (Fig. 4 D,F). The “normal” expression of ITCH explains why sheep do not experience the spectrum of immunological conditions evident in the “itchy” mouse, in which both ITCH and ASIP expression is disrupted due to a genomic inversion affecting both loci (Perry et al. 1998). ASIP and ITCH expression levels in the skin and other tissues was confirmed by real-time quantitative PCR analysis of three biological replicates each of Merino white and black sheep tissue (data not shown).
Figure 4.
RT-PCR end-point gene expression analysis of ovine ASIP and ITCH in five tissues of a white and a self-color black Merino sheep. The position of primers used to amplify each product is shown in a schematic to the right of the corresponding 1.5% agarose gel image. It, It′, and IE are noncoding exons. Open boxes Ex2, Ex3 and Ex4 (A, B, and C) and open boxes Ex17, Ex18, Ex1, and Ex2 (D, E, and F) represent coding exons of the ASIP or ITCH genes, respectively. Other alternatively spliced transcripts of the agouti gene have also amplified as evident from the bands in panel B. The most abundant ASIP transcripts are those with exon IE (∼368 bp as shown in the panel B schematic). It and It′ were both present in ITCH transcripts and generated bands of the expected size (based on similarity to the bovine sequence), but no alternatively spliced forms were evident. 3′ RACE (panel C) showed ASIP transcripts in all tissues studied to have the same polyadenylation site.
Alternatively spliced ASIP transcripts
RT-PCR identified several ASIP transcripts from the skin of white Merino sheep. Primer Agt9 (positioned in the first noncoding exon It) and primer Agt6 (in the 3′ UTR of ASIP) (Supplemental Table S2) were used to amplify cDNA derived from the skin of white Merino sheep. These products were cloned and sequenced, and seven alternative transcripts (GenBank accession nos. EU420024–EU420030) were obtained (Fig. 5A). Significant differences were evident for the sequences of the ovine skin noncoding exons compared with those reported for cow, mouse, and pig (Vrieling et al. 1994; Leeb et al. 2000; Girardot et al. 2005). Every ovine skin transcript had the two noncoding exons that we have labeled It and It′ in addition to one or two other noncoding exons designated IA to IE (Fig. 5A). A terminal G nucleotide on the sequence of ASIP transcripts derived by 5′ RACE determined exon It to be the most 5′ noncoding exon. The noncoding exons It and It′ were also part of the 5′ region of the downstream ITCH gene (Fig. 5A) and were 67% identical to the noncoding exons in ASIP transcripts from the sienna yellow (Asy) mouse (Duhl et al. 1994).
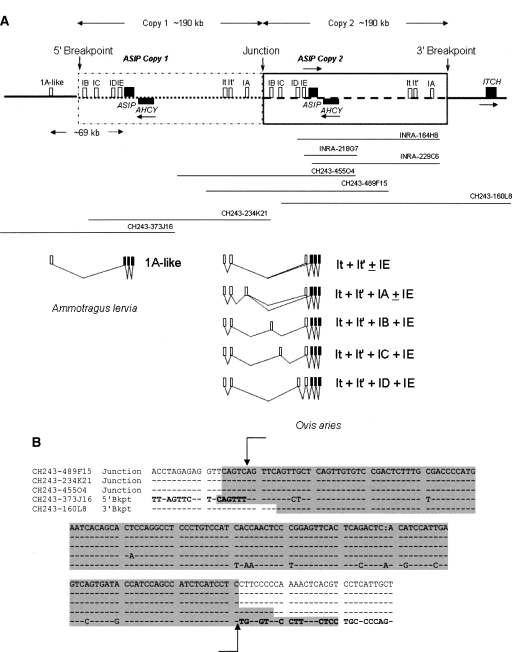
Figure 5.
Schematic showing the sequenced ovine BAC clones below the structure of the ovine genomic tandem duplication and an alignment of the breakpoint and junction point sequences. (A) Noncoding exons are shown as open boxes. Protein coding exons are shown as solid black boxes. Arrows above or below the genes indicate the direction of transcription. Clones INRA-164H8, INRA-218G7, and INRA-229C6 were from the INRA Romanov Sheep BAC library (Vaiman et al. 1999), and clones CH243-160L8, CH243-234K21, CH243-455O4, CH243-373J16, and CH243-489F15 were from the CHORI-243 Texel sheep BAC library (Dalrymple et al. 2007). The three INRA BAC clones did not span the junction, 5′ or 3′ breakpoints. Clones CH243-455O4, CH243-489F15, and CH243-234K21 spanned the junction between copies; clone CH243-160L8 spanned the 3′ breakpoint and clone CH243-373J16 the 5′ breakpoint. Seven ASIP transcripts identified from the skin of a white Merino sheep (Ovis aries) and one transcript identified from the ventral skin of a Barbary sheep (Ammotragus lervia) are shown in the lower section. The coding exons 2, 3, and 4 and Barbary sheep noncoding exon 1A-like are numbered according to the nomenclature of Bultman et al. (1992) and Vrieling et al. (1994). All other noncoding exons are named alphabetically, IA to IE. Exons It and It′ are noncoding exons of both ITCH and ASIP Merino transcripts (see also Fig. 4). The positions of the 5′ and 3′ breakpoints are located 5′ of the ASIP and ITCH coding sequence regions, respectively. The ITCH promoter, including noncoding exons It, It′, and IA, was duplicated and positioned upstream of the duplicated ASIP noncoding exons IB to IE, creating a new ovine hybrid ITCH/ASIP promoter. The complete ASIP and AHCY coding exons were also within the ∼190-kb duplicated segment. Not drawn to scale. (B) DNA sequences from five Texel sheep BAC clones comprising the regions spanning the 5′ and 3′ breakpoints and the junction point between the gene duplication. Sequence identity to the master sequence is shown with a dash. The boundaries of the 143 bp of sequence similarity are marked with vertical arrows. Flanking sequences unique to the 5′ and 3′ breakpoint regions, respectively, are in bold. The highlighted (gray) regions of clone CH243-160L8 sequence of the 3′ Breakpoint was identified by Repbase (Kohany et al. 2006) as having 82% identity to a region of Bovidae non-LTR/RTE BDDF2 repetitive SINE sequence. The highlighted (gray) regions of clones CH243-373J16 (of the 5′ breakpoint) and clones CH243-489F15, CH243-234K21, and CH243-455O4 (all of the junction point) were identified as having 87%–88% identity to a region of Bovidae non-LTR BOV2 repetitive SINE sequence.
The expression and RT-PCR results indicated that the dominant white phenotype of Merino sheep is caused by high levels of expression of functional ASIP transcripts driven by an ITCH promoter region. Comparative analysis of the human, dog, and mouse genomes (http://genome.ucsc.edu/) places the ITCH gene downstream of ASIP in these species. The ITCH-ASIP hybrid transcripts therefore suggested the presence of a tandem duplication or rearrangement and deletion in this region of the ovine genome.
Identification of a duplication encompassing the ASIP, AHCY, and ITCH regions
As the ovine genome sequence was not yet determined, to characterize the ovine ASIP-ITCH genomic region, we obtained and sequenced three Romanov sheep and five Texel sheep BAC clones. Sequencing of these ovine BAC clones identified a large (∼190 kb) tandem genomic duplication (Fig. 5A; Supplemental Fig. S1). Alignment of ASIP noncoding exons It (77 bp), It′ (78 bp), and IA (54 bp) sequences identified from alternatively spliced transcripts from the skin of white Merino sheep to BAC clone INRA-164H8 and INRA-229C6 sequences from a Romanov sheep positioned them ∼119, 123, and 131 kb, respectively, 3′ of the ASIP start codon (Fig. 5A). The ID (56 bp) and IE (88 bp) noncoding exons were located ∼6.6 kb and 298 bp, respectively, 5′ of the ASIP start codon (Fig. 5A). The IB (178 bp) and IC (97 bp) noncoding exons were not present in INRA-164H8 sequence, which ended ∼10 kb 5′ of the ASIP start codon. Comparative alignment of the noncoding exon IB and IC sequences to the dog, human, and cow genome sequences placed these exons further 5′ of the ASIP start codon in these species (data not shown).
