Abstract
A regional analysis of nucleotide substitution rates along human genes and their flanking regions allows us to quantify the effect of mutational mechanisms associated with transcription in germ line cells. Our analysis reveals three distinct patterns of substitution rates. First, a sharp decline in the deamination rate of methylated CpG dinucleotides, which is observed in the vicinity of the 5′ end of genes. Second, a strand asymmetry in complementary substitution rates, which extends from the 5′ end to 1 kbp downstream from the 3′ end, associated with transcription-coupled repair. Finally, a localized strand asymmetry, an excess of C→T over G→A substitution in the nontemplate strand confined to the first 1–2 kbp downstream of the 5′ end of genes. We hypothesize that higher exposure of the nontemplate strand near the 5′ end of genes leads to a higher cytosine deamination rate. Up to now, only the somatic hypermutation (SHM) pathway has been known to mediate localized and strand-specific mutagenic processes associated with transcription in mammalia. The mutational patterns in SHM are induced by cytosine deaminase, which just targets single-stranded DNA. This DNA conformation is induced by R-loops, which preferentially occur at the 5′ ends of genes. We predict that R-loops are extensively formed in the beginning of transcribed regions in germ line cells.
Understanding the processes that lead to spontaneous DNA mutations is important for studies of genome evolution and the genesis of noninherited genetic diseases such as cancer. New mutations arise as a result of numerous processes that damage DNA and the unsuccessful repair of this damage by cellular repair pathways. Besides a certain level of background activity of these processes, mutagenic sources may additionally be influenced by the DNA sequence, the DNA structure, or processes of cellular metabolism like transcription and replication (Maki 2002). Up to now, little is known about the in vivo properties of these additional sources and less is known about the relative contribution of each source to the mutation patterns in the genome.
Replication, and to a lesser extent, transcription, have been shown to affect nucleotide mutation rates in a variety of genomes. Hallmarks of these processes are particular strand asymmetries. In bacteria, the replication of the genome from one unique origin of replication promotes certain nucleotide substitutions within the two strands, generating heterogeneities in the base composition along the circular genome and strand asymmetries between the leading and lagging strand (Lobry 1996; Kano-Sueoka et al. 1999). In addition, transcription itself and transcription-coupled repair (TCR) processes (Svejstrup 2002) also lead to strand asymmetries between the template and nontemplate strand (Rocha et al. 2006). In particular, transcription can lead to an elevation of cytosine deamination rates on the nontemplate strand (Beletskii and Bhagwat 1996, 1998).
In higher eukaryotes, processes coupled to replication and transcription also affect the genomic sequence. In contrast to bacteria, DNA replication in higher eukaryotes initiates from multiple origins (Francino and Ochman 2000; Aladjem 2007). On an evolutionary timescale, segments of DNA might be replicated from the next 5′ or 3′ origin lowering or canceling strand asymmetries of the replication process. In a similar fashion, transcription occurs from both strands and from different origins. Yet, the impact of transcription is easier to detect, since transcription usually starts from well-defined transcription start sites and proceeds only in one direction to synthesize the RNA message. Moreover, most of the transcription start sites (TSSs) tend to be conserved, even between mammals (Frith et al. 2006; Taylor et al. 2006). This stability over long periods of time enables the accumulation of mutations associated with transcription, which can be observed by analyzing genomic sequence data.
Biases in the nucleotide composition of single-stranded DNA (ssDNA) have been used as evidence for biases in mutational processes or selection. According to Chargaff’s second parity rule, an asymmetry in the frequencies of complementary nucleotides on ssDNA, such as an excess of T over A, implies that mutation rates are not identical on the complementary strands (Lobry 1996). Green et al. (2003) have observed an excess of G+T over A+C on the nontemplate strand in human genes, which has later been found to be correlated with transcription levels (Majewski 2003). To quantify violations of Chargaff’s second parity rule, one introduces the TA skew = (T − A)/(T + A) and GC skew = (G − C)/(G + C). Both skews are found to be positive in mammalian nontemplate strands, but are close to zero in the 5′ flanking sequences of genes (Touchon et al. 2004).
Previous studies further revealed that the nucleotide composition varies along transcribed DNA (Louie et al. 2003; Touchon et al. 2004). The TA and GC skews are found to be maximal at the immediate downstream region from the 5′ end of genes; further nucleotide densities are observed to be dependent on the distance from the 5′ end and 3′ end of introns (Touchon et al. 2003; Aerts et al. 2004; Fujimori et al. 2005). Apart from skews of complementary nucleotides, the GC content monotonically decreases with the increasing distance from the TSS in both directions (Saxonov et al. 2006). This regional behavior of the nucleotide composition in genes and their flanking regions imply corresponding regional behaviors of substitution processes. Yet, it is not clear whether and how mutational mechanisms vary along the transcripts.
The signatures of mutational processes associated with transcription are particular strand asymmetries with respect to the template/nontemplate strand. For instance, using 1.5 Mbp of orthologous regions in chimpanzee and human, Green et al. (2003) calculated the rates of intronic nucleotide transitions, and found out that in the nontemplate strand the purine transitions (A→G and G→A) occurred at a higher rate than their complementary pyrimidine transitions. Later, a similar bias of A→G vs. T→C was reported in an analysis of single nucleotide polymorphisms (SNPs) in introns and fourfold degenerates (FFD) sites (Qu et al. 2006). This strand bias in substitution rates has been hypothesized to be a result of TCR and of different misinsertion rates for the four nucleotides (Green et al. 2003). Further on, Hwang and Green (2004) carried out an analysis of context dependence of mutation rates in 1.7-Mbp genomic regions across the phylogeny of 19 mammalians species. Beside the known asymmetries of complementary transition rates in transcribed regions, they also found similar (but weaker) asymmetries in the transversion rates. Some of these processes also showed signatures of neighbor dependencies.
