Abstract
Studies of ancient DNA have been hindered by the preciousness of remains, the small quantities of undamaged DNA accessible, and the limitations associated with conventional PCR amplification. In these studies, we developed and applied a genomewide adapter-mediated emulsion PCR amplification protocol for ancient mammalian samples estimated to be between 45,000 and 69,000 yr old. Using 454 Life Sciences (Roche) and Illumina sequencing (formerly Solexa sequencing) technologies, we examined over 100 megabases of DNA from amplified extracts, revealing unbiased sequence coverage with substantial amounts of nonredundant nuclear sequences from the sample sources and negligible levels of human contamination. We consistently recorded over 500-fold increases, such that nanogram quantities of starting material could be amplified to microgram quantities. Application of our protocol to a 50,000-yr-old uncharacterized bone sample that was unsuccessful in mitochondrial PCR provided sufficient nuclear sequences for comparison with extant mammals and subsequent phylogenetic classification of the remains. The combined use of emulsion PCR amplification and high-throughput sequencing allows for the generation of large quantities of DNA sequence data from ancient remains. Using such techniques, even small amounts of ancient remains with low levels of endogenous DNA preservation may yield substantial quantities of nuclear DNA, enabling novel applications of ancient DNA genomics to the investigation of extinct phyla.
A central objective of ancient DNA genomics is to obtain large amounts of nuclear DNA sequence from the remains of long-deceased organisms. Such sequences have the unique potential to provide insights into the biology of extinct organisms and the evolution of living species (Hofreiter et al. 2001). The limited availability of ancient remains has severely impeded advances toward this goal, with ancient DNA studies largely restricted to PCR and sequencing of mitochondrial DNA or isolated nuclear loci, for example, from mammoth (Rompler et al. 2006) and Neanderthals (Krause et al. 2007; Lalueza-Fox et al. 2007). Recent application of high-throughput sequencing technologies to ancient DNA has allowed more thorough characterization of the properties of ancient DNA molecules (Briggs et al. 2007; Gilbert et al. 2007a) and sequencing of entire mitochondrial genomes from well-preserved samples (Gilbert et al. 2007b). These technologies, coupled with the availability of reference genomes for DNA sequence analysis, have allowed the recovery of multiple nuclear DNA sequences from the remains of ancient cave bear (Noonan et al. 2005), mammoth (Poinar et al. 2006), and Neanderthal (Green et al. 2006; Noonan et al. 2006). However, the methods used in these studies provided relatively small amounts of sequence at the expense of the destruction of precious ancient remains.
The scarcity of ancient remains, particularly from hominids, puts considerable constraint on the availability of material for the destructive sampling required for DNA isolation. Recent technological advances in molecular biology and genomics offer new potential ways to maximize the amount of DNA sequence obtained from limited quantities of ancient material available. One such opportunity is application of genomewide amplification to ancient DNA extracts. While numerous approaches to genomewide amplification exist (Hawkins et al. 2002), many are susceptible to potential biases that would make them unsuitable for use with ancient DNA, such as preferential amplification of contaminating modern human or environmental sequences over ancient sequences and changes in the composition of sequences from the sample source (Meyerhans et al. 1990; Polz and Cavanaugh 1998; Qiu et al. 2001). Emulsion PCR (Williams et al. 2006) theoretically escapes these biases by isolating individual template molecules in aqueous PCR reaction droplets, offering a potentially advantageous strategy for the processing of DNA from ancient remains. Here we describe the development of an emulsion PCR protocol for the amplification of 45,000- to 69,000-yr-old ancient DNA extracts, and the subsequent use of high-throughput sequencing of an amplified ancient DNA extract to generate sufficient nuclear sequences for the phylogenetic identification of unknown remains.
Results
Ancient DNA extraction from mammalian remains
As a substrate for evaluating novel ancient DNA methodologies, DNA extracts were prepared from ancient mammalian remains from Denisova Cave in the Siberian Altai (Derevianko et al. 2003). These included the remains of a horse long bone and wolf mandible from Layer 11, dated by radiothermoluminescence to 40,000–50,000 yr before present (BP) and an incisor from Layer 12 dated to 69,000 yr BP whose origin could not be unambiguously established by morphological evidence. Decontamination and DNA extraction were performed in a dedicated ancient DNA facility with strict contamination controls and vigorous decontamination measures for each sample using previously established methods (Yang et al. 1998, 2004). The average yield of ancient DNA from the remains was ∼50 ng of DNA per gram of bone used for the extraction (Supplemental Table 1).
