Abstract
Segmental duplications (SDs) play an important role in genome rearrangement, evolution, and the copy-number variation (CNV) of primate genomes. Such sequences are difficult to detect, a priori, because they share no defining sequence features that distinguish them from unique portions of the genome. Current sequence annotation of segmental duplications requires computationally intensive, genome-wide self-comparisons that cannot be easily implemented on new data sets. Based on the successful implementation of RepeatMasker, we developed a new genome annotation tool, DupMasker. The program uses a library of nonredundant consensus sequences of human segmental duplications, wherein a majority of the ancestral origins have been determined based on comparisons to mammalian outgroup genomes. Using DupMasker, new human and nonhuman primate (NHP) sequences may be readily queried to provide details on the origin and degree of sequence identity of each duplicon. This program can be applied to delineate the order and orientation of duplicons within complex duplication blocks and used to characterize structural variation differences between sequenced human haplotypes. We predict this tool will be valuable in the annotation of large-insert sequence clones, allowing putative unique and duplicated regions of the genomes to be annotated prior to whole genome assembly comparisons.
Initial analysis of the human genome and other primate genomes reveals that 4%–6% of each genome is composed of segmental duplications (Bailey et al. 2001, 2002; Cheung et al. 2003; Chen et al. 2004; She et al. 2004, 2006; Sainz et al. 2006). We now know that segmental duplications are hot spots for non-allelic homologous recombination (NAHR), copy-number variations (CNVs), and genomic rearrangements, leading to more than two dozen genomic diseases (Lupski 1998). The organization of human segmental duplications is complex. They are arranged into duplication blocks of mosaic architecture made up of many independent duplication events (termed duplicons) that have both shared and independent evolutionary histories (Jiang et al. 2007). These patterns are difficult to discern based solely on pairwise alignments and usually require detailed hand curation to delineate the evolutionary breakpoint boundaries.
Current methods used to detect segmental duplications are based on a self-comparison of the entire genome or based on comparison of whole genome shotgun sequence data against a reference genome (Bailey et al. 2001, 2002). These methods have two notable limitations. First, the existing pipelines are computationally intensive and are not easily implemented on new genome assemblies or incomplete data sets. Second, the output of these available methods provides limited information regarding the substructure, relationship, or ancestral origin of the segmental duplications (Jiang et al. 2007). As a result, cross-comparison between loci or species is limited to a series of pairwise alignments and is complicated by the difficulty of mapping between incompletely sequenced paralogs.
Taking advantage of the consensus sequence library and ancestral state information provided by our previous study (Jiang et al. 2007), we developed the software DupMasker, which (1) defines the orientation of individual duplicons for a given primate genomic sequence, (2) delineates the fine mosaic substructure for a given complex duplication block, and (3) provides information regarding the ancestral origin for 70% of human segmental duplications.
Results
We developed DupMasker in three basic steps. We first constructed a library (duplib) of consensus sequences for duplication subunits (size ≥100 bp) (Jiang et al. 2007), which captures 97.2% of the sequence information within the human set of segmental duplications (≥90% identity and ≥1 kb in length). Previously, we decomposed all human 28,856 pairwise alignments into a nonredundant set of 12,087 duplication subunits using a modified “A-Bruijn” graph algorithm. Of these, the ancestral origin could be determined for 67.3% by comparison with mammalian outgroup species. We generated a representative consensus sequence for each of these duplicons and identified each duplicon by its ancestral map location in the human genome, adding biological definition to the library. We note, however, that the ancestral location for ∼30% of the duplicated base pairs (particularly those organized as tandem clusters) is currently impossible to resolve due to gene conversion (a.k.a. concerted evolution) or ambiguous mapping to ancestral mammalian species. In these cases, the duplications are simply represented as human duplication subunit consensus as opposed to ancestral duplicons.
The next step integrates the duplication library into a modified version of RepeatMasker, which performs a sequence comparison of query sequence and consensus sequences within duplib. The procedure initially excludes common primate repeat sequences using RepeatMasker libraries specific for each species. Seed alignments are then generated based on comparing the remaining input sequence to the human duplication library. Next, duplicons are clustered according to ID and consensus agreement, and edges are extended along the query sequence until either a consensus is exhausted or a region of nonrepeat masked sequence (>7 kb) is encountered. This length boundary was selected as the upper bound for most retrotransposon L1 insertions. The clusters with similar IDs are then grouped, and groups of consensus and bounded query regions are realigned using WUBLAST2 (Washington University BLAST version 2.0).