In this study we also use a comparative genomics approach to investigate a nucleotide substitution pattern in association with transcription. In contrast to previous studies, we especially want to investigate the spatial variations in these substitution patterns along the transcripts. The availability of genome-wide human–chimpanzee–rhesus alignments enabled us to estimate 12 single nucleotide substitution frequencies as well as the deamination rate of CpG dinucleotides surrounding the 5′ and 3′ ends of genes. In order to be able to resolve variations of substitution rates on a high spatial resolution, we had to keep the number of parameters in our model small (Arndt and Hwa 2005) and just included one neighbor-dependent substitution process, the CpG methylation deamination process (CpG→CpA and CpG→TpG), which is known to be the predominant substitution process in mammals (Arndt et al. 2003). In order to minimize the effects due to selection, we analyzed only intronic parts of genes and their 5′ and 3′ flanking intergenic sequences.
We establish the regional patterns (with respect to the TSS) of nucleotide substitution rates in the human linage since the human–chimpanzee divergence. Our analysis reveals three types of regional patterns of substitution rates. The most pronounced behavior is a sharp decrease of CpG deamination in the proximity of the TSS. Further, we show that in intronic regions most of the substitutions rates are strand asymmetric. This strand asymmetry is not observed in intergenic regions upstream to the TSS, showing that transcription-coupled mechanisms are involved in this breaking of the strand symmetry. The first type of strand asymmetric pattern extends along the whole transcript: We measure an excess of A→G over T→C, A→T over T→A, C→G over G→C, and G→T over C→A on the nontemplate strand consistently along the whole transcript (naturally, the opposite asymmetries are found on the template strand). The second type of asymmetry is an excess of C→T over G→A transitions restricted to the immediate 1–2-kbp region downstream from the TSS. This localized asymmetry is found to be dependent both on the distance from the TSS and on local GC content. We argue that the mechanisms that are involved in the formation of local asymmetry might be coupled to transcription initiation and be involved in the somatic hypermutation pathway, which targets the first 1–2 kbp of genes. Finally, we show that these regional patterns of nucleotide substitutions can lead to the known regional patterns of nucleotide composition.
Results and Discussion
Analysis of nucleotide substitutions in the vicinity of genes
The main goal of our analysis is to investigate the dependence of nucleotide substitution rates on transcription and the distance from the TSS. To do so, one is tempted to estimate substitution rates at different distances from the TSS on a single gene level. However, the low divergence between human and chimpanzee prevents us from getting reliable estimates of mutation rates in small windows; in particular, because we wish to estimate 14 different mutation rates. To overcome this problem, we estimate mutation rates in genome-wide pooled 200-bp long nonoverlapping windows, which are located at fixed distances from individual TSS (for details, see Fig. 1 and Methods). Such analysis is possible with the availability of genome-wide human–chimpanzee–rhesus alignments that cover about 85% of the human genome. In order to minimize the effects of selection, we analyzed only the intronic parts of genes and their 5′ and 3′ flanking intergenic sequences. Finally, we estimated the strand dependency by estimating substitution rates in the nontemplate strand only.
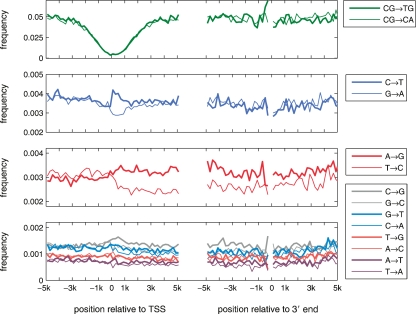
Figure 1.
Regions of analysis around the 5′ end of genes. The substitution analysis was done in the 10,000-bp-long regions centered on the 5′ end of gene (denoted by two vertical lines). This region of analysis was further truncated if the next upstream gene was closer than 10,000 bp, or the 3′ end of the gene was closer than 5000 bp. Further, we excluded all exons. Bold lines depict the finally analyzed sequences.
Substitution frequencies have been estimated from pooled triple alignments of genomic sequences from human, chimp, and rhesus. We used a maximum likelihood approach that correctly handles effects due to back-mutations and is able to reliably estimate substitution frequencies from given aligned sequences as described in the Methods section. A similar regional analysis was also done in the surrounding regions of the 3′ end of genes.
CpG loss rate declines near the TSS
Our analysis revealed three types of regional substitution patterns (Fig. 2). The strongest regional behavior is the reduction of loss of CpG’s. In our framework, the cytosine in a CpG dinucleotide may undergo two mutual independent processes to become a T. The first is the common C→T transition, irrespective of the neighboring bases; the second is the neighbor-dependent CpG methylation deamination process CpG→TpG (CpG→CpA on the reverse strand). The frequencies of the latter processes are reduced by a factor of 15 near the TSSs. This decrease in the CpG loss rate occurs symmetrically down- and upstream of the TSS, and both processes, CpG→TpG and its reverse complement substitution CpG→CpA, are affected in the same way.
Figure 2.
Substitution rates in introns and in intergenic regions in the vicinity of 5′ and 3′ ends of human genes. The plots show the estimated 12 single-nucleotide substitution rates and the CpG deamination rates in nonoverlapping 200-bp-long windows along the nontemplate strand. The distances of the windows’ centers from the 5′ or 3′ end are indicated on the X-axes. The estimation of substitution frequencies has been performed using the nontemplate strand.