Each ancient DNA extract was first characterized by PCR and sequencing of the mitochondrial variable region using species-specific primers. Mitochondrial sequences were obtained from both horse and wolf extracts, indicating the presence of ancient DNA sequences. In contrast, mitochondrial PCR of the incisor extract following tentative identification of the remains as sheep or goat yielded no products, suggesting either extensive degradation of this sample or misidentification of the remains (Supplemental Table 1). We did not attempt additional mitochondrial PCR of this sample using more general primers as this would require further destructive use of the DNA extract, with no guarantee that the reactions would be successful.
Emulsion PCR amplification of ancient DNA
We next developed a system for adapter-mediated emulsion PCR (Williams et al. 2006) amplification of small quantities of ancient DNA extract such that the resulting material could be transferred directly to high-throughput sequencing platforms. A shared feature of amplification technologies and new high-throughput sequencing platforms, such as the 454 Life Sciences (Roche) GS20 (Margulies et al. 2005) and Illumina, is the initial creation of DNA libraries by ligation of specific adapter sequences. Emulsion PCR using adapters and primers identical to those used in sequencing applications therefore allows transfer of amplified material directly to DNA sequencing platforms.
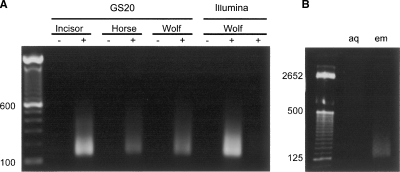
Emulsion PCR using GS20 adapters resulted in ∼1000-fold amplification of ancient incisor DNA, 560-fold amplification of ancient horse DNA, and 600-fold amplification of ancient wolf DNA as determined by fluorescence-based quantification of pre- and post-amplification DNA extracts (Fig. 1). Illumina sequencing (formerly Solexa sequencing) was performed exclusively on the ancient wolf DNA sample. Emulsion PCR using Illumina adapters resulted in 700-fold amplification of ancient wolf DNA, with amplification possible from as little as 0.1 ng of this extract (Fig. 1). For all samples, the majority of the amplified DNA fragments were in the size range 40–150 bp, consistent with degraded ancient DNA (Fig. 1A). In contrast, PCR amplification of the same extracts in aqueous solution (lacking the generation of the individual DNA molecule emulsion generated reactors) resulted in DNA fragments with an average size of several hundred bases, presumably the result of chimeras formed during the PCR reaction (Fig. 1B).
Figure 1.
Emulsion PCR amplification of ancient DNA extracts. (A) PAGE gel electrophoresis of ancient DNA extracts pre- (−) and post-amplification (+), using GS20 or Illumina adapters. Each pre-amplification lane contains 1 ng of ancient DNA. Each post-amplification lane contains one-fifth the amplification product from 1 ng (* = 0.1 ng, Wolf, right-hand lane) of starting DNA; 100-bp DNA ladder is shown for size comparison. The lengths of GS20 adapters (88 bp) and Illumina adapters (90 bp) must be taken into consideration when calculating amplified fragment size range. (B) Emulsion PCR (em) and aqueous nonemulsion PCR (aq) amplification of ancient wolf DNA. Nonemulsion PCR consistently results in higher molecular weight products, presumably due to chimeric sequences formed during the amplification process.