The current version of the program uses a human library consisting of 12,087 duplication subunits and generates two standard outputs. These outputs include a file containing the duplication seed information and a second file that contains the information of locally chained duplicons, including duplication subunit ID and orientation in respect to consensus sequence and ancestral locus information.
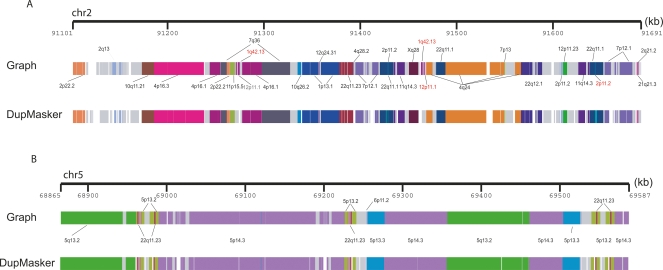
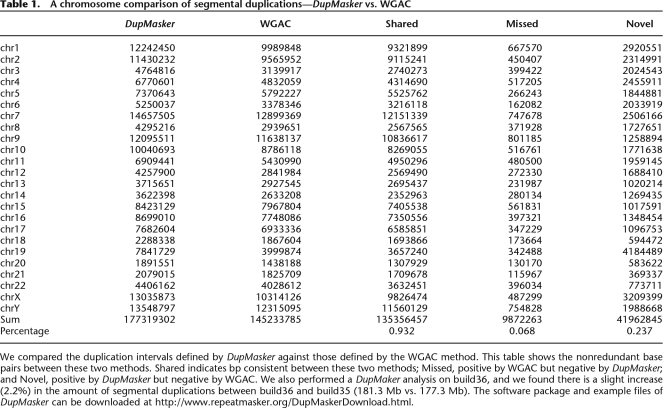
Based on this design, we constructed a prototype version of DupMasker and assessed its efficacy by benchmarking it against previously annotated human segmental duplications mapping to chromosome 2p11 and 5q13.2 regions (Fig. 1A,B). A comparison of previously validated duplication structures with those determined by DupMasker shows very good correspondence (33/36 duplication subunits correctly identified with previous annotated sequences) (Horvath et al. 2000; Jiang et al. 2007). Several limitations were noted, especially in the treatment of repeats within or near the boundaries of segmental duplications. For example, some smaller subunits were not identified simply due to overlap with low-complexity repeat sequence. More importantly, the enrichment of common repeats at the boundaries of the duplicons significantly limited our precision in defining the edges of each duplicon using the initial prototype. To eliminate potential repeat-induced artifacts, we excluded all duplicons that contained <50 bp of nonrepeat sequence. Finally, we empirically assessed differential weighting schemes to improve junction detection. Based on these modifications, we estimate that ∼93.2% of human input sequence can now be correctly annotated as segmental duplication (Table 1).
Figure 1.
DupMasker defines the substructure of human segmental duplication blocks. Human segmental duplications are organized into complex duplication blocks where individual duplicons originate from different regions of the genome. We assessed the ability of DupMasker to accurately define these ancestral duplicons in this context by comparing results from two regions studied previously in detail (Horvath et al. 2000; Jiang et al. 2007). We schematically display (PARASIGHT: http://eichlerlab.gs.washington.edu/jeff/parasight/index.html) duplicons detected using the A-Bruijn graph approach (top) versus DupMasker (bottom) for (A), an ∼600-kbp region on chromosome 2p11 and (B), an ∼700-kbp region on chromosome 5q13.2. The different duplicons are illustrated as color-coded blocks; different colors correspond to different cytogenetic band locations of the ancestral loci. We found 33/36 nonredundant duplicons blocks are consistent between these two results. The three mismatched blocks are relatively small in length (length <1.5 kb, highlighted in red).
Table 1.