The rates of CpG-loss (Fig. 2) are anticorrelated with the C+G density (Fig. 3). The high GC content near the TSS is related to the fact that the majority of TSSs in human genes are part of CpG Islands (CGIs) (Saxonov et al. 2006). CGIs are sequence segments rich in C+G nucleotides and CpG dinucleotides with respect to the other parts of the genome (Ioshikhes and Zhang 2000). The methylation status of CGIs is suggested to have an important role in regulation: being unmethylated allows transcription, but being methylated suppresses transcription (Bird 2002).
Figure 3.
Ratios between complementary transition rates and the GC content plotted against distance from the 5′ end of genes calculated in 200-bp-long windows along the nontemplate strand, combined with information from all genes and presented by six different genomic contexts. (Intronic) Genes that were used in Figure 2; (introns w/o 200bp) the 200 bp in introns’ edges were excluded; (GC-rich windows and AT-rich windows) DNA sequences with GC content of >50% and <41%, respectively. Windows that contained less than 100 kbp-long DNA sequences were omitted (see Supplemental Fig. S15 for the amount of sequence in each window).
The observed reduction of the CpG methylation deamination rate near the TSS can therefore be explained by two mechanisms. First, the lack of methylation in CGIs in germ line cells (Weber et al. 2007) decreases the probability of C→T transitions (in CpGs). Second, purifying selection might counteract the loss of CpG in order to preserve the existence of a CGI for regulatory processes in somatic cells (Majewski and Ott 2002). A loss of CpGs due to mutations would lead eventually to the loss of the CGI property of a gene promoter and change the gene expression pattern (Jaenisch and Bird 2003).
An excess of C→T over G→A substitutions restricted to the first 1–2 kbp downstream from the TSS
For the neighbor-independent single nucleotide substitutions, we find much richer patterns by estimating and comparing the complementary substitutions on the nontemplate strand. First, reverse complement substitution processes do not occur at the same rates on the nontemplate strand, i.e., the transcription process singles out one strand breaking the symmetry between the two strands in untranscribed regions. Second, this breaking of the symmetry occurs only downstream from the TSS. Strikingly, the first such asymmetry, an excess of the C→T over G→A, is confined to the first 1–2-kbp-long region downstream from the TSS (Fig. 2). For this localized asymmetry we observe an elevation of the C→T transition rate by ∼20% in the first kilobase pair of the transcript compared with the rate in promoter regions upstream of the TSS, whereas the G→A rate decreases by about the same percentage in transcribed regions. The difference between these two rates reaches up to 40% and the gap between these rates is closed at a distance of ∼1.5 kbp to the TSS (Fig. 3). This asymmetry is specific to the 5′ end of genes, and it is not detected in the vicinity of the 3′ end (Supplemental Fig. S1), which confirms the localized nature of this strand bias. The trend of the localized asymmetry that we found is opposite to the one that had been reported by Green et al. (2003). This discrepancy between the results may very well be explained by the fact that, in contrast to the previous study, we surveyed the whole genome and included a broader spectrum of transcripts into our analysis (see below).
Global asymmetry: A→G exceed T→C transitions along the transcript
In contrast to the above process, other processes show a global asymmetry defined as a bias in complementary nucleotide substitutions that extends along the whole transcript. There are four pairs of nucleotide substitution processes that show global asymmetries: A→G/T→C, C→G/G→C, A→T/T→A, and G→T/C→A. In Figure 2, it is shown that the asymmetry in the substitution frequencies extends from the TSS down to the end of the analyzed region (5 kbp downstream from the TSS). In order to check whether the (global) asymmetries extend along the whole transcript, we also analyzed the 10-kbp-long region centered on the 3′ end of genes (Fig. 1). Interestingly, these asymmetries extended not only until the 3′ end, but, on average, also into the 1000-bp downstream region to the 3′ end of genes (Fig. 1). This extension is similar to the extension of the TA bias in the nucleotide composition as far as 1000 downstream to the 3′ end, which reflects the fact that the termination position of the transcription process is not always at the annotated 3′ end of genes and might continue several hundreds of bases further (Dye and Proudfoot 2001; Louie et al. 2003). The bias in the transition frequencies A→G over T→C was also reported previously by Green et al. (2003), who analyzed 1.5 Mbp of human chromosome 7. Our genome-wide analysis reveals similar biases for three out of the four transversions (Fig. 2; Supplemental Fig. S1), which confirms a prediction from theoretical considerations (Touchon et al. 2003).
Surprisingly, we found opposite biases in the A→G/T→C substitution frequencies in the 5′ upstream regions compared with those in 5′ downstream regions (see Fig. 2), which might be indicative of frequent antisense transcription of the reverse strand, upstream of the 5′ end of a gene on the forward strand. This asymmetry eventually vanishes at about 10 kbp upstream of the TSS (Supplemental Fig. S2).
The current single nucleotide substitution rates lead to observed TA and GC skews
As was mentioned above, GC and TA skews are observed in human introns and have been suggested to be a result of a bias in substitution rates. Using a genome evolution model (see Supplemental Methods), we show that the estimated nucleotide substitution rates in human introns can lead to the observed TA and GC skews in these regions (Supplemental Fig. S3; Supplemental Results and Discussion). Since it takes a long time to build up such skews, this also implies that the molecular mechanisms that shape the mutational patterns in transcribed DNA are not recent in primate evolution, but rather ancient.