The Merino ASIP intron and exon sequence data and the INRA BAC clone sequence data were used to search the CHORI Texel sheep library BAC-end sequences to identify BAC clones in the region of the ovine ASIP gene. In addition, the mapping of the full set of sheep BACs to the cow, dog, and human genomes (Dalrymple et al. 2007) was used to identify BACs potentially located across the putative duplication and flanking genomic regions. Twenty-nine clones that spanned ∼2 Mb of sequence encompassing the ASIP, S-adenosylhomocysteine hydrolase (AHCY), ITCH, and flanking genomic regions of ovine chromosome 13 was obtained for analysis. The BAC clones were then ordered across the region and five BACs predicted to span the complete region including the junction point, and 5′ and 3′ breakpoints were sequenced. We obtained ∼500 kb of contiguous sequence that contained a 190-kb tandem duplication, including the complete ASIP and AHCY coding regions and the ITCH promoter and noncoding exon sequences It, It′, and IA (Fig. 5A). Sequence data from clones CH243-455O4, CH243-234K21, and CH243-489F15 spanned the junction between the tandem duplicated copies. BAC clone CH243-160L8 spanned the 3′ breakpoint, and clone CH243-373J16 spanned the 5′ breakpoint.
Romanov, Texel, and Merino sheep haplotypes
Sequencing of the Romanov and Texel Sheep BAC clones and Merino sheep PCR products from genomic DNA identified seven ovine ASIP haplotypes (Supplemental Table S1). Haplotypes 1 and 2 were determined by cloning and sequencing the PCR products from the genomic DNA of a white (AWt) and a self-color black (Aa) Merino sheep. The white animal contained haplotypes 1 and 2, and the self-color black animal contained only haplotype 2. Ovine ASIP haplotypes 3 and 4 were determined by sequencing of INRA BAC clones INRA-218G7, INRA-229C6, and INRA-164H8 from Romanov sheep. Ovine ASIP haplotypes 5, 6, and 7 were identified from the Texel sheep BAC clone sequences. Haplotype 5 was designated ASIP copy 1 as it was positioned 5′ to haplotypes 6 and 7, both designated ASIP copy 2 (Fig. 5A). Haplotypes 6 and 7 (ASIP copy 2) were 99.9% identical to the functional haplotypes 1 and 3 from Merino and Romanov sheep, while haplotype 5 (ASIP copy 1) showed greatest identity (99.7%) to the nonfunctional A allele containing haplotype 4 from Romanov sheep (see also Fig. 6). The position of the sheep functional haplotypes 6 and 7 at ASIP copy 2 places them 3′ to, and therefore under, the regulation of the duplicated ITCH promoter (Fig. 5A).
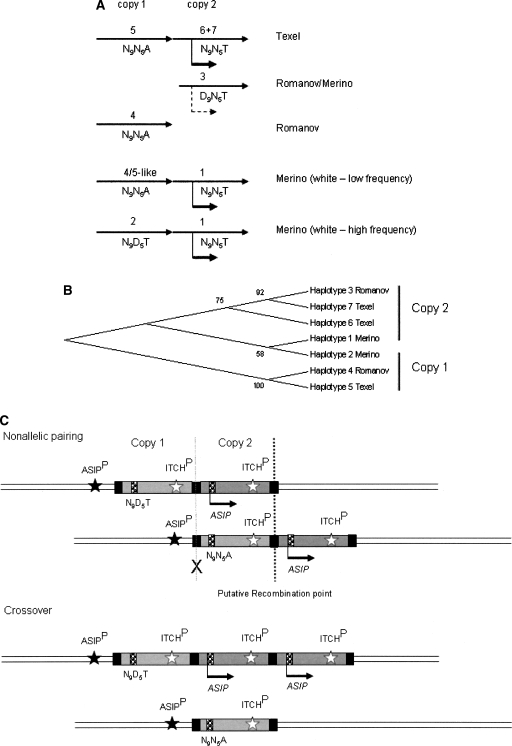
Figure 6.
A model for the organization and evolution of the ovine ASIP locus in dominant white and recessive black domestic sheep. The proposed positions (A) and clustering (B) of copy 1 (inactive) and copy 2 (active) haplotypes identified from Texel, Romanov, and Merino sheep are shown. Solid arrows indicate high levels of expression of haplotypes at copy 2. A dashed arrow indicates expression of a haplotype 3-like D9 allele detected in Merino skin. The two haplotypes identified in the Romanov BAC library may not be derived from a duplicated ASIP-ITCH region as pure Romanov sheep have a recessive black-like phenotype. However, haplotype 4 clusters with haplotype 5 in copy 1 of the Texel and haplotype 3 clusters with haplotype 6 and 7 from the Texel and haplotype 1 from the Merino (B). This clustering suggests that the Romanov haplotypes are derived from the two different copies of the putative original gene duplication. The low frequency presence of a haplotype 4/5-like copy 1 region in the Merino sheep population is deduced from the identification of N9N5A genotypes in the black Merinos and is consistent with the organization of the region in the Texel sheep CHORI-243 library BACs. (C) Schematic showing resolution of nonallelic pairing between duplicated ASIP-ITCH copies with crossover products showing reciprocal deletion and triplication. The 190-kb segments of copy 1 and copy 2 of a white Merino are shown as light and dark gray boxes, respectively. Black boxes indicate the similar SINE sequence regions at the junction, 5′ and 3′ breakpoints. In this example, the ASIP genotype shown represents a white animal heterozygous at copy 1 for the nonfunctional alleles (N9D5T/N9N5A) and homozygous at copy 2 for the functional alleles (N9N5T/N9N5T). Arrows below the ASIP genes indicate the direction of transcription driven by the ITCH promoter (ITCHP). Nonallelic pairing and crossover between the junction and 3′ breakpoints (dark dashed line) would result in creation of a single copy nonfunctional (N9N5A) allele, as shown. The resulting single ASIP copy is not expressed as the ancestral ASIP promoter (ASIPP) is silent. A crossover point (gray cross) that would result in the positioning of a nonfunctional (N9N5A) allele under the regulation of the duplicated ITCH promoter is unlikely to occur as expression of “A” alleles was not detected in Merino sheep. Variable positioning of functional haplotypes at copy 1 and 2 combined with nonallelic pairing and crossover explains the different single copy alleles identified in black Merinos. Mutations and gene conversion events could also contribute to the diversity at the locus.
Haplotypes 1, 6, and 7 all contained the proposed functional N9, N5, and T alleles. Haplotype 2 contained the predicted nonfunctional D5 and functional T alleles and was otherwise very similar to Haplotype 1. Haplotype 3 contained the D9, N5, and T alleles and was also otherwise very similar to the functional haplotypes 1, 6, and 7. Haplotypes 4 and 5 both contained the predicted nonfunctional A allele and were quite different to all other haplotypes. Using haplotype 1 as the reference sequence, haplotypes 4 and 5 had 35 and 40 nucleotide substitutions, respectively. Transcripts from haplotypes 1, 3, 6, and 7 would be predicted to produce functional agouti protein, while transcripts from haplotypes 2, 4, and 5 would be predicted to produce nonfunctional agouti protein.