A chromosome comparison of segmental duplications—DupMasker vs. WGAC
We compared the duplication intervals defined by DupMasker against those defined by the WGAC method. This table shows the nonredundant base pairs between these two methods. Shared indicates bp consistent between these two methods; Missed, positive by WGAC but negative by DupMasker; and Novel, positive by DupMasker but negative by WGAC. We also performed a DupMaker analysis on build36, and we found there is a slight increase (2.2%) in the amount of segmental duplications between build36 and build35 (181.3 Mb vs. 177.3 Mb). The software package and example files of DupMasker can be downloaded at http://www.repeatmasker.org/DupMaskerDownload.html.
In order to assess the validity of DupMasker as a stand-alone program to accurately identify segmental duplications, we analyzed the entire human genome (build35) using DupMasker and compared the consistency between DupMasker results versus Whole Genome Assembly Comparison (WGAC) data (Table 1). Overall, 93.2% of duplications (135.35 Mb/145.23 Mb) are consistent (shared) between these two methods. A relatively small fraction (6.9% or 9.87 Mb) was identified by WGAC but not detected by DupMasker as a segmental duplication. Sequence analysis of these “missed” duplications showed that the majority (8.99 Mb/9.87 Mb = 91.1% by base pair composition) corresponded to common repeat sequences. Such losses are expected for segmental duplications enriched in common repeats due to the initial triage design of DupMasker, which excludes repeat regions.
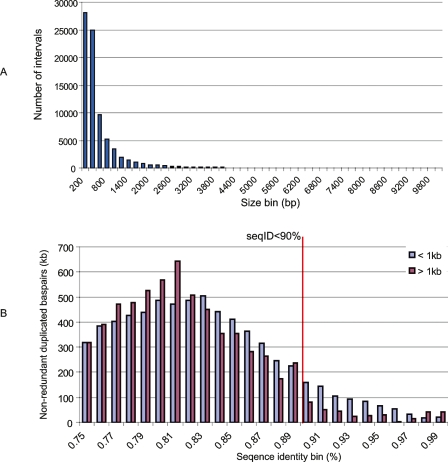
In contrast, DupMasker predicted 41.96 Mb of duplications that were not originally classified using the WGAC method. We termed these DupMasker-only duplications as “novel” segmental duplications. Similar to RepeatMasker, DupMasker has the ability to detect smaller and more divergent duplications (>75% identity with respect to the consensus and less than 1 kbp in length). The WGAC procedure operationally defines segmental duplications as pairwise alignments 1 kbp or more and 90% or more sequence identity. We therefore assessed the length and percentage of identity distribution of these putative “novel” SDs. We found that 91.0% (38.17/41.96 Mb) of these duplications were less than 1 kb (Fig. 2A). More than half of these novel intervals are common repeats (21.95/41.96 Mb) due to the imprecision of boundary definition within repeat-rich regions. We also performed a modified WGAC analysis on the 41.96 Mb using a more relaxed threshold (nonrepetitive sequence alignment size 100 bp or more and BLAST-sequence identity 75% or more) than that of standard WGAC. This modified WGAC analysis identified alignments for 31% (13.1/41.96 Mb) of these “novel” SDs. Among these 13.1 Mb alignments, 97.7% (12.8/13.1 Mb) represent either small (size < 1 kbp) or relatively ancient duplications (sequence identity < 90%) (Fig. 2B; Supplemental Table 1).
Figure 2.
The size and sequence identity distribution of “novel” duplications. (A) The length distribution of DupMasker duplications not detected by WGAC (termed “novel” SDs) reveals that the majority (99% by number of intervals, 91% by base pair) of these intervals are small fragments (size <1 kb). (B) We found 52.3% (21.9 Mb) of these small intervals are common repeats due to imprecision of boundary definition within repeat-rich regions. We performed a modified WGAC analysis using a relaxed threshold (require nonrepeat alignment ≥100 bp and sequence identity ≥75%) on these “novel” SDs. The analysis revealed alignments for 31% (13.1/41.96 Mb) of these “novel” SDs. Among the 13.1-Mb alignments, 97.7% (12.8/13.1 Mb) represent either small (size <1 kbp) or relatively ancient duplications (sequence identity <90%).
We tested three different applications of DupMasker: the analysis of regions flanking genomic disorders, the analysis of sequence from regions of structural variation, and a genome-wide analysis of a nonhuman primate genome assembly. Results from these various applications illustrate the utility of this software tool.