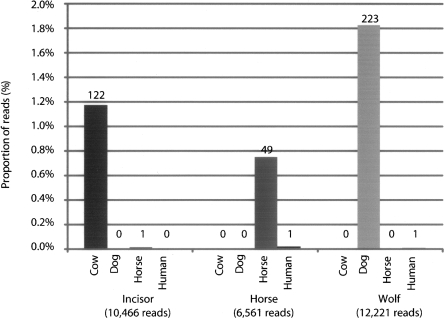
Sequencing of each of the GS20 adapter-amplified ancient DNA extracts by two lanes of a GS20 titration run yielded 10,446 sequences (754,190 bp) from ancient incisor DNA, 6561 sequences (498,856 bp) from ancient horse DNA, and 12,221 sequences (923,888 bp) from ancient wolf DNA. The sequence composition of the amplified extracts was determined by BLAST searches against cow, horse, dog, and human reference genomes and the nonredundant nucleotide sequence databases. Of sequences from the ancient horse extract, 0.7% (49/6561) had a best hit to the horse genome, while 1.8% (223/12,221) of sequences from the ancient wolf DNA extract had a best hit to the dog genome. The ancient incisor extract which gave negative mitochondrial PCR results nonetheless contained 1.1% (122/10,446) of sequences with a best hit to the cow genome (Fig. 2), suggesting that the negative mitochondrial PCR results were due to species misidentification rather than degradation of ancient DNA. For all samples, at most one sequence had a best alignment to a different mammalian reference genome, indicating correct classification of the samples and lack of contamination between samples. The number of sequences with best alignments to human was in each case less than 0.015%, with the ratio of ancient mammalian sequences to contaminating human sequences at least 49:1 (Fig. 2). The remaining sequences from each sample were either microbial (11.9% on average) or had no similarity to any currently known sequences (86.8% on average) (Supplemental Table 2).
Figure 2.
Sequence composition of amplified ancient DNA extracts. Extracts were sequenced by GS20 and aligned to mammalian genomes using BLAST. Sequences aligning to microbial sequences in NR or with no alignment are not shown.
For each sample, the average length of endogenous ancient sequences was less than the average length of all reads in the sample (Supplemental Fig. 2). Furthermore, we observe an excess of C > T and G > A mismatches in alignments of ancient sequences to the appropriate reference genome (Supplemental Fig. 3). These results show that emulsion PCR amplification retains the characteristic signatures of reduced fragment length and cytidine deamination associated with ancient DNA.
Evaluation of bias in amplified ancient DNA extracts
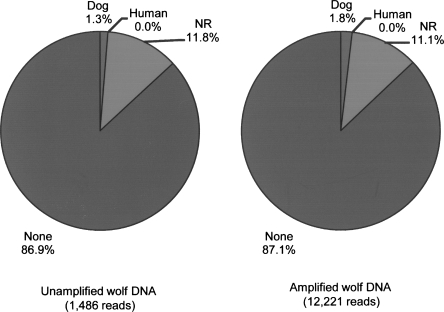
Several lines of evidence suggest that the amplification of ancient DNA is unbiased. For each amplified sample, alignments of ancient DNA sequences to the reference genomes are distributed among the chromosomes (Supplemental Fig. 1) and the proportion of sequences derived from annotated coding sequences and transposable elements is consistent with the genome wide distribution of these features (Supplemental Table 3). Furthermore, comparison of the amplified wolf DNA GS20 sequences with 1486 clones from a DNA library prepared from the unamplified ancient wolf DNA extract revealed no substantial change in sequence composition following amplification (Fig. 3).
Figure 3.
Sequence composition of amplified and unamplified ancient wolf DNA. Sequence composition was determined by BLAST analysis of 1486 capillary sequencing reads of clones from an unamplified ancient wolf DNA and 12,221 reads generated by GS20 sequencing of the amplified wolf DNA extract. The NR category includes any sequences that had a hit to the nonredundant nucleotide database, including those that aligned ambiguously to multiple database sequences.
A potential problem of any amplification procedure is reduction in sequence complexity. To evaluate sequence complexity, all sequences from each data set were aligned to themselves using BLAST, and clustering of the results was used to identify groups of similar sequences. For all amplified samples, over 93% of sequences are present only once, indicating a high degree of sequence complexity (Supplemental Table 3).