The strand asymmetries are not restricted to introns
The substitution patterns that were reported so far are also found in nonintronic parts of transcripts: in 5′ UTR, the 3′ UTR, and fourfold degenerate (FFD) sites (see Supplemental Results and Discussion; Supplemental Figs. S4–S6). Presumably. these parts evolved under stronger selection constraints than introns. The existence of the local and global asymmetries in different parts of the transcripts, which are under different levels of evolutionary constraints, implies that the asymmetries are most likely either invoked by a bias in the molecular mutational processes or as a result of selection acting on functional elements, which are common to all different parts of the transcript. Candidates for such functional elements that are common to introns, UTR, and FFD sites would be splicing elements that are supposed to be enriched in intronic edges or in first introns (Chamary and Hurst 2004; Touchon et al. 2004). However, the strand asymmetries were also found excluding either intronic edges or first introns (see Fig. 3; Supplemental Results and Discussion; Supplemental Fig. S7). In addition, in introns the substitution patterns are formed both in nonrepetitive parts and repeats, which are assumed to contain less functional elements than any other parts of the transcripts (see Fig. 3; Supplemental Results and Discussion; Supplemental Figs. S8, S9).
Strand asymmetries are correlated with transcription and transcription initiation in embryonic stem cells
So far, our results suggest that the substitution rates are shaped by mutational molecular mechanisms. The asymmetry A→G vs. T→C, was suggested to be the result of strand specificity of TCR and a biases in misinsertion of A→G over T→C during replication, which is not attributed to transcribed/nontranscribed differences of strands (Green et al. 2003). In similar fashion, TCR and bias in misinsertion rates between complementary transversions can lead to the asymmetries that are observed in Supplemental Figure S1, i.e., biases in misinsertions of C→G over G→C, G→T over C→A, and A→T over T→A. However, the fact that TCR acts on the whole transcript rules out its role in the restriction of the C→T over G→A bias to the first 1000 bp. Therefore, we suggest that other mutational mechanisms are responsible for this bias. It is known that there are several mutagenic processes that target ssDNA, which is a by-product of RNA polymerase activity. Hence, higher frequencies of ssDNA in this region could lead to higher transition rates of C→T in the beginning of a transcript, rather than in the rest of the transcript.
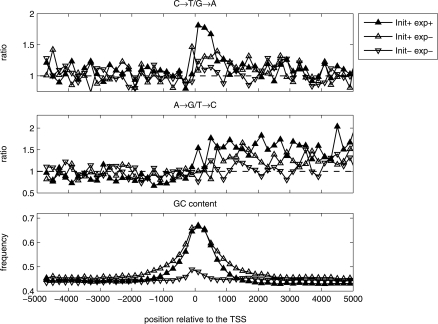
Recent works show that RNA polymerase II (pol II) activity is not homogenously distributed along transcripts (Kim et al. 2005; Muse et al. 2007); moreover, in many genes there are marks of transcription initiation but not of complete elongation. In about 80% of genes in embryonic stem cells (ESC), there is an initiation of transcription, even though just 50% of the genes are fully transcribed (Guenther et al. 2007). We used this recent data set, in which genes were tested for their initiation and expression states in embryonic stem cells. We divided the genes in this data set into four groups according to their expression status in ESC (denoted by exp+/−), and their initiation status (init+/−, see Methods section). We calculated the substitution rates in the 5 kbp upstream and downstream regions of the 5′ end of genes in three groups, init+exp+, init+exp−, and init−exp− (the number of genes in the last init−exp+ was too small for analysis). The results show that the local excess of C→T over G→A is strongest in genes that are classified as init+exp+, a weaker bias (half as strong as for the init+exp+ group) was found in the group of init+exp− genes. In the set of init−exp− genes, the local asymmetry is actually absent (Fig. 4). Similar behavior is also observed for the global asymmetry (the excess of A→G over T→C). Hence, we concluded that initiation of transcription (in ESC) is correlated with formation of the local and global asymmetries, although these asymmetries are weaker in init+exp− than in init+exp+ genes.
Figure 4.
Correlation between strand asymmetry and transcription status of genes in embryonic stem cells (ESC). The ratios between complementary transition rates and the GC content are calculated in three gene classes (Guenther et al. 2007): genes that experienced initiation and transcription (exp+init+); genes that experienced initiation but not complete transcription (exp−init+); genes that experienced initiation but not complete transcription (exp−init−). The length and the nucleotides’ composition properties of the sequences that were used for substitution estimation in each window are presented in Supplemental Figure S16. The estimation of substitution frequencies has been performed using the nontemplate strand.
The average GC content near the TSS of genes in init+exp− and init+exp+ sets is much higher than that of genes in the init−exp− class (Fig. 4); this might imply that the local asymmetry is related to high GC content in introns independent of the distance from the TSS. In order to rule this out, we estimated the substitution rates in DNA sequences with different GC content surrounding the 5′ end of genes (see Fig. 3; Supplemental Results and Discussion; Supplemental Figs. S10, S11). Our analysis shows that the local strand asymmetry is dependent both on the distance from the TSS and the GC content in the vicinity of the TSS. Since both sets of genes, exp+init+ and exp+init−, have, on average, a high GC content in promoter regions but different levels of strand asymmetries, we suggest that GC content is indirectly correlated with the asymmetry level due to higher transcription activity of GC-rich promoters in germ line cells. Clearly, a limitation of this analysis is that although it was carried out in genes that are expressed in ESC, we cannot be sure that they are also expressed in germ line cells, where mutations have to occur in order to be passed on to the next generation.