Analysis of regions associated with genomic disorders
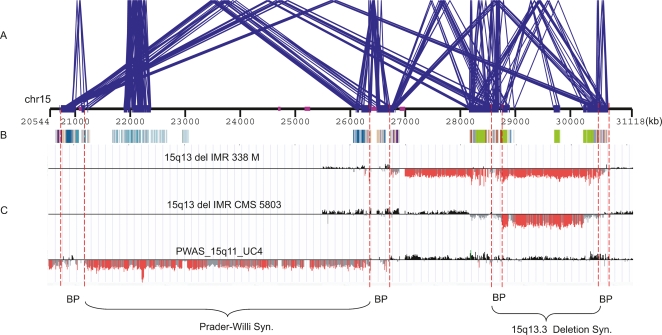
Duplication-rich regions of the human genome are hotspots of NAHR, leading to many human diseases, known as genomic disorders (Lupski 1998; Sharp et al. 2005, 2006, 2008; Mefford et al. 2007). Delineating the duplication architecture of those regions and their underlying LCRs (low copy repeats) or duplicons is important for understanding not only the evolutionary origin but likely sequences that promote non-allelic homologous recombination. DupMasker allows the duplication architecture flanking these regions to be decoded and provides information regarding the divergence and orientation of each individual fragment. Figure 3 is a schematic showing the architecture, as predicted by DupMasker, one of the most unstable regions of the human genome associated with Prader-Willi syndrome, and a recently described mental retardation syndrome (Sharp et al. 2008). DupMasker identifies candidate duplicons of high-sequence identity and proper orientation (color-coded boxes). The duplication architecture corresponds to breakpoints defined by arrayCGH experimental results (highlighted by dashed lines in Fig. 3). These results highlight the utility of DupMasker to predict regions of potential instability associated with NAHR-mediated microdeletion syndromes.
Figure 3.
Duplication architecture flanking genomic disorders. This figure shows the duplication architecture defined by DupMasker for one of the most unstable regions of the human genome (15q11–15q13). (A) Blue lines delineate intrachromosomal duplications of high-sequence identity (size ≥10 kb and sequence identity ≥95%) within this region (WGAC) and identify four breakpoint regions associated with Prader-Willi/Angelman Syndrome and the 15q13.3 deletion syndrome. (B) The duplication substructures defined by DupMasker are depicted as color-coded boxes with different colors representing different cytogenetic band locations of duplicons. (C) ArrayCGH data from one patient with Prader-Willi syndrome (bottom) and two patients with chr15q13.3 deletion (Sharp et al. 2008) indicate the patients’ deletion breakpoints overlap with the duplicons defined by DupMasker. The locations of the breakpoint intervals are highlighted by red dashed lines.
Analysis of sequenced clones
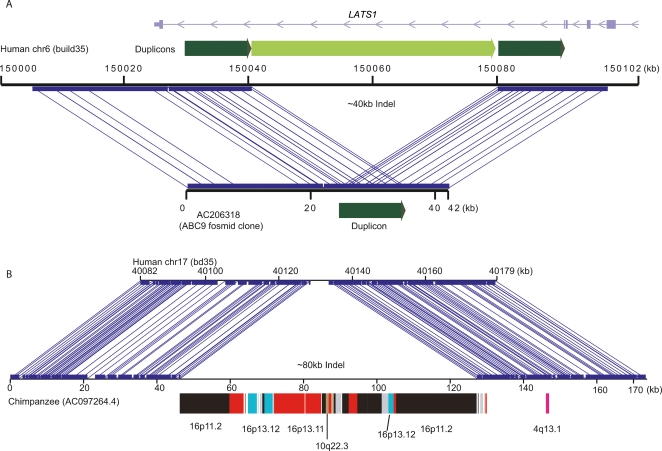
Another application for DupMasker is to annotate the duplication composition of sequenced clones, such as fosmid or BAC clones. This can be used to readily exclude certain regions for PCR or oligonucleotide design based on the underlying copy-number and sequence identity of the duplications. Moreover, regions of copy-number variation are particularly enriched in segmental duplications (Sharp et al. 2005; Redon et al. 2006), and annotated duplication maps of two sequences can be used to reconstruct the series of rearrangements that have occurred between any two human haplotypes. Since many of the segmental duplications are shared between humans and other nonhuman primate species, this is particularly valuable when characterizing nonhuman primate sequences that appear rearranged compared with the human genome. Figure 4 shows examples of structural variation between human haplotypes and between species that can be characterized using DupMasker. Figure 4A reveals a large deletion in human individual (ABC9) mediated by an NAHR between flanking duplicons, while Figure 4B depicts a lineage-specific segmental duplication insertion event in chimpanzee compared with the corresponding human sequence. We predict that DupMasker will be particularly valuable in annotating the breakpoints of CNVs and speciation chromosomes, which are significantly enriched for segmental duplications (Armengol et al. 2003; Bailey et al. 2004).