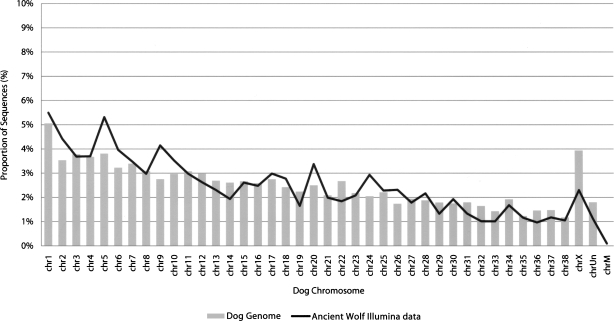
In order to verify the applicability of our method to different sequencing platforms, we performed Illumina sequencing of Illumina adapter-amplified ancient wolf DNA. A partial (one-eighth) run performed on 12 ng of amplified wolf DNA yielded 2,733,613 reads of 40 bp, comprising over 109 Mb of sequence in total. BLAT alignment of these sequences to the dog, human, cow, and horse reference genomes identified 2.1% (57,360/2,733,613) with a best hit to the dog genome, representing over 2.2 Mb of wolf nuclear DNA. In contrast, only 128 (0.005%) sequences had a best alignment to the human genome, confirming our previous observations of low levels of human contamination in this sample (Supplemental Table 1). Alignments to other mammalian genomes were similarly small (<0.006%). The wolf sequence alignments are distributed among the dog chromosomes and show a clear correlation with chromosome length (Fig. 4), providing further evidence that amplification is unbiased. Analysis of mismatches in Illumina reads aligned to the dog genome reveals an excess of DNA damage associated C > T mismatches, particularly near the starts of the sequences (Supplemental Fig. 4). These observations confirm the presence of the damaged DNA signature in emulsion PCR amplified DNA and are consistent with recent findings that damage accumulates near the ends of ancient DNA molecules and may be associated with DNA strand breaks (Briggs et al. 2007). Self-alignment of Illumina sequences revealed that the vast majority (98%) are unique, confirming our previous finding that sample complexity is maintained by emulsion PCR (Supplemental Table 3).
Figure 4.
Distribution of ancient wolf Illumina sequences on the dog genome. The proportion of the dog genome contained on each chromosome (bars) is shown along with the proportion of ancient wolf sequences aligning to each dog chromosome (black line).
Identification of a 69,000-yr-old ancient incisor
Analysis of ancient DNA sequences from the 69,000-yr-old incisor revealed 122 sequences with close similarity to the cow reference genome. This observation raised the possibility that the incisor may be bovine in origin. In order to define the origins of the remains more precisely, we used the nuclear sequences identified from comparison of the ancient incisor to the cow genome to perform comparative sequencing of extant ruminants.
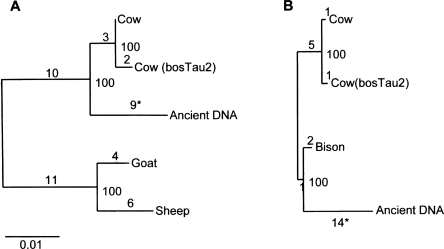
Of the 122 nuclear sequences from the incisor, 44 were over 50 bp in length and aligned to nonrepetitive regions of the cow genome. Nine out of 44 of these fragments, comprising 654 bp, were successfully amplified from orthologous regions of modern deer, goat, sheep, and cow genomic DNA. Estimation of the phylogeny of the aligned orthologous sequences by maximum likelihood resulted in the expected topology for sheep, goat, and cow, and with the ancient incisor sequences closely related to modern cow (Fig. 5A). Notably, seven out of nine substitutions observed in the incisor sequences were C > T or A > G, consistent with ancient DNA damage. To further refine the classification of the ancient incisor, we amplified a total of 17 nuclear fragments, comprising 1153 bp of orthologous sequences from extant cow, bison, and sheep DNA. The resulting phylogenetic analysis suggests that the incisor sequences are more closely related to modern bison than to cattle, suggesting that the remains are derived from bison (Fig. 5B). We subsequently confirmed this classification by PCR amplification of a mitochondrial fragment from the amplified ancient DNA extract using bovine specific primers, revealing sequences with a best match to steppe bison (Bison priscus) (Supplemental Fig. 3).
Figure 5.
Maximum-likelihood estimation of the phylogenetic relationship of ancient incisor DNA sequences to extant ruminants (including orthologous sequences from the cow reference genome bosTau2). (A) Phylogenetic reconstruction using 645-bp orthologous sequence from several ruminant species. Deer sequence was used to root the trees. (B) Phylogenetic reconstruction using a 1153-bp orthologous sequence from bovine species. Sheep sequence was used to root the trees; (*) seven out of nine and 12 out of 14 substitutions on the ancient incisor branch are C > T or A > G, consistent with ancient DNA damage. The total number of substitutions is shown for each branch, along with percent support for internal nodes.