Localization of mutations
Which mechanisms are responsible for the localization of cytosine deamination in the nontranscribed strand near the TSS? One possible process coupled to transcription is the activity of the activation induced (cytosine) deamination (AID, currently known as AICDA) enzyme as part of the somatic hypermutation (SHM) pathway during B-cell development (Odegard and Schatz 2006). This enzyme induces both single nucleotide mutations and rearrangement at the immunoglobulin gene loci. Especially, it promotes cytosine deamination in ssDNA during transcription, but just at 1–2 kbp downstream from the TSS (Odegard and Schatz 2006). Its activity is thought to be focused on so-called AID hotspots (Pham et al. 2003). However, our analysis showed that the rate of C→T does not exceed G→A in AID-hot-spots (Supplemental Fig. S12). Therefore, we could not substantiate the involvement of AID in the generation of the localized asymmetry on a genome-wide scale (see Supplemental Text).
Although we could not directly associate DNA modifiers like AID with the local asymmetry, the similarity between the localization of the strand asymmetry in substitution rates and the restrictions of SHM might be the result of similar mechanisms.
Possible mechanisms, which can elevate mutagenesis, are the ones that lead to formation of a single-stranded DNA (ssDNA) of the nontranscribed strand. A ssDNA structure has been proposed to occur in variable (V-) regions during SHM in order to provide the substrate for AID, which mutates only ssDNA. Further, the nucleotides in ssDNA are subjected to higher spontaneous DNA damage than nucleotides in double-stranded DNA (dsDNA). Cytosine in ssDNA is more prone to deamination than in dsDNA (Frederico et al. 1990; Beletskii and Bhagwat 1996). Hence, if a localized ssDNA of the nontranscribed strand is formed near the TSS, it might explain the higher rate of C→T transitions on this strand. Interestingly, the observed excess of C→T over G→A in bacterial genomes has been also suggested to be mediated by ssDNA on the nontranscribed strand (Francino and Ochman 2001), since the average length of a bacterial gene in 1–2 kb of the induced range of the asymmetry along bacterial genes is similar to the asymmetry pattern we observed for human genes (Francino and Ochman 2001). Hence, we propose that localized conformation of ssDNA near the TSS can explain the observed strand asymmetry
There are a some mechanisms that have been suggested to invoke ssDNA in V-regions. One of these mechanisms is the formation of RNA/DNA hybrids (R-loops) on the template strand in the first 1–2 kbp of the V-regions of immunoglobulin genes (Yu et al. 2003, 2005; Huang et al. 2006). R-loops are formed on the transcribed strand, leaving the nontranscribed strand in ssDNA formation at higher frequency than its complementary strand (Li and Manley 2005; Ronai et al. 2007). Using a biophysical model (Carlon and Heim 2006) we calculated the differences between the free-energy levels of RNA/DNA hybrids and DNA/DNA (i.e., dsDNA) structures in various distances of the TSS for genes in our data set. According to this model, for most genes the RNA/DNA hybrid in the first 200 bp downstream from the TSS is more stable than DNA/DNA confirmation (Supplemental Fig. S13). Moreover, the averaged difference between the energies of these two structures is peaked at the immediate 200-bp-long region downstream from the TSS (Supplemental Fig. S13). The predicted higher stability can be attributed to higher GC content and the GC skew near the TSS of human genes, since GpG dinucleotides are the main contributors to the energetic difference between RNA/DNA hybrids and DNA/DNA structures (Carlon and Heim 2006). It is important to note that until now it was not fully understood what the conditions for R-loops formations were. A couple of recent studies have suggested that R-loops are initiated from 50-bp-long regions, which contain the motif GGGGC TGGGG and comprise at least 50% Gs (Huang et al. 2007; Roy et al. 2008). We found ∼300 genes that, in their first 1000 bp, contain such regions. Estimating substitution rates in 1-kbp windows along these genes, indeed, shows a higher degree of asymmetry (Supplemental Fig. S14), but the small number of genes is not enough to establish a significant association. A recent study suggests that, on top of the higher stability of RNA/DNA compared with the DNA/DNA structure, a capping enzyme can promote formation of transcriptional R-loops in vitro (Kaneko et al. 2007). Since the formation of a cap is a necessary process at early stages of transcription, this finding implies that R-loops are indeed found at higher frequencies near the TSS than in the rest of the transcript.
Beside R-loops, the non-B DNA conformations, G-quadruplexes (G4), also might be formed near the TSS of human genes on the nontranscribed strand (Du et al. 2008). A formation of G4 structure in the nontemplate strand and, in parallel, a formation of R-loop on the template strand, is often called G-loops and has been also observed in different situations (Duquette et al. 2004). It is not known when and where such structures are formed, but several DNA sequence motifs have been suggested to have higher probability to form G-quadruplexes (Yadav et al. 2008). Recent studies reveal an enrichment of such motifs in promoter regions and in the first 500 bp of human genes (Du et al. 2008). Therefore, these G4 conformations have the potential to create a gradient of ssDNA of the nontemplate strand along human transcripts.
In summary, we propose the following model for the generation of the localized substitution bias. We assume that the nontranscribed strand at the start of genes is in ssDNA formation at a higher frequency than in regions further downstream. There are several mechanisms that may induce a localized formation of ssDNA of the transcribed strand. These mechanisms include either the higher occupation time of pol II near the TSS (Mikkelsen et al. 2007) or the formation of G/R-loops. As a consequence of these mechanisms, the transcribed strand near the TSS is protected either by RNA pol II complex or by the RNA/DNA hybrid; whereas the nontranscribed strand is left in ssDNA formation, which is prone to higher rates of cytosine deamination. This is due to spontaneous chemical processes (Frederico et al. 1990) or due to the enzymatic activity of DNA deaminases like AID or APOBEC3 (Larson and Maizels 2004; Rosenberg et al. 2007). These processes eventually can invoke the observed higher rate of C→T transition on the nontranscribed strand.