Figure 4.
Genomic comparisons by DupMasker. DupMasker facilitates the characterization of duplication-mediated genomic rearrangements. (A) Miropeats (Parsons 1995) comparison between human reference genome (build35, top) against a fosmid clone (bottom) from a Japanese individual (ABC9) identifies a ∼40-kbp deletion. DupMasker on this region identified a pair of tandem duplications (dark green) flanking the internal duplicon (light green), which was likely deleted by NAHR in this Japanese individual. The deletion removes part of the intron of the LATS1 gene. (B) A similar comparison between sequences from a chimpanzee BAC clone (AC097264.4) and its orthologous locus on human chromosome 17 predicts a large (∼80 kbp) chimpanzee-specific insertion. DupMasker analysis suggests that the insertion is the result of a duplicative transposition event composed of segmental duplications that originated from human–chimpanzee ancestral chromosome 16.
Analysis of nonhuman primate genomes
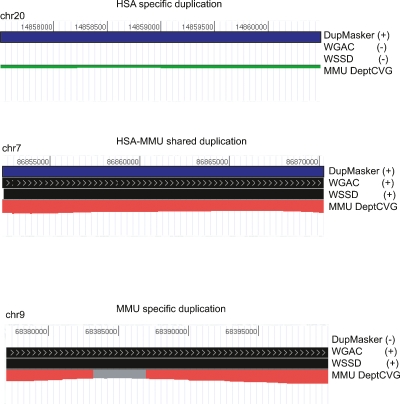
Since the consensus sequence library is based on human sequence, it will be necessary to update the library to include species-specific duplications from other nonhuman primate genomes as they are identified. In this regard, DupMasker greatly facilitates the identification of lineage-specific duplications. For example, if we apply DupMasker (human duplib) to a nonhuman primate genome assembly, we can compare DupMasker regions in the NHP genome (duplicated in human) against regions predicted to be duplicated by independent analyses of those genomes (predicted to be duplicated within the NHP by WGAC/WSSD). Such analyses will readily distinguish three types of duplications: duplications shared between human and the NHP, duplications specific to human, and duplications specific to the NHP. Figure 5 illustrates the way different types of duplication (e.g., lineage-specific or shared duplications) can be identified through a comparison of different duplication analyses on the macaque genome (Gibbs et al. 2007) compared with those detected by DupMasker. This comparison of the macaque genome predicts that 22.3 Mb are shared duplication between macaque and human, while 122.9 Mb emerged within the human lineage and only 24.3 Mb emerged within the macaque lineage since divergence.
Figure 5.
Assigning lineage-specific and shared duplications in primates. We applied DupMasker (standard default settings) to the macaque genome (RheMac2) and readily identified shared and lineage-specific duplications by comparing the results with duplication maps of the Rhesus Macaque Genome (Gibbs et al. 2007). We found that 84% (121.0/143.3 Mb) of duplications in the human genome are human-lineage specific. There are 22.3 Mb of duplications shared between human and macaque, and 24.3% (24.3/46.4 Mb) of duplications defined in the macaque genome that are macaque-lineage specific.
Discussion
We have developed an annotation tool that allows the complex duplication structure of regions to be deciphered and compared without the need for initiating a genome-wide self-comparison. The annotation provides insight into the origin, degree of sequence identity, and orientation of duplicons embedded within sequence. Since many segmental duplications recurrently duplicate (Johnson et al. 2006) or have been shared among species closely related to human, the distribution of this tool will enhance the sequence and assembly of complex regions of great ape genomes by allowing annotators within the sequence centers to distinguish unique from duplicated regions. “DupMasking” of BACs will flag potential regions of new insertion that can then be further characterized. Similar to RepeatMasker, distribution of this tool will have other more pragmatic uses to genetics and genome research, ranging from enhancing oligonucleotide PCR design to improving genotyping assays. Many commercial/customized platforms for SNP genotyping wish to avoid highly duplicated regions of the genome. Our tool not only allows such regions to be identified but also provides information on the copy number of each segment within the reference genome assembly (Supplemental Table 2).