Nonallelic pairing, crossover, and gene conversion events arguably occur more frequently between almost identical tandem duplications (Lindsay et al. 2006; Myers and McCarroll 2006), and thus, the tandem duplications of the ASIP gene are likely candidates for these events. Indeed, the sequences of the ASIP copy 1 and 2 haplotypes suggest mutations, recombination, and gene conversion events possibly have occurred between them. It is conceivable that Haplotype 5 (ASIP copy 1) could be derived from recombination and/or gene conversion between Texel sheep equivalents of haplotypes 1 and 4. In addition, Texel sheep haplotype 6 (ASIP copy 2), which was otherwise identical to haplotype 7 (ASIP copy 2), shared two nucleotides (at positions 4124 bp and 5051 bp) with haplotype 5 (ASIP copy 1), which are likely a result of mutation or gene conversion events. Haplotype 5 also had an A nucleotide at position 3096 bp not present in any other haplotypes that is likely a result of a mutation.
Analysis of the duplication junction and breakpoint sequences
We analyzed the duplication junction and breakpoint sequences for regions that could facilitate duplication events. By comparing the DNA sequence of each of the BAC clones containing the junction, 5′ and 3′ breakpoints, a region of sequence similarity of ∼143 bp was identified at all three sites (Fig. 5B). These sequences were identified by Repbase (Kohany et al. 2006) as having 82%–88% identity to Bovidae non-LTR BOV2 and BDDF2 repetitive SINE sequence regions. The duplication may have been facilitated by homologous recombination between these repetitive elements.
Analysis of ASIP copy number variation in domestic Merino sheep
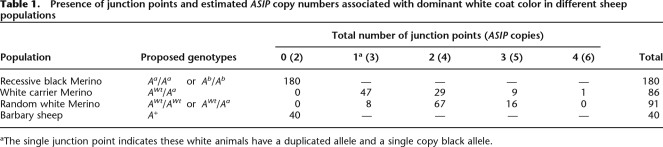
Sequence flanking the shared 143-bp sequence at the junction and 5′ breakpoint was used to develop an asymmetric competitive PCR copy number assay (Supplemental Fig. S2). The junction point and 5′ breakpoint PCR products (see Methods) were used initially to assess the presence and absence of duplicated copy alleles in the genomes of white and recessive black Merino sheep. All white Merinos successfully assayed (n = 177) amplified both a junction point and 5′ breakpoint PCR product and therefore had duplicated ASIP alleles (Table 1). In contrast, all of the successfully assayed recessive black Merinos (n = 180) amplified a 5′ breakpoint product only and thus had only single copy silent (refer expression data) ASIP alleles.
Table 1.
Presence of junction points and estimated ASIP copy numbers associated with dominant white coat color in different sheep populations
aThe single junction point indicates these white animals have a duplicated allele and a single copy black allele.
To calibrate the ABI3130xl copy number assay, PCR products of the junction point and 5′ breakpoint were cloned into pCR 2.1-TOPO, and a standard curve was established using a dilution series of plasmid DNA containing the junction point mixed with plasmid DNA containing the 5′ breakpoint from 0%–100%. The resulting standard curve was linear (Supplemental Fig. S3). The genomic DNA of 16 white Merino sheep (eight that were confirmed as carriers of a single copy recessive black-allele by pedigree analysis) was next analyzed (in quadruplicate) in an initial evaluation of the ABI3130xl copy number assay. Three of the confirmed carriers assayed contained a single junction point allele as would be predicted for a carrier animal (Supplemental Fig. S4, lanes 2,3,8). However, the five other carriers contained two junction points, which indicated triplicated alleles and suggested even greater diversity in copy number could occur at the ovine ASIP locus. The ABI3130xl copy number test was used to assess ASIP copy number variation in the white sheep of the Merino sheep coat color panel. The copy number genotypes of the assayed carrier and random white sheep indicated that white Merino sheep contain multiple copy ASIP alleles (three to six copies) per genome (Table 1).
Comparative analysis of ASIP copy number variation and expression in Barbary sheep (Ammotragus lervia), an ancient Caprinae species
Barbary sheep (Fig. 2) have a wild-type agouti coat color pattern—tan body and pale belly—and are assumed to be homozygous for the wild-type ASIP allele (A+). PCR analysis (n = 40) amplified only the 5′ breakpoint product, indicating single copy ASIP alleles (Table 1). The 5′RACE experiment determined that unlike the silent single copy ASIP of recessive black Merino sheep, the single copy Barbary sheep ASIP has a functional promoter and is responsible for the pale belly phenotype. The partial ASIP transcript (GenBank accession no. EU420031) from Barbary sheep ventral skin had a single noncoding 1A-like exon and coding exons 2, 3, and partial exon 4 sequence (Fig. 5A) with high similarity to the homologous ventral-specific mouse (Bultman et al. 1992; Vrieling et al. 1994) and pig 1A (Drogemuller et al. 2006) transcripts. Analysis of the Texel sheep BAC clone genomic sequence positioned the Barbary sheep noncoding 1A-like exon sequence ∼69 kb 5′ of ASIP copy 1 exon 2 in domestic sheep, 27 kb further upstream of the 5′ breakpoint (Fig. 5A). Our data support a functional ASIP promoter driving expression of a single ASIP gene determining coat color patterns in this ancient species of the Caprinae (goat–antelope) subfamily.
Discussion
It has taken more than a century of mouse classical genetics and two decades of molecular genetic analysis to begin to understand the molecular mechanisms regulating yellow coat color and the associated pleiotropic effects from deregulated murine ASIP expression (Wolff 2003). The molecular regulation of coat color of other mammalian species at the ASIP locus, however, still remains comparatively unknown. The expression of the mouse ASIP gene ordinarily is tightly spatially and temporally regulated and restricted primarily to the skin (Bultman et al. 1992). In humans, (Voisey et al. 2002), cattle (Girardot et al. 2005, 2006), and, as we have now shown, in domestic sheep, ASIP is more widely expressed with various transcripts identified from several tissues. While there are no apparent negative effects of this ubiquitous ASIP expression in sheep, the possible consequences of the wider tissue expression is still to be investigated. Interestingly, in Icelandic sheep, pleiotropic effects of the dominant white or tan (AWt) allele result in reduced fecundity and greater seasonality in reproduction (Adalsteinsson 1975). However, in mice ubiquitous ASIP expression results in adult onset obesity, suggesting that the signaling pathways that normally regulate reproduction and body weight may be different in these mammalian species.
It was also recently shown that the ASIP coding sequence has been completely deleted from the gibbon genome (Nakayama and Ishida 2006) with no apparent detrimental effect, while it remains intact in all other investigated primate genomes (Mundy and Kelly 2006). Nakayama and Ishida (2006) speculate that the gibbon ASIP deletion may affect its energy homeostasis and so contribute to the smaller body mass of the gibbon relative to other primates. Studies in humans and animal models have elucidated a role for components of the melanocortin pathway in immunity, energy homeostasis, and reproduction (Henry 2003; Carroll et al. 2005). ASIP regulation of lipid metabolism in adipose tissue has been examined in mouse and humans (Voisey et al. 2002), but to date, no studies have investigated the potential impact of increased or deregulated ASIP expression on ovine energy balance or fertility. However, lean sheep fed ad libitum can become obese and have proven to be a useful animal model for human obesity (McCann et al. 1991; Henry 2003). Investigating the impact of ASIP copy number variation on key production traits of leanness, body fat, and fertility could provide new insights into the physiology of energy metabolism and reproduction of livestock and other mammalian species.