Final remarks
Today it is assumed that >90% of the human genome is transcribed (The ENCODE Project Consortium 2007). Therefore, our results imply that the majority of the genome evolved under mutational processes, which are not strand symmetric, in contrast to the current common assumption. As the number of sequenced genomes will increase, these mutational signatures could also be used for detecting novel transcripts (Green et al. 2003; Glusman et al. 2006) and to a lesser extent, to identify novel TSSs. The asymmetry in the mutation patterns can also contribute to the understanding of the transcription process itself. The reversal of the global symmetry in regions upstream of the TSS suggests frequent antisense transcription from the reverse strand; the presence of the asymmetry on the forward strand beyond the 3′ end of genes is indicative of the continuation of transcription beyond their annotated 3′ ends. Moreover, taking into account the present knowledge about repair and mutagenic processes in humans, we suggest that the local mutational pattern is a result of transcriptional-mediated ssDNA structure.
The ssDNA conformation has been suggested to be prone to double-strand breaks and to mediate genomic rearrangements as a consequence of the activity of the repair mechanisms that fix these breaks (Li and Manley 2006; Aguilera and Gomez-Gonzalez 2008). Therefore, we suggest that transcription of the first 1–2 kb of genes might be a novel mechanism for genome-wide instability and a driver of rearrangements.
Finally, the substitution asymmetries can be considered as phenotype derived, which, in this case, is gene expression, i.e., genes that are expressed are mutated by transcriptional-related mechanisms. This raises the question as to whether these mutational processes have some beneficial impact. Especially, we think that the transcription-related mutational patterns observed in this article are not limited to germ line cells and they can be active in somatic cells (as well as stem cells). In particular, transcription can be a major source of mutations in nondividing cells. Recent studies show that mutational processes can promote diversity of cells in somatic tissues and even yield a mechanism for differentiation of cells, as has been suggested for neural tissues (Muotri and Gage 2006).
Methods
Sequence data and annotation
Triple human–chimp–rhesus alignments were retrieved from Ensembl database, version 41 from October 2006 (Hubbard et al. 2007). They are based on the releases Homo sapiens (41, 36c), Pan troglodytes (41, 21), and Macaca mulatta (41, 10a), and were generated by MLAGAN (Brudno et al. 2003). The annotation for genes, exons, and translatable exons are according to Ensembl version 41, which uses the NCBI36 annotation of the human genome.
The position of the 5′ end of a Ensembl gene, which is coded on the forward (backward) strand, is defined as the lowest (highest) position among all 5′ chromosomal locations of its transcripts. To ensure a high quality of the TSS annotation, a gene is only included into the analysis if one of the transcripts defining the 5′ end is also in the RefSeq transcript or peptide database (Wheeler et al. 2006). We further included only the first (most 5′) TSS if a gene has multiple TSSs. In this way we minimized the effects of transcription on intergenic regions upstream of the 5′ ends of genes. In addition, genes that were located on sex chromosomes were filtered out. After applying all of these filters, we are left with 15,552 genes that are used for estimation of the substitution rates.
For regional analysis of the 3′ end of the gene, we chose genes with transcripts that were longer than 50 kbp, in order to minimize the effects of distance from the 5′ end on mutation rates.
Regional analysis
The primary analysis was done on DNA sequences in the vicinity of the 5′ ends of genes. For each gene, we determined a region of analysis, which was defined as the region 5000 bp upstream of and downstream from the TSS. However, in order to avoid the twofold analysis of one region, the upstream region was truncated to the middle position between two genes if the next upstream gene was closer than 10,000 bp (Fig. 1). For genes shorter than 5 kbp, the downstream region was also truncated and included the sequence up to the 3′ end of the gene (Fig. 1). A similar procedure has been applied to determine sequences for the regional analysis surrounding the 3′ end of genes.
The next step involved retrieving the human–chimp–rhesus triple alignments for each region of analysis. In order to reduce effects of selection, we excluded all exons using the annotation of the human genome. Masking out exonic sequences, we kept the positions of intronic sequence segments relative to the TSS unchanged (Fig. 1). The resulting sequences have been further partioned into nonoverlapping 200-bp-long windows, where the reference point was the 5′ end of the gene (Fig. 1). For each window, we extracted the appropriate triple alignment for all genes and concatenated them to estimate substitution frequencies as outlined below. The length of these triple-alignments in different windows varies due to different restrictions on the gene sets or sequence characteristics (Supplemental Fig. S15).
Substitution analysis
We measure the nucleotide substitution frequencies from multiple alignments. During the measurement process we do not assume the stationarity of the nucleotide composition, the time reversibility of the nucleotide substitution process, and most importantly, that reverse complement substitution processes are coupled. These three assumptions are often made during a phylogenetic analysis, but would foreclose our analysis. We therefore use a recently introduced methodology, which does not make these assumptions (Duret and Arndt 2008).
This method uses a maximum likelihood approach to estimate substitution rates from a multiple alignment of contemporary sequences from three or more species with known phylogenetic relationships. Substitution processes are assumed to be homogeneous in time along each branch in the phylogenetic tree, but may vary from one branch to another. Our method is able to reliably estimate the 14 frequencies (for the 12 neighbor-independent substitutions and the two neighbor-dependent CpG methylation deamination processes) along all branches that are not connected to the root node (Duret and Arndt 2008). In our current setting, the root node would represent the last common ancestor of human and rhesus. Here, however, we will only analyze the substitution frequencies along the terminal branch that connect the last common ancestor of human and chimp with contemporary humans.