Further enhancements will entail the modification of the duplication library specifically for each nonhuman primate species. We anticipate the discovery of a significant number of lineage-specific duplications (and deletions) in different primate genomes (Cheng et al. 2005). As these regions are discovered, the human duplication library will be modified accordingly to include chimpanzee-specific and macaque-specific duplications. The annotation of BAC sequences will be particularly useful in this regard since we recognize that lineage-specific duplications will occur nonrandomly in the genome (i.e., in the vicinity of shared duplication blocks). Thus, as BAC insert sequences are annotated using WSSD (Bailey et al. 2002) and DupMasker, new regions of duplication will be identified. These sequences can be extracted and added to the species-specific duplib as part of the reiterative process of modifying the human duplib. Ultimately, a duplication library specific for each of the primates will emerge.
In addition, we now know that duplication regions are hotspots for extensive copy-number and structural variation. Considering that duplication-mediated NAHR is the most common mechanism leading to copy-number variation (Kidd et al. 2008), we predict that DupMasker will aid in characterizing the duplication architecture of these regions as more copy-number variant regions become sequenced (Fig. 4).
Methods
Duplication library
We developed a library of consensus sequences (duplib) based on the WGAC human segmental duplication data set. The initial data set consisted of 28,856 pairwise alignments (sequence identity ≥90% and size ≥1 kb) defined by the WGAC method (build35) (Bailey et al. 2001). We applied a modified A-Bruijn graph approach (Pevzner et al. 2004; Jiang et al. 2007) to convert pairwise alignments into nonredundant duplication subunits (n = 12,087, size ≥ 100 bp), as described previously. A set of consensus sequences was generated for each duplication by identifying the majority-rule nucleotide within each multiple sequence alignment. The available ancestral state information (102.4 Mb/67.2% of all duplications) for duplication subunits was defined by a reciprocal best-hit between human and outgroup mammalian genomes (Jiang et al. 2007).
DupMasker design
The program initially screens input sequences for all common interspersed repeats using standard RepeatMasker settings (primate library). Repeat-masked base pairs are replaced with Ns, and seed alignments are identified between duplib and the masked test sequence using WUBLAST2 (minimal BLAST score = 300). These seed alignments are stored as part of the *.dupout file. We extend seed alignments by combining local fragments. Local collinear seeds (adjacent seeds from the same duplicon, in the same orientation, and within a default gap length of ≤7 kbp) are first chained. Next, the chained query sequence is realigned against the unmasked consensus sequence in the library. The realignment results are stored as part of the *.duplicons output file. The program uses a simple UNIX command line format: segdupmask [-options] [input DNA sequence file]. There are four basic options: (1) –maxDiv restricts the maximal divergence (sequence identity) between the seeds and the consensus sequence; (2) –maxWidth restricts the maximum nonrepetitive/nonseed realign gaps (default is 7 kb) for chaining; (3) –forceSearch forces the program to perform all steps despite the presence of previous result files (by default the program will select previous *.dupout and *.out for a given input sequence, omitting the first two steps of the procedure); and (4) –align option generates alignments as part of the standard output. The input file for DupMasker is a single text file containing the DNA sequence in FASTA format. After the execution, DupMasker creates two standard output files: (1) a text file containing information of all seed alignments (*dupout) and (2) a text file containing information of all chained duplicons (*duplicons) with ancestral state information.
Acknowledgments
We thank Pavel Pevzner and Haixu Tang for assistance in implementation of the modified A-Bruijn graph theory algorithm; Heather Mefford, Jeffrey Kidd, and Tonia Brown for useful comments; and Lin Chen for computational assistance. This work was supported by an NIH grant GM058815 to E.E.E. and a Rosetta Inpharmatics fellowship (Merck Laboratories) to Z.J. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078477.108.