Mammalian species thus far investigated have had a single copy of the ASIP gene identified within their genomes. The genetic cause of coat color patterns in domestic sheep at the ASIP locus has been difficult to determine due in part to the prospect of a second ASIP-like locus, which was proposed by Smit et al. (2002). Further, with the exception of the mouse (Bultman et al. 1992, 1994) and the pig (Drogemuller et al. 2006), mutations in the coding sequence that disrupt functional ASIP protein were associated with the recessive black phenotypes of these species (Vage et al. 1997; Kuramoto et al. 2001; Rieder et al. 2001; Eizirik et al. 2003; Kerns et al. 2004). We have shown that the regulation of the recessive black coat color phenotypes by the ovine ASIP gene is not attributable to simple coding mutations as described for other mammals and that the dominant white phenotype involves variation in gene copy number and deregulated expression.
Analysis of the ovine ASIP coding regions of recessive self-color black and badgerface Merino sheep identified four mutations, two of which (a 5-bp deletion and a g.5172T>A SNP) would be predicted to disrupt the functional protein. Both these coding sequence mutations, however, failed to completely associate with the recessive black Merino phenotypes. Of the recessive black Merinos, only ∼60% and 11% were homozygous for either the D5 or A allele, respectively, and none were homozygous for both. All white Merinos investigated were either homozygous for the normal alleles or heterozygous for one or the other of these mutations. Additionally all white Merinos had at least one duplicated ASIP allele, while all of the recessive black Merinos contained only single copy alleles.
ASIP expression was detected in all tissues examined from white sheep, with multiple transcripts identified from both the skin and internal tissues. Expression was not detected from any tissues of recessive self-color black sheep. Further, all transcripts identified from white sheep skin began with the ITCH noncoding exons It and It′, and none contained the 5-bp or g.5172T>A predicted nonfunctional mutations. These data indicated that expression in the white sheep was driven from the duplicated copy of an ITCH promoter positioned upstream of ASIP and that the identified functional mutations are not present in these expressed ASIP copies. The data also indicated that in domestic sheep the progenitor ASIP promoter, unlike the promoter of the single copy ASIP of ancient Barbary sheep, was not functional. Because ASIP expression was not detected in the single copy alleles of the recessive black sheep, the likely causative mutation of the recessive black phenotype is an as yet unidentified regulatory mutation of the progenitor gene promoter region.
In mice, the molecular events associated with an unusually high reversion rate of agouti recessive black coat color pattern mutations to more dominant alleles were identified to be as a result of insertions and deletions mediated by homologous recombination between repetitive elements (Bultman et al. 1994). In the gibbon, the removal of a 100-kb region including the ASIP coding sequence was mediated by Alu repeat elements (Nakayama and Ishida 2006). A large duplication involving the KIT locus, causing dominant white skin color in pigs, has originated by homologous recombination between LINE elements (Marklund et al. 1998; Giuffra et al. 2002). The sequences at the borders of the duplicated ovine ASIP gene segments contain repeat elements identified as BOV2 and BDDF2 non-LTR SINE sequences (Kohany et al. 2006). Thus, it seems likely that the ovine dominant white and recessive black phenotypes are mediated at least in part by homologous recombination involving these repeat elements. The sequencing and subsequent characterization of the human genome revealed that 5% of the human genome consists of long highly similar duplicated sequences known as low copy repeats (LCRs) or segmental duplications (Lindsay et al. 2006). It is now emerging that such segmental duplications are hotspots for allelic and nonallelic homologous recombination events that can result in genomic disorders causing clinical diseases (Bischof et al. 2006).
The exchange of sequence variations between duplicated loci known as gene conversion events mediated through allelic and nonallelic recombination (Myers and McCarroll 2006) can lead to a high degree of similarity between duplicated genes (Lindsay et al. 2006). A high degree of nucleotide sequence similarity (>99%) occurred between the 5379 bp of the multiple ASIP gene copies of each of the BAC clone sequences from the start of exon 2 to the end of the 3′-untranslated region (Supplemental Table S1). Gene conversion events together with allelic and nonallelic homologous recombination between repetitive sequences are likely to be contributing to variation and evolution of ovine ASIP sequences.
Based on our results, we propose a model for the evolution of the ovine ASIP locus (Fig. 6). The model proposes that a duplication, mediated by homologous sequences in SINES flanking the ASIP coding and ITCH promoter regions, occurred in an ancestor of domestic sheep, positioning a second copy of the ITCH promoter upstream of a second ASIP coding sequence, deregulating its expression. Subsequently, possibly with the creation of the duplicated segment, or by other successive mutation events, the progenitor ASIP promoter was inactivated. Sheep with genotypes that have at least one duplicated ASIP allele are always white as strong (artificial) selection pressure for white coat color has maintained the functional (N5, T) ASIP alleles at the expressed (copy 2) position (Fig. 6A). With the expressed (copy 2) ASIP functional coding sequence under strong selection pressure, the two ASIP regions subsequently diverged in sequence, generating functional and nonfunctional haplotype clusters (Supplemental Table S1; Fig. 6B). The most likely major route of generation of single copy loci was nonallelic pairing and recombination between duplicated alleles (Fig. 6C). The single copy recessive black causing alleles, with functional or nonfunctional coding sequence haplotypes, subsequently resulted from nonalleic recombination and/or gene conversion events (Fig. 6C). Our data from asymmetric competitive PCR copy number assays of recessive black and white Merinos show that while recessive black Merinos always have a single ASIP copy, white Merinos can have two to four and possibly even five ASIP copy alleles, further supporting this model.
Tandem gene duplication represents an underinvestigated source of molecular variation in livestock species. The duplication/deletion of the agouti gene is the first characterized example in sheep of the involvement of gene duplication in the creation of a genetic variation that contributes to a major breed and production trait—coat color phenotypes. This gene copy-number polymorphism introduces variation into the sheep genome and causes a phenotypic trait to which natural or artificial selection may then apply. The ovine chromosome 13 tandem duplication, encompassing the ASIP and AHCY genes and promoter region of the ITCH gene, is an ideal locus for the investigation of the consequences of duplication on sequence variation and evolution via natural and artificial selection in commercially important livestock species.
Classical genetic analysis first proposed alleles of the agouti gene as a major determinant of coat color phenotypes in sheep more than a half a century ago (Rendel 1957; Brooker and Dolling 1969). With a major goal of identifying the molecular cause of unwanted recessive black sheep phenotypes, the molecular genetic investigation of the ovine agouti gene in the Australian Merino sheep was begun a decade ago (Parsons et al. 1997, 1999a, b). Until now, without the benefit of an ovine genome sequence, progress in understanding the molecular mechanisms regulating coat color in domestic and wild sheep populations at the ASIP locus has been elusive. The characterization of a large tandem duplication encompassing the ASIP, AHCY, and ITCH genes provides an explanation for this difficulty. In the future, further comparative investigation of the agouti architecture in domestic sheep breeds and extant wild sheep populations will, in evolutionary terms, determine when this dominant agouti mutation first occurred and its impact on and evolution since the domestication of sheep.
Methods
Panel of sheep coat color phenotypes
Samples of 6 mL blood (EDTA tubes) and 0.9-mm skin biopsies were collected from Australian Merino sheep. All experiments were approved by and performed according to the guidelines of the CSIRO Chiswick NSW Animal Ethics Committee (AEC approval nos. 01/19, 02/03, 03/62, 04/30). Color photographs displaying the dorsal, ventral, sides, and face were taken of each pigmented animal for determination of the self-color black or badgerface Merino phenotypes. Sheep were sourced from a wide geographic distribution throughout Australia, and animals from different producers were presumed to be unrelated. The Merino DNA panel consisted of samples from 94 baldy self-color black sheep (from 28 different producers), 48 spotted self-color black sheep (from 16 producers), 48 badgerface animals (from 12 producers), 87 white carrier animals (57 confirmed to be carriers by commercial pedigree testing and 30 suspected carriers) from a total of 20 different producers, and 96 random white animals (of unknown carrier status) from 22 producers. Barbary sheep (A. lervia) blood and ventral skin samples were sourced from the Western Plains Zoo (Dubbo, NSW Australia).