All frequencies are measured per base pair and they estimate the (fractional) number of nucleotide exchanges per base pair in a given time interval, i.e., along a branch in the phylogenetic tree. We may compute the corresponding substitution rates (measured per base pair and time) by dividing these frequencies by the physical time that passed along a branch, i.e., in our case, the time period after the human–chimp split. However, since we only compare various rates and are not interested in their absolute values, we may just work with these frequencies.
In contrast to the work of Hwang and Green (2004) we do not include more neighbor-dependent processes in order to keep the number of parameters in our model low and to be able to perform our analysis on a fine spatial resolution along the transcripts. Note that we showed previously that the inclusion of more neighbor-dependent processes is likely not to be a significant enhancement of the model (Arndt and Hwa 2005). We convinced ourselves of that fact again and also included transversions on CpG sites for testing purposes. The final results did not show significant deviations from the simpler model (data not shown), and therefore we used the model with 14 substitution frequencies.
Acknowledgments
We thank Nina F. Papavasiliou and David G. Schatz for the useful discussions on AID activity and its sequence specificity.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076570.108.
References
- Aerts S., Thijs G., Dabrowski M., Moreau Y., De Moor B., Thijs G., Dabrowski M., Moreau Y., De Moor B., Dabrowski M., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics. 2004;5:34. doi: 10.1186/1471-2164-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Gomez-Gonzalez B., Gomez-Gonzalez B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- Arndt P.F., Hwa T., Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21:2322–2328. doi: 10.1093/bioinformatics/bti376. [DOI] [PubMed] [Google Scholar]
- Arndt P.F., Petrov D.A., Hwa T., Petrov D.A., Hwa T., Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 2003;20:1887–1896. doi: 10.1093/molbev/msg204. [DOI] [PubMed] [Google Scholar]
- Beletskii A., Bhagwat A.S., Bhagwat A.S. Transcription-induced mutations: Increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A., Bhagwat A.S., Bhagwat A.S. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem. 1998;379:549–551. [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Davydov E., Green E.D., Sidow A., Batzoglou S., Green E.D., Sidow A., Batzoglou S., Sidow A., Batzoglou S., Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlon E., Heim T., Heim T. Thermodynamics of RNA/DNA hybridization in high-density oligonucleotide microarrays. Phys. A: Stat. Mech. Appl. 2006;362:433–449. [Google Scholar]
- Chamary J.-V., Hurst L.D., Hurst L.D. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: Evidence for selectively driven codon usage. Mol. Biol. Evol. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhao Y., Li N., Zhao Y., Li N., Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N., Handa P., Vincent J.A., Taylor A.F., Maizels N., Vincent J.A., Taylor A.F., Maizels N., Taylor A.F., Maizels N., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes & Dev. 2004;18:1618–1829. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Arndt P.F., Arndt P.F. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye M.J., Proudfoot N.J., Proudfoot N.J. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino M.P., Ochman H., Ochman H. Strand symmetry around the β-globin origin of replication in primates. Mol. Biol. Evol. 2000;17:416–422. doi: 10.1093/oxfordjournals.molbev.a026321. [DOI] [PubMed] [Google Scholar]
- Francino M.P., Ochman H., Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol. 2001;18:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- Frederico L.A., Kunkel T.A., Shaw B.R., Kunkel T.A., Shaw B.R., Shaw B.R. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayshizaki Y., Sandelin A., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayshizaki Y., Sandelin A., Fredman D., Kai C., Kawai J., Carninci P., Hayshizaki Y., Sandelin A., Kai C., Kawai J., Carninci P., Hayshizaki Y., Sandelin A., Kawai J., Carninci P., Hayshizaki Y., Sandelin A., Carninci P., Hayshizaki Y., Sandelin A., Hayshizaki Y., Sandelin A., Sandelin A. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16:713–722. doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori S., Washio T., Tomita M., Washio T., Tomita M., Tomita M. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genomics. 2005;6:26. doi: 10.1186/1471-2164-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G., Qin S., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F.A., Qin S., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F.A., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F.A., Siegel A.F., Roach J.C., Hood L., Smit A.F.A., Roach J.C., Hood L., Smit A.F.A., Hood L., Smit A.F.A., Smit A.F.A. A third approach to gene prediction suggests thousands of additional human transcribed regions. PLoS Comput. Biol. 2006;2:e18. doi: 10.1371/journal.pcbi.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., Ewing B., Miller W., Thomas P.J., Green E.D., Ewing B., Miller W., Thomas P.J., Green E.D., Miller W., Thomas P.J., Green E.D., Thomas P.J., Green E.D., Green E.D. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A., Levine S.S., Boyer L.A., Jaenisch R., Young R.A., Boyer L.A., Jaenisch R., Young R.A., Jaenisch R., Young R.A., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.-T., Yu K., Hsieh C.-L., Lieber M.R., Yu K., Hsieh C.-L., Lieber M.R., Hsieh C.-L., Lieber M.R., Lieber M.R. Downstream boundary of chromosomal R-loops at murine switch regions: Implications for the mechanism of class switch recombination. Proc. Natl. Acad. Sci. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.-T., Yu K., Balter B.B., Selsing E., Oruc Z., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Yu K., Balter B.B., Selsing E., Oruc Z., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Balter B.B., Selsing E., Oruc Z., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Selsing E., Oruc Z., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Oruc Z., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Khamlichi A.A., Hsieh C.