Discussion
Using adapter-mediated emulsion PCR, we reproducibly achieved over 500-fold amplification of ancient DNA, with several lines of evidence indicating that our emulsion PCR amplification protocol is unbiased and that the complexity of the original DNA extracts is maintained. In the most successful amplification, 100 pg of starting DNA was amplified to 103 ng. Illumina sequencing produced 2.1-Mb of wolf nuclear DNA sequences from ∼12 ng of an amplified 40,000-yr-old wolf DNA extract. Extrapolation of these results suggests that, in principle, single-fold coverage of the wolf genome could be achieved from less than 20 ng of starting DNA. These results also suggest that ancient DNA extracts may be amplified to quantities sufficient for downstream approaches, such as capture of targeted sequences of interest (Noonan et al. 2006; Hodges et al. 2007).
We show that emulsion PCR amplified ancient DNA is suitable for analysis by several sequencing methods, depending on the questions being asked and the resources available. Amplified ancient DNA may be cloned into bacteria and sequenced by widely available conventional capillary-sequencing technologies. For deeper sequencing analyses, ancient DNA amplified using the appropriate adapters may be analyzed directly on a number of high-throughput sequencing platforms. An advantage of technology such as the GS20 sequencer to ancient DNA studies is that average read length (∼200 bp) is greater than the length of the average ancient DNA molecule (<100 bp). This allows read-length analyses to be used to evaluate DNA preservation and potential contamination, a feature that may be particularly useful in sequencing DNA from ancient hominids. In other cases, generation of millions of short reads using technology such as the Illumina Genome Analyzer provides, allows identification of substantial quantities of endogenous nuclear DNA despite the majority of sequences being from environmental contamination (for example, the endogenous DNA from the samples used in this study is between 0.7% and 1.8%). These studies will become increasingly feasible and economical with increase in throughput and reduction in cost of DNA sequencing.
In this study, we examined ancient DNA from non-hominid mammalian remains. This allowed us to accurately evaluate the amount of contaminating human DNA in each sample. Overall, we found evidence for only small amounts of contamination from human DNA (<0.02%), with the ratio of contaminating human sequences to ancient sequences at most 0.018%–0.7% for the samples used in this study. For ancient DNA from hominids, the high degree of sequence similarity between ancient and modern sequences makes evaluation of contamination very difficult. Currently, contamination of Neanderthal ancient DNA extracts is evaluated by PCR amplification of the mitochondrial variable region. However, single-locus mitochondrial PCR is susceptible to potential biases and quantitative errors, such as preferential amplification of modern undamaged sequences over damaged ancient sequences. In the current study, the amount of human contamination from our genomic analyses appears to be consistent between samples from the same site processed under the same conditions. These results suggest that, assuming clean handling of samples pre- and post-DNA extraction, the extent of modern human contamination of Neanderthal samples may be reasonably estimated from the level of human DNA contamination of adjacent non-human remains taken from the same site and handled in the same way.
Our studies included an ancient DNA extract from an incisor that could not be identified by morphological analysis and for which no mitochondrial sequences could be generated from PCR amplifications. In contrast, a single emulsion PCR amplification using our method generated sufficient nuclear sequences to clearly demonstrate that the remains belong to the genus Bos. Informed by these results, we were able to repeat mitochondrial sequencing using the appropriate primers and confirm this classification. Our amplification and random shot-gun sequence sampling approach to identification of unknown remains is conservative of material and will rapidly improve in efficiency and precision as more mammalian reference genome sequences become available. This novel application of ancient DNA technology may be of value in identifying morphologically indistinct faunal remains, and it provides new opportunities for ancient DNA researchers to revisit samples that have previously tested negative based on mitochondrial PCR sequencing results alone.
Methods
Ancient DNA extraction
DNA was generated from Siberian samples using the silica-spin column method (Yang et al. 1998). Samples underwent outer surface decontamination and ∼1 g of sample was ground using a liquid nitrogen grinding mill. The bone powder was then incubated in lysis buffer (0.5 M EDTA at pH 8.0, 0.5% SDS, and 0.5 mg/mL proteinase K) in a rotating hybridization oven at 50°C overnight. After centrifugation, 2–3 mL of lysis supernatant was concentrated using an Amicon Ultra-4 centrifugal filter (Billerica). Approximately 100 μL of concentrated supernatant was passed through a QIAquick column (QIAGEN) for final purification.