Preparation of gDNA and total RNA
QiAmp DNA Mini kit (Qiagen) was used to extract genomic DNA from whole blood. Skin biopsies were collected in RNA Later (Ambion) and stored at −80°C. Total RNA was prepared from the skin of white and recessive black Merino sheep and from the ventral skin of Barbary sheep using TRIzol (Invitrogen) in accordance with the manufacturer’s recommendation. In each extraction, 5 mL of TRIzol was used to extract 200–500 mg of skin. To ensure removal of contaminating DNA, total RNA was treated with DNase1 (Ambion DNA-free). RNA quality was visually assessed using agarose gel electrophoresis and a UV transilluminator and quantified by spectrophotometry.
ASIP PCR, cloning, and sequencing
PCR primers (Supplemental Table S2 online) for the amplification of intron sequences from genomic DNA templates were designed from sheep ASIP EST sequence. PCR primers were designed to amplify intron 1 regions between noncoding exon 1E and coding exon 2, intron 2 regions between exon 2 and exon 3, and intron 3 regions between exons 3 and 4. PCR reactions (20 μL) contained 50 ng gDNA, 200 μM dNTPs, 10 pmole of each primer, 1× Q solution, 1.5 mM MgCl2, 0.5 units Taq polymerase (Qiagen), and 1× reaction buffer (Qiagen). PCR conditions were 3 min at 94°C; 30 sec at 94°C, 1 min at 57°C, and 2 min at 72°C for 35 cycles; and 5 min at 72°C. PCR products were amplified from a white Merino ram and a self-color black Merino lamb. PCR products were cloned using a TOPO TA Cloning Kit (Invitrogen) and sequenced using Big Dye Terminator 3.1 reaction mix. PCR products spanning exons 2, 3, and 4 were generated (see Supplemental Table S2 primer sequences) and sequenced directly to identify coding mutations in the DNA of selected animals from the panel of Merino coat color phenotypes. For direct sequencing, PCR products were processed with ExoSAP-IT (USB Corporation) according to the manufacturer’s recommendations. All sequencing was done using ABI Prism Big Dye Terminator 3.1 chemistry and either an ABI 377 Prism DNA autosequencer or ABI 3130xl Genetic Analyser (Applied Biosystems). Sequence data were imported into Sequencher v4.2 (Gene Codes Corp.) for analysis. Sequences from the white and black Merino identified as Haplotypes 1 and 2 in Supplemental Table S1 have been deposited in GenBank (accession nos. EU420022 and EU420023).
Genotyping polymorphisms
The presence of D9 and D5 polymorphisms in exon 2 of the ASIP coding region was analyzed by PCR spanning both indels (see Supplemental Table S2 primer sequences) in a 20 μL PCR reaction volume with Qiagen Taq performed according to the manufacturer’s recommendations. Alleles were visualized by autoradiography and/or fluorescence on an ABI 377 Prism DNA autosequencer or ABI 3130xl Genetic Analyser (Applied Biosystems). A TaqMan MGB genotyping assay was developed for the g.5172T>A SNP in exon 4 of ovine ASIP (Assay by Design, Applied Biosystems). Genotypes of the panel of Merino coat color phenotypes were automatically assigned with a quality score of 90, using the Applied Biosystems 7900HT sequence detector
Characterization of Ovis aries BAC library clones
The three INRA Romanov Sheep BAC library clones (Vaiman et al. 1999) were obtained from the BAC-YAC Resource center of the Animal Genetics Department of INRA (http://dga.jouy.inra.fr/grafra/INRA_libraries_database_simplified.htm). Shotgun plasmid libraries were prepared by the Australian Genome Research Facility (AGRF; http://www.agrf.org.au/) from each of three INRA ovine BAC clones INRA-164H8, INRA-218G7, and INRA-229C6 DNA that had tested positive for ASIP coding sequence by PCR. Clones INRA-164H8 and INRA-218G7 were sequenced to 8× coverage and clone INRA-229C6 to 2× coverage by the AGRF (GenBank accession nos. EU185098–EU185100). Sequence data were imported into Sequencher v4.2 (Gene Codes) for analysis. Clones INRA-218G7 (∼75 kb) and INRA-229C6 (∼150 kb) partially shared the same sequence and were aligned to the longer clone INRA-164H8 (∼150 kb). Twenty-nine Texel Sheep BAC clones (Dalrymple et al. 2007) predicted to span the region within two Megabase pairs of ASIP were ordered from the Children’s Hospital and Research Center at Oakland (CHRCO; http://bacpac.chori.org/home.htm). The 29 BAC clones were PCR analyzed for the presence or absence of both coding and noncoding regions of ASIP, AHCY, and ITCH genes. Primers used in this analysis are listed in Supplemental Table S2 online. BAC clone INRA-164H8 was used as the ovine reference sequence. BAC-end sequences from clones CH243-455O4 and CH243-489F15 that aligned to the reference sequence in a “head-to-head” orientation (with 5′ ends closest) were predicted to span a junction between duplicated DNA copies. BAC clones with end sequences that aligned to the reference sequence in a “tail-to-tail” orientation (with 3′ ends closest) were predicted to be fully contained within the reference sequence. Clone CH243-373J16 was predicted to provide sequence data further 5′ and clone CH243-160L8 further 3′ of the INRA-164H8 sequence. Clones CH243-234K21 and CH243-373J16 contained the putative nonfunctional “A” ASIP allele. These five clones of the 29 CHORI Texel sheep BAC clones—CH243-455O4, CH243-489F15, CH243-373J16, CH243-160L8, CH243-234K21—were selected for complete sequencing to 6× coverage (Macrogen; http://www.macrogen.com/eng/macrogen/macrogen_main2.jsp). Sequences were analyzed using Sequencher software v4.2 (GeneCodes). Sequences have been deposited in GenBank (GenBank accession nos. EU185093–EU185097).
Haplotypes phylogenetic analysis
The ovine ASIP haplotype sequences from this study were aligned and phylogenetic analyses performed using Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0 (Tamura et al. 2007). The p-distance was used to estimate genetic distances and neighbor joining used to construct the phylogeny. Testing of inferred phylogeny was by bootstrap with 500 replications.
RT-PCR analysis
First-strand oligo(dT)15 primed cDNA was generated using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. PCR products were amplified, labeled with 33P-dCTP, and separated by electrophoresis on a 5% polacrylamide gel and analyzed by autoradiography or visualized with ethidium bromide staining following electrophoresis on a 1.0% agarose gel.
5′ and 3′ RACE
For 5′ and 3′ RACE experiments respectively, oligo(dT)18 primed or UAP primed cDNA was prepared from 1–2 μg of total RNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. oligo(dT)18 primed cDNA was poly A-tailed with dATP using Terminal Transferase (NEB) and excess primer and dNTPs removed using Qiaquick PCR Purification columns (Qiagen). 5′ RACE products were amplified in touchdown PCR reactions (68°C –63°C) with primers GSP1 5′ and UAP (Supplemental Table S2) and Qiagen Taq according to the manufacturer’s instructions, followed by a nested PCR with primer GSP2 5′ and UAP. Nested PCR was also used to amplify 3′ RACE products. Primers GSP1 3′ and UAP were used in the first PCR reaction followed by primers GSP2 3′ and UAP in the second PCR reaction. PCR products were visualized by electrophoresis and ethidium bromide staining on 1.5% agarose gels. Products were gel purified and cloned for sequencing using TOPO TA Cloning (Invitrogen).