References
- Armengol L., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Cheung J., Scherer S.W., Estivill X., Scherer S.W., Estivill X., Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Massa H.F., Trask B.J., Eichler E.E., Trask B.J., Eichler E.E., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Kent W.J., Haussler D., Eichler E.E., Haussler D., Eichler E.E., Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.C., Saarela J., Clark R.A., Miettinen T., Chi A., Eichler E.E., Peltonen L., Palotie A., Saarela J., Clark R.A., Miettinen T., Chi A., Eichler E.E., Peltonen L., Palotie A., Clark R.A., Miettinen T., Chi A., Eichler E.E., Peltonen L., Palotie A., Miettinen T., Chi A., Eichler E.E., Peltonen L., Palotie A., Chi A., Eichler E.E., Peltonen L., Palotie A., Eichler E.E., Peltonen L., Palotie A., Peltonen L., Palotie A., Palotie A. Segmental duplications flank the multiple sclerosis locus on chromosome 17q. Genome Res. 2004;14:1483–1492. doi: 10.1101/gr.2340804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Church D., DeJong P., Wilson R.K., Paabo S., DeJong P., Wilson R.K., Paabo S., Wilson R.K., Paabo S., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Cheung J., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Lau K., Tsui L.C., Scherer S.W., Tsui L.C., Scherer S.W., Scherer S.W. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Strausberg R.L., Venter J.C., Wilson R.K., Venter J.C., Wilson R.K., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Horvath J., Schwartz S., Eichler E., Schwartz S., Eichler E., Eichler E. The mosaic structure of a 2p11 pericentromeric segment: A strategy for characterizing complex regions of the human genome. Genome Res. 2000;10:839–852. doi: 10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E., She X., Pevzner P.A., Eichler E.E., Pevzner P.A., Eichler E.E., Eichler E.E. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- Johnson M.E., Cheng Z., Morrison V.A., Scherer S., Ventura M., Gibbs R.A., Green E.D., Eichler E.E., Cheng Z., Morrison V.A., Scherer S., Ventura M., Gibbs R.A., Green E.D., Eichler E.E., Morrison V.A., Scherer S., Ventura M., Gibbs R.A., Green E.D., Eichler E.E., Scherer S., Ventura M., Gibbs R.A., Green E.D., Eichler E.E., Ventura M., Gibbs R.A., Green E.D., Eichler E.E., Gibbs R.A., Green E.D., Eichler E.E., Green E.D., Eichler E.E., Eichler E.E. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc. Natl. Acad. Sci. 2006;103:17626–17631. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., Hansen N., Teague B., Alkan C., Antonacci F., Teague B., Alkan C., Antonacci F., Alkan C., Antonacci F., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J.R. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Mefford H.C., Clauin S., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Clauin S., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., Cooper G.M., Ventura M., Ropers H.H., Ventura M., Ropers H.H., Ropers H.H., et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.D. Miropeats: Graphical DNA sequence comparisons. Comput. Appl. Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Pevzner P.A., Tang H., Tesler G., Tang H., Tesler G., Tesler G. De novo repeat classification and fragment assembly. Genome Res. 2004;14:1786–1796. doi: 10.1101/gr.2395204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fiegler H., Shapero M.H., Carson A.R., Chen W., Shapero M.H., Carson A.R., Chen W., Carson A.R., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz J., Rovensky P., Gudjonsson S.A., Thorleifsson G., Stefansson K., Gulcher J.R., Rovensky P., Gudjonsson S.A., Thorleifsson G., Stefansson K., Gulcher J.R., Gudjonsson S.A., Thorleifsson G., Stefansson K., Gulcher J.R., Thorleifsson G., Stefansson K., Gulcher J.R., Stefansson K., Gulcher J.R., Gulcher J.R. Segmental duplication density decrease with distance to human-mouse breaks of synteny. Eur. J. Hum. Genet. 2006;14:216–221. doi: 10.1038/sj.ejhg.5201534. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Clark R.A., Schwartz S., Segraves R., Schwartz S., Segraves R., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Stewart H., Price S.M., Blair E., Hennekam R.C., Price S.M., Blair E., Hennekam R.C., Blair E., Hennekam R.C., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., Schroer R.J., Novara F., De Gregori M., Ciccone R., Novara F., De Gregori M., Ciccone R., De Gregori M., Ciccone R., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Sutton G., Halpern A.L., Eichler E.E., Halpern A.L., Eichler E.E., Eichler E.E. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- She X., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., Rocchi M., Green E.D., Archidiacano N., Green E.D., Archidiacano N., Archidiacano N., et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]