Ancient DNA quantification
Ancient DNA extracts were quantified using Quant-iT dsDNA HS assay kit (Q32851, Invitrogen), following the manufacturer’s manual. For each sample, 1 μL of the DNA extracts was used for the quantification.
Mitochondrial PCR from ancient DNA extracts
Three genus-specific primer sets were used to amplify fragments of the hypervariable control region within the horse, sheep/goat, and wolf mitochondrial DNA genome. Due to the degraded nature of the DNA, short fragments of mtDNA were targeted, with primers designed to amplify a 122-bp fragment for the horse, a 139-bp fragment for the sheep/goat, and a 149-bp fragment for the wolf. All PCR amplifications were conducted in a Mastercycler Personal (Eppendorf) in a 30-μL reaction volume containing 50 mM KCl and 10 mM Tris-HCl, 2.5 mM MgCl2, 0.2 mM dNTP, 1.5 mg/mL BSA, 0.3 μM each primer, 3 μL of DNA sample, and 2.5 U of AmpliTaq Gold. The conditions of PCR amplification were as follows: The initial denaturing took place at 95°C for 12 min, followed by 60 cycles at 95°C for 30 sec, 52°C for 30 sec, 70°C for 40 sec, followed by a final 7-min extension at 72°C. Five microliters of PCR product was separated by electrophoresis on 2% agarose gel and visualized using SYBR-Green on a Dark Reader Box. PCR products were purified using QIAGEN’s MinElute and then sent to the Central Facility of the Institute for Molecular Biology and Biotechnology Laboratory at McMaster University for sequencing on an ABI 3100 apparatus.
End repair
One nanogram of each ancient mammalian DNA extract was end-repaired using the End-It DNA End Repair Kit (ER0720, EPICENTRE Biotechnologies) according to the manufacturer’s instructions. End-repaired DNA was extracted using phenol/chloroform/isoamyl alcohol (Ambion), precipitated in ethanol, and resuspended in 5 μL of TE buffer.
Linker ligation
Ligation reactions comprised 5 μL of end-repaired DNA, 1 μL of PEG (50%), 1 μL of 10× ligation buffer, 1 μL of T4 DNA ligase (5 U/μL, Fermentas), and either 1 μL of GS20 adapter-1 and 1 μL of GS20 adapter-2 (20 μM each) or 1 μL of Illumina P1 adapter and 1 μL of Illumina P2 adapter (20 μM each), depending on the downstream sequencing application. Adapter sequences are described in the Supplemental Material, available online. Ligation was performed at 22°C for 1 h, and linker-ligated DNA was recovered using a MiniElute Reaction Cleanup Kit (QIAGEN), following the manufacturer’s protocol. The DNA was eluted in 10 μL of elution buffer (QIAGEN).
Emulsion PCR amplification
The emulsion PCR protocol was a modification of a previously described method (Williams et al. 2006), optimized to increase the yield and specificity of the PCR reaction from ancient DNA extracts. To 10 μL of end-repaired and linker-ligated ancient DNA were added 26 μL of 10× Pfu PCR buffer, 26 μL of BSA (100 mg/mL), 5.2 μL of dNTP (10 mM), and either 13 μL of GS20 forward primer and 13 μL of GS20 reverse primer (10 μM each) or 13 μL of Illumina PCR forward primer and 13 μL of Illumina PCR reverse primer (10 μM each; primer sequences are described in Supplementary material), and water to 254.8 μL. The mix was incubated at 72°C for 3 min followed by addition of 5.2 μL of Pfu Turbo DNA polymerase (Stratagene), further incubated at 72°C for another 5 min, and then left to cool to room temperature. Emulsions were formed by dropwise addition of 200 μL of PCR mix to 400 μL of oil surfactant mixture as described elsewhere (Williams et al. 2006); 50-μL aliquots were transferred to 200-μL PCR tubes, topped with mineral oil, and subjected to PCR (one cycle of 94°C for 5 min; 40 cycles of 72°C for 3 min and 94°C for 1 min; and one cycle of 72°C for 8 min). PCR products were frozen (−20°C 1 h) and then returned to room temperature. Pooled PCR product was centrifuged (14,000 rpm for 5 min), and the upper organic phase was discarded. The aqueous phase was extracted twice by addition of 500 μL of water-saturated diethyl ether (Sigma) followed by vortexing for 10 sec, centrifugation (14,000 rpm for 5 min), and disposal of upper solvent phase. Following a further extraction with 1 mL of water-saturated chloroform, vortexing for 10 sec, and centrifugation (14,000 rpm for 5 min), DNA was recovered from the upper aqueous phase using a MiniElute PCR Purification Kit (QIAGEN) and elution in 30 μL of elution buffer. Emulsion PCR products were examined by electrophoresis using Ready Gel TBE Gel (4%–20%, Bio-Rad Laboratories) followed by staining with SYBR Gold (Molecular Probes) in TBE buffer.