Junction point and 5′ breakpoint assay
To estimate ovine ASIP copy numbers, an asymmetric competitive PCR protocol was developed following modification to the PCR method of Pielberg et al. (2003). Both the junction between duplicated DNA copies and the 5′ breakpoint sequences were amplified in the one reaction tube. Primers Agt16 and Agt18 (Supplemental Table S2) spanning the junction between the duplicated copies produced a unique 242-bp product, while Agt16 and Agt17 spanning the 5′ breakpoint sequence produced a 238-bp product (Supplemental Fig. S2). The products were amplified by asymmetric competitive PCR in a 5 μL reaction volume in 384-well plates containing 5–50 ng gDNA, 500 μM dNTP’s, 3 mM MgCl2, 1.25× reaction buffer (Qiagen), 5 pmole of specific forward primers (Agt17 and Agt18), 0.05 pmole of shared reverse primer (Agt16), and 0.15 units of HotStar Taq polymerase (Qiagen). PCR conditions were 15 min at 95°C; 30 sec at 95°C, 30 sec at 60°C, and 1 min at 72°C for 40 cycles; and 5 min at 72°C. A 1 μL aliquot of a 1:10 dilution of PCR products was analyzed on an ABI 3130xl Genetic Analyser and the data analyzed using GeneMapper software (Liz 600 as size standard). The number of junction points (and ASIP copies) is estimated from the standard curve (Supplemental Fig. S3) using the ratio of the area under the 242-bp (junction point) peak compared with the total area represented by both peaks (junction point and breakpoint). To deduce discrete copy number categories for the data, peak areas were analyzed using the mean and standard deviation for each expected category (one, two, three, or four copies) and the normdist statistical function.
Acknowledgments
We thank S. McWilliam for assistance in the identification of the CHORI-243 BACs and management of BAC clone sequences and B. Dalrymple for demonstrating how to use the virtual sheep genome and related resources to identify sheep BACs and for his advice during the progress of the project. We thank D. Maxwell for sheep industry advice, assistance with industry liaison, the Merino photograph and together with G. Uphill for assistance with organization and collection of Merino Sheep industry animal samples. We thank the Australian sheep industry producers who provided sheep samples. Sincere thanks to T. Portas and the staff of Western Plains Zoo Dubbo, NSW Australia for the Barbary sheep samples and photograph. We thank J. McEwan for the Texel and J. Kirts for the Romanov sheep photographs. Thank you to I. Franklin and Y. LI for stimulating discussions and suggestions on genetics, agouti and sheep pigmentation. Thank you to G. Davies, R. Forage, W. Barendse, and J. van der Werf for their support. We are grateful to SheepGenomics, an initiative of Australian Wool Innovation Limited and Meat and Livestock Australia for bioinformatics support. Thank you to G. Wijffels, B. Dalrymple, W. Barendse, and F. Nicholas for comments on the manuscript. We gratefully acknowledge the BAC-YAC Resource centre of the Animal Genetics Department of INRA in providing the INRA Romanov sheep BAC clones and the Children’s Hospital and Research Centre at Oakland for the Texel sheep BAC clones. This work was a project of the Co-operative Research Centre for the Australian Sheep Industry, Armidale, Australia.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. EU185093–EU185100 and EU420022–EU420031.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.072090.107.
References
- Adalsteinsson S. Colour inheritance in Icelandic sheep and relation between colour, fertility and fertilisation. J. Agric. Res. Iceland. 1970;2:3–135. [Google Scholar]
- Adalsteinsson S. Depressed fertility in Icelandic sheep caused by a single colour gene. Ann. Genet. Sel. Anim. 1975;7:445–447. doi: 10.1186/1297-9686-7-4-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.C., Lamoreux M.L., Lamoreux M.L. The colour loci of mice—A genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Bischof J.M., Chiang A.R., Scheetz T.E., Stone E.M., Casavant T.L., Sheffield V.C., Braun T.A., Chiang A.R., Scheetz T.E., Stone E.M., Casavant T.L., Sheffield V.C., Braun T.A., Scheetz T.E., Stone E.M., Casavant T.L., Sheffield V.C., Braun T.A., Stone E.M., Casavant T.L., Sheffield V.C., Braun T.A., Casavant T.L., Sheffield V.C., Braun T.A., Sheffield V.C., Braun T.A., Braun T.A. Genome-wide identification of pseudogenes capable of disease-causing gene conversion. Hum. Mutat. 2006;27:545–552. doi: 10.1002/humu.20335. [DOI] [PubMed] [Google Scholar]
- Bonilla C., Boxill L.A., Donald S.A., Williams T., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Boxill L.A., Donald S.A., Williams T., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Donald S.A., Williams T., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Williams T., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Dios S., Norton H.L., Shriver M.D., Kittles R.A., Norton H.L., Shriver M.D., Kittles R.A., Shriver M.D., Kittles R.A., Kittles R.A. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin colour in African Americans. Hum. Genet. 2005;116:402–406. doi: 10.1007/s00439-004-1251-2. [DOI] [PubMed] [Google Scholar]
- Brooker M.G., Dolling C.H.S., Dolling C.H.S. Pigmentation of sheep. II. The inheritance of coloured patterns in black Merino. Aust. J. Agric. Res. 1969;20:387–394. [Google Scholar]
- Bultman S.J., Michaud E.J., Woychik R.P., Michaud E.J., Woychik R.P., Woychik R.P. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–1204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- Bultman S.J., Klebig M.L., Michaud E.J., Sweet H.O., Davisson M.T., Woychik R.P., Klebig M.L., Michaud E.J., Sweet H.O., Davisson M.T., Woychik R.P., Michaud E.J., Sweet H.O., Davisson M.T., Woychik R.P., Sweet H.O., Davisson M.T., Woychik R.P., Davisson M.T., Woychik R.P., Woychik R.P. Molecular analysis of reverse mutations from nonagouti (a) to black-and-tan (at) and white-bellied agouti (Aw) reveals alternative forms of agouti transcripts. Genes & Dev. 1994;8:481–490. doi: 10.1101/gad.8.4.481. [DOI] [PubMed] [Google Scholar]
- Carroll L., Voisey J., van Daal A., Voisey J., van Daal A., van Daal A. Gene polymorphisms and their effects in the melanocortin system. Peptides. 2005;26:1871–1885. doi: 10.1016/j.peptides.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Dalrymple B.P., Kirkness E.F., Nefedov M., McWilliam S., Ratnakumar A., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., Kirkness E.F., Nefedov M., McWilliam S., Ratnakumar A., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., Nefedov M., McWilliam S., Ratnakumar A., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., McWilliam S., Ratnakumar A., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., Ratnakumar A., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., Barris W., Zhao S., Shetty J., Maddox J.F., O Grady M., Zhao S., Shetty J., Maddox J.F., O Grady M., Shetty J., Maddox J.F., O Grady M., Maddox J.F., O Grady M., O Grady M., et al. Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biol. 2007;8:R152. doi: 10.1186/gb-2007-8-7-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C., Giese A., Martins-Wess F., Wiedemann S., Andersson L., Brenig B., Fries R., Leeb T., Giese A., Martins-Wess F., Wiedemann S., Andersson L., Brenig B., Fries R., Leeb T., Martins-Wess F., Wiedemann S., Andersson L., Brenig B., Fries R., Leeb T., Wiedemann S., Andersson L., Brenig B., Fries R., Leeb T., Andersson L., Brenig B., Fries R., Leeb T., Brenig B., Fries R., Leeb T., Fries R., Leeb T., Leeb T. The mutation causing the black-and-tan phenotype of Mangalitza pigs maps to the porcine ASIP locus but does not affect its coding sequence. Mamm. Genome. 2006;17:58–66. doi: 10.1007/s00335-005-0104-1. [DOI] [PubMed] [Google Scholar]