-L., Lieber M.R., Hsieh C.-L., Lieber M.R., Lieber M.R. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Sμ class switch region. Mol. Cell. Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T.J.P., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.G., Green P., Green P. Inaugural article: Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I.P., Zhang M.Q., Zhang M.Q. Large-scale human promoter mapping using CpG islands. Nat. Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Bird A., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Chu C., Shatkin A.J., Manley J.L., Chu C., Shatkin A.J., Manley J.L., Shatkin A.J., Manley J.L., Manley J.L. Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc. Natl. Acad. Sci. 2007;104:17620–17625. doi: 10.1073/pnas.0708866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano-Sueoka T., Lobry J.R., Sueoka N., Lobry J.R., Sueoka N., Sueoka N. Intra-strand biases in bacteriophage T4 genome. Gene. 1999;238:59–64. doi: 10.1016/s0378-1119(99)00296-6. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson E., Maizels N., Maizels N.2004Transcription-coupled mutagenesis by the DNA deaminase AID Genome Biol. 5211 . http://genomebiology.com/2004/5/3/211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Manley J.L., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li X., Manley J.L., Manley J.L. Cotranscriptional processes and their influence on genome stability. Genes & Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Louie E., Ott J., Majewski J., Ott J., Majewski J., Majewski J. Nucleotide frequency variation across human genes. Genome Res. 2003;13:2594–2601. doi: 10.1101/gr.1317703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Ott J., Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H. Origins of spontaneous mutations: Specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Alvarez P., Brockman W., Kim T.-K., Koche R.P., Brockman W., Kim T.-K., Koche R.P., Kim T.-K., Koche R.P., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A.R., Gage F.H., Gage F.H. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Grissom S.F., Zeitlinger J., Adelman K., Zeitlinger J., Adelman K., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard V.H., Schatz D.G., Schatz D.G. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F., Bransteitter R., Petruska J., Goodman M.F., Petruska J., Goodman M.F., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Qu H.-Q., Lawrence S., Guo F., Majewski J., Polychronakos C., Lawrence S., Guo F., Majewski J., Polychronakos C., Guo F., Majewski J., Polychronakos C., Majewski J., Polychronakos C., Polychronakos C. Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: Evidence for functional effects of synonymous polymorphisms. BMC Genomics. 2006;7:213. doi: 10.1186/1471-2164-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E.P.C., Touchon M., Feil E.J., Touchon M., Feil E.J., Feil E.J. Similar compositional biases are caused by very different mutational effects. Genome Res. 2006;16:1537–1547. doi: 10.1101/gr.5525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronai D., Iglesias-Ussel M.D., Fan M., Li Z., Martin A., Scharff M.D., Iglesias-Ussel M.D., Fan M., Li Z., Martin A., Scharff M.D., Fan M., Li Z., Martin A., Scharff M.D., Li Z., Martin A., Scharff M.D., Martin A., Scharff M.D., Scharff M.D. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Environ. Monit. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg B.R., Papavasiliou F.N., Papavasiliou F.N. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv. Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg B.R., Papavasiliou F.N., Frederick W.A., Tasuku H., Papavasiliou F.N., Frederick W.A., Tasuku H., Frederick W.A., Tasuku H., Tasuku H. Advances in immunology. Academic Press; New York: 2007. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection; pp. 215–244. [DOI] [PubMed] [Google Scholar]
- Roy D., Yu K., Lieber M.R., Yu K., Lieber M.R., Lieber M.R. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D.L., Berg P., Brutlag D.L., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J.Q. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- Taylor M.S., Kai C., Kawai J., Carninci P., Hayashizaki Y., Semple C.A.M., Kai C., Kawai J., Carninci P., Hayashizaki Y., Semple C.A.M., Kawai J., Carninci P., Hayashizaki Y., Semple C.A.M., Carninci P., Hayashizaki Y., Semple C.A.M., Hayashizaki Y., Semple C.A.M., Semple C.A.M. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M., Nicolay S., Arneodo A., d'Aubenton-Carafa Y., Thermes C., Nicolay S., Arneodo A., d'Aubenton-Carafa Y., Thermes C., Arneodo A., d'Aubenton-Carafa Y., Thermes C., d'Aubenton-Carafa Y., Thermes C., Thermes C. Transcription-coupled TA and GC strand asymmetries in the human genome. FEBS Lett. 2003;555:579–582. doi: 10.1016/s0014-5793(03)01306-1. [DOI] [PubMed] [Google Scholar]
- Touchon M., Arneodo A., d'Aubenton-Carafa Y., Thermes C., Arneodo A., d'Aubenton-Carafa Y., Thermes C., d'Aubenton-Carafa Y., Thermes C., Thermes C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 2004;32:4969–4978. doi: 10.1093/nar/gkh823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D., Ramos L., Paabo S., Rebhan M., Schubeler D., Paabo S., Rebhan M., Schubeler D., Rebhan M., Schubeler D., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Church D.M., DiCuccio M., Edgar R., Federhen S., DiCuccio M., Edgar R., Federhen S., Edgar R., Federhen S., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V.K., Abraham J.K., Mani P., Kulshrestha R., Chowdhury S., Abraham J.K., Mani P., Kulshrestha R., Chowdhury S., Mani P., Kulshrestha R., Chowdhury S., Kulshrestha R., Chowdhury S., Chowdhury S. QuadBase: Genome-wide database of G4 DNA occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R., Hsieh C.L., Wilson T.E., Lieber M.R., Wilson T.E., Lieber M.R., Lieber M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu K., Roy D., Bayramyan M., Haworth I.S., Lieber M.R., Roy D., Bayramyan M., Haworth I.S., Lieber M.R., Bayramyan M., Haworth I.S., Lieber M.R., Haworth I.S., Lieber M.R., Lieber M.R. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol. Cell. Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]