- Duhl D.M., Vrieling H., Miller K.A., Wolff G.L., Barsh G.S., Vrieling H., Miller K.A., Wolff G.L., Barsh G.S., Miller K.A., Wolff G.L., Barsh G.S., Wolff G.L., Barsh G.S., Barsh G.S. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Eizirik E., Yuhki N., Johnson W.E., Menotti-raymond M., Hannah S.S., Obrien S.J., Yuhki N., Johnson W.E., Menotti-raymond M., Hannah S.S., Obrien S.J., Johnson W.E., Menotti-raymond M., Hannah S.S., Obrien S.J., Menotti-raymond M., Hannah S.S., Obrien S.J., Hannah S.S., Obrien S.J., Obrien S.J. Molecular genetics and evolution of melanism in the cat family. Curr. Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., Baya M., Beal J.S., Bello S.M., Beal J.S., Bello S.M., Bello S.M., et al. The Mouse Genome Database (MGD): From genes to mice—A community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot M., Martin J., Guibert S., Leveziel H., Julien R., Oulmouden A., Martin J., Guibert S., Leveziel H., Julien R., Oulmouden A., Guibert S., Leveziel H., Julien R., Oulmouden A., Leveziel H., Julien R., Oulmouden A., Julien R., Oulmouden A., Oulmouden A. Widespread expression of the bovine Agouti gene results from at least three alternative promoters. Pigment Cell Res. 2005;18:34–41. doi: 10.1111/j.1600-0749.2004.00195.x. [DOI] [PubMed] [Google Scholar]
- Girardot M., Guibert S., Laforet M.P., Gallard Y., Larroque H., Oulmouden A., Guibert S., Laforet M.P., Gallard Y., Larroque H., Oulmouden A., Laforet M.P., Gallard Y., Larroque H., Oulmouden A., Gallard Y., Larroque H., Oulmouden A., Larroque H., Oulmouden A., Oulmouden A. The insertion of a full-length Bos taurus line element is responsible for a transcriptional deregulation of the Normande Agouti gene. Pigment Cell Res. 2006;19:346–355. doi: 10.1111/j.1600-0749.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- Giuffra E., Tornsten A., Marklund S., Bongcam-Rudloff E., Chardon P., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Tornsten A., Marklund S., Bongcam-Rudloff E., Chardon P., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Marklund S., Bongcam-Rudloff E., Chardon P., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Bongcam-Rudloff E., Chardon P., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Chardon P., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Kijas J.M., Anderson S.I., Archibald A.L., Andersson L., Anderson S.I., Archibald A.L., Andersson L., Archibald A.L., Andersson L., Andersson L. A large duplication associated with dominant white colour in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome. 2002;13:569–577. doi: 10.1007/s00335-002-2184-5. [DOI] [PubMed] [Google Scholar]
- Hayman R.H., Cooper D.W., Cooper D.W. The frequency of pigmented sheep in the Australian Merino. Wool Technol. Sheep Breeding. 1965;12:81–85. [Google Scholar]
- Henry B.A. Links between the appetite regulating systems and the neuroendocrine hypothalamus: Lessons from the sheep. J. Neuroendocrinol. 2003;15:697–709. doi: 10.1046/j.1365-2826.2003.01049.x. [DOI] [PubMed] [Google Scholar]
- Kanetsky P.A., Swoyer J., Panossian S., Holmes R., Guerry D., Rebbeck T.R., Swoyer J., Panossian S., Holmes R., Guerry D., Rebbeck T.R., Panossian S., Holmes R., Guerry D., Rebbeck T.R., Holmes R., Guerry D., Rebbeck T.R., Guerry D., Rebbeck T.R., Rebbeck T.R. A polymorphism in the agouti signalling protein gene is associated with human pigmentation. Am. J. Hum. Genet. 2002;70:770–775. doi: 10.1086/339076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns J.A., Newton J., Berryere T.G., Rubin E.M., Cheng J.F., Schmutz S.M., Barsh G.S., Newton J., Berryere T.G., Rubin E.M., Cheng J.F., Schmutz S.M., Barsh G.S., Berryere T.G., Rubin E.M., Cheng J.F., Schmutz S.M., Barsh G.S., Rubin E.M., Cheng J.F., Schmutz S.M., Barsh G.S., Cheng J.F., Schmutz S.M., Barsh G.S., Schmutz S.M., Barsh G.S., Barsh G.S. Characterization of the dog Agouti gene and a nonagouti mutation in German Shepherd dogs. Mamm. Genome. 2004;15:798–808. doi: 10.1007/s00335-004-2377-1. [DOI] [PubMed] [Google Scholar]
- Kohany O., Gentles A., Hankus L., Jurka J., Gentles A., Hankus L., Jurka J., Hankus L., Jurka J., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto T., Nomoto T., Sugimura T., Ushijima T., Nomoto T., Sugimura T., Ushijima T., Sugimura T., Ushijima T., Ushijima T. Cloning of the rat Agouti gene and identification of the rat nonagouti mutation. Mamm. Genome. 2001;12:469–471. doi: 10.1007/s003350020010. [DOI] [PubMed] [Google Scholar]
- Kwon H.Y., Bultman S.J., Loffler C., Chen W.J., Furdon P.J., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Bultman S.J., Loffler C., Chen W.J., Furdon P.J., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Loffler C., Chen W.J., Furdon P.J., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Chen W.J., Furdon P.J., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Furdon P.J., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Powell J.G., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Usala A.L., Wilkison W., Hansmann I., Woychik R.P., Wilkison W., Hansmann I., Woychik R.P., Hansmann I., Woychik R.P., Woychik R.P. Molecular structure and chromosomal mapping of the human homolog of the Agouti gene. Proc. Natl. Acad. Sci. 1994;91:9760–9764. doi: 10.1073/pnas.91.21.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb T., Deppe A., Kriegesmann B., Brenig B., Deppe A., Kriegesmann B., Brenig B., Kriegesmann B., Brenig B., Brenig B. Genomic structure and nucleotide polymorphisms of the porcine Agouti signalling protein gene (ASIP) Anim. Genet. 2000;31:335–336. doi: 10.1046/j.1365-2052.2000.00656.x. [DOI] [PubMed] [Google Scholar]
- Lindsay S.J., Khajavi M., Lupski J.R., Hurles M.E., Khajavi M., Lupski J.R., Hurles M.E., Lupski J.R., Hurles M.E., Hurles M.E. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund S., Kijas J., Rodriguez-Martinez H., Ronnstrand L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L., Kijas J., Rodriguez-Martinez H., Ronnstrand L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L., Rodriguez-Martinez H., Ronnstrand L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L., Ronnstrand L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L., Moller M., Lange D., Edfors-Lilja I., Andersson L., Lange D., Edfors-Lilja I., Andersson L., Edfors-Lilja I., Andersson L., Andersson L. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998;8:826–833. doi: 10.1101/gr.8.8.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J.P., Bergman E.N., Beermann D.H., Bergman E.N., Beermann D.H., Beermann D.H. Dynamic and static phases of severe dietary obesity in sheep: Food intakes, endocrinology and carcass and organ chemical composition. J. Nutr. 1991;122:496–505. doi: 10.1093/jn/122.3.496. [DOI] [PubMed] [Google Scholar]
- Mundy N.I., Kelly J., Kelly J. Investigation of the role of the Agouti signalling protein gene (ASIP) in coat colour evolution in primates. Mamm. Genome. 2006;17:1205–1213. doi: 10.1007/s00335-006-0056-0. [DOI] [PubMed] [Google Scholar]
- Myers S.R., McCarroll S.A., McCarroll S.A. New insights into the biological basis of genomic disorders. Nat. Genet. 2006;38:1363–1364. doi: 10.1038/ng1206-1363. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Ishida T., Ishida T. Alu-mediated 100-kb deletion in the primate genome: the loss of the Agouti signalling protein gene in the lesser apes. Genome Res. 2006;16:485–490. doi: 10.1101/gr.4763906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons Y.M., Fleet M.R., Cooper D.W., Fleet M.R., Cooper D.W., Cooper D.W. Investigation of the gene responsible for recessive pigmentation in Australian Merino sheep. Proc. Assoc. Advanc. Anim. Breed. Genet. 1997;12:447–451. [Google Scholar]
- Parsons Y.M., Fleet M.R., Cooper D.W., Fleet M.R., Cooper D.W., Cooper D.W. The Agouti gene: A positional candidate for recessive self-colour black pigmentation in Australian Merino sheep. Aust. J. Agric. Res. 1999a;50:1099–1103. [Google Scholar]
- Parsons Y.M., Fleet M.R., Cooper D.W., Fleet M.R., Cooper D.W., Cooper D.W. Isolation of the ovine Agouti coding sequence. Pigment Cell Res. 1999b;12:394–397. doi: 10.1111/j.1600-0749.1999.tb00524.x. [DOI] [PubMed] [Google Scholar]
- Perry W.L., Hustad C.M., Swing D.A., O’Sullivan T.N., Jenkins N.A., Copeland N.G., Hustad C.M., Swing D.A., O’Sullivan T.N., Jenkins N.A., Copeland N.G., Swing D.A., O’Sullivan T.N., Jenkins N.A., Copeland N.G., O’Sullivan T.N., Jenkins N.A., Copeland N.G., Jenkins N.A., Copeland N.G., Copeland N.G. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- Pielberg G., Day A.E., Plastow G.S., Andersson L., Day A.E., Plastow G.S., Andersson L., Plastow G.S., Andersson L., Andersson L. A sensitive method for detecting variation in copy numbers of duplicated genes. Genome Res. 2003;13:2171–2177. doi: 10.1101/gr.1188203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendel J. Heredity of colour and white designs in farm animals. Kunglishe Hogskole Landbruksadamie Tidskrift. 1957;96:207–263. [Google Scholar]
- Rieder S., Taourit S., Mariat D., Langlois B., Guerin G., Taourit S., Mariat D., Langlois B., Guerin G., Mariat D., Langlois B., Guerin G., Langlois B., Guerin G., Guerin G. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat colour phenotypes in horses (Equus caballus) Mamm. Genome. 2001;12:450–455. doi: 10.1007/s003350020017. [DOI] [PubMed] [Google Scholar]
- Smit M.A., Shay T.L., Beever J.E., Notter D.R., Cockett N.E., Shay T.L., Beever J.E., Notter D.R., Cockett N.E., Beever J.E., Notter D.R., Cockett N.E., Notter D.R., Cockett N.E., Cockett N.E. Identification of an agouti -like locus in sheep. Anim. Genet. 2002;33:383–385. doi: 10.1046/j.1365-2052.2002.00896_5.x. [DOI] [PubMed] [Google Scholar]
- Sponenberg D.P. Genetics of colour and hair texture. In: Piper L.R., Ruvinsky A., Ruvinsky A., editors. The genetics of sheep. New York, NY: 1997. pp. 51–86. [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., Dudley J., Nei M., Kumar S., Nei M., Kumar S., Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vage D.I., Lu D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Lu D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Lien S., Adalsteinsson S., Cone R.D., Adalsteinsson S., Cone R.D., Cone R.D. A non-epistatic interaction of agouti and extension in the fox, Vulpes vulpes. Nat. Genet. 1997;15:311–315. doi: 10.1038/ng0397-311. [DOI] [PubMed] [Google Scholar]
- Vaiman D., Billault A., Tabet-Aoul K., Schibler L., Vilette D., Oustry-Vaiman A., Soravito C., Cribiu E.P., Billault A., Tabet-Aoul K., Schibler L., Vilette D., Oustry-Vaiman A., Soravito C., Cribiu E.P., Tabet-Aoul K., Schibler L., Vilette D., Oustry-Vaiman A., Soravito C., Cribiu E.P., Schibler L., Vilette D., Oustry-Vaiman A., Soravito C., Cribiu E.P., Vilette D., Oustry-Vaiman A., Soravito C., Cribiu E.P., Oustry-Vaiman A., Soravito C., Cribiu E.P., Soravito C., Cribiu E.P., Cribiu E.P. Construction and characterization of a sheep BAC library of three genome equivalents. Mamm. Genome. 1999;10:585–587. doi: 10.1007/s003359901049. [DOI] [PubMed] [Google Scholar]
- Voisey J., van Daal A., van Daal A. Agouti: From mouse to man, from skin to fat. Pigment Cell Res. 2002;15:10–18. doi: 10.1034/j.1600-0749.2002.00039.x. [DOI] [PubMed] [Google Scholar]
- Voisey J., Imbeault P., Hutley L., Prins J.B., van Daal A., Imbeault P., Hutley L., Prins J.B., van Daal A., Hutley L., Prins J.B., van Daal A., Prins J.B., van Daal A., van Daal A. Body mass index-related human adipocyte agouti expression is sex-specific but not depot-specific. Obes. Res. 2002;10:447–452. doi: 10.1038/oby.2002.62. [DOI] [PubMed] [Google Scholar]
- Voisey J., Gomez-Cabrera M.D.C., Smit D.J., Leonard J.H., Sturm R.A., van Daal A., Gomez-Cabrera M.D.C., Smit D.J., Leonard J.H., Sturm R.A., van Daal A., Smit D.J., Leonard J.H., Sturm R.A., van Daal A., Leonard J.H., Sturm R.A., van Daal A., Sturm R.A., van Daal A., van Daal A. A polymorphism in the Agouti signalling protein (ASIP) is associated with decreased levels of mRNA. Pigment Cell Res. 2006;19:226–231. doi: 10.1111/j.1600-0749.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- Vrieling H., Duhl D.M., Millar S.E., Miller K.A., Barsh G.S., Duhl D.M., Millar S.E., Miller K.A., Barsh G.S., Millar S.E., Miller K.A., Barsh G.S., Miller K.A., Barsh G.S., Barsh G.S. Differences in dorsal and ventral pigmentation result from regional expression of the mouse Agouti gene. Proc. Natl. Acad. Sci. 1994;91:5667–5671. doi: 10.1073/pnas.91.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G.L. Regulation of yellow pigment formation in mice: A historical perspective. Pigment Cell Res. 2003;16:2–15. doi: 10.1034/j.1600-0749.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- Wolff G.L., Roberts D.W., Mountjoy K.G., Roberts D.W., Mountjoy K.G., Mountjoy K.G. Physiological consequences of ectopic agouti gene expression: The yellow obese mouse syndrome. Physiol. Genomics. 1999;1:151–163. doi: 10.1152/physiolgenomics.1999.1.3.151. [DOI] [PubMed] [Google Scholar]