Sequencing of emulsion PCR amplified ancient DNA
GS20 sequencing was performed according to the manufacturer’s instructions (454 Life Sciences [Roche]) and as described previously (Margulies et al. 2005) except that the initial bead-based emulsion PCR amplification was omitted and amplified ancient DNAs containing the GS20 sequencing adapter were hybridized directly to DNA capture beads (approximately four DNA molecules per bead). Beads were enriched for those containing bound DNA, and sequencing of two lanes, each containing ∼36,000 enriched beads, was performed for each sample.
Illumina sequencing was performed according to the manufacturer’s instructions (Illumina) except that emulsion PCR amplified ancient DNA containing the Illumina sequencing adapter were applied directly to the cluster station for bridge amplification. The resulting flow cell was sequenced for 40 cycles to generate 40-bp reads.
Direct cloning and capillary sequencing of unamplified ancient wolf DNA
End repair of 10 ng of wolf DNA was performed as described above and resuspended in 6 μL of water. Vector ligation was performed as described for linkers except 1 μL (3 ng) of linearized blunt-ended pPET3h vector was used in place of vectors, and final elution was in 10 μL of water. One microliter of ligated DNA was electroporated into One Shot TOP10 Electrocomp Escherichia coli (Invitrogen). Transformed cells were recovered at 37°C for 1 h in 500 μL of SOC medium (Invitrogen) and grown on LB agar bioassay plates containing 100 μg/mL of ampicillin and 50 mg/mL of X-gal at 37°C overnight. Individual white recombinant colonies were selected and picked into 384-well microtiter plates containing LB/glycerol (7.5%) media supplemented with 100 μg of ampicillin/mL. Clones were sequenced from both directions from the vector using forward primer (5′-GTTTTCCCAGTCACG ACGTTGTA-3′) and reverse primer (5′-AGGAAACAGCTATGAC CAT-3′). For details on production sequencing protocols, see http://www.jgi.doe.gov/sequencing/protocols/prots_production.html.
Alignment of capillary and GS20 sequences
Forward and reverse capillary sequences were assembled using phred/phrap (Ewing and Green 1998; Ewing et al. 1998), and vector sequences were masked using cross_match. Both capillary and GS20 sequences with fewer than 30-bp unmasked sequence and with less than 80% bases with quality score >20 were rejected. Sequences were aligned to human (hg18), dog (canFam2), cow (bosTau2), and horse (equCab1) reference genome sequences (downloaded from the UCSC genome browser; Kuhn et al. 2007) and the nonredundant nucleotide sequence database (nt) using megaBLAST with an E-value threshold of 0.001. Alignments less than 30 bp or with a bit score less than 50 were rejected. The top-scoring alignment for each query sequence across all databases was identified by comparison of bit scores. Sequences aligning to multiple mammalian genomes was considered to be “ambiguous mammalian” if (1) there was a second alignment with equal or greater percent identity to the best-scoring alignment and an alignment length >95% of the length of the top-scoring alignment; or (2) there was a second alignment with length equal or greater than the top-scoring alignment and percent identity >95% of that of the best-scoring alignment. Sequences which only had ambiguous alignments to the same genome were considered as ambiguous but were assigned to that genome.
Alignment of Illumina sequences
Wolf Illumina sequences were aligned to the dog, human, horse, and cow reference genomes using BLAT (Kent 2002). The best alignment to each genome was determined by comparison of BLAT scores. Alignments with a BLAT score of <35, with >3-bp insertion or deletion, or with >50% simple repeat were considered unreliable alignments and discarded. The overall top alignment for each sequence was determined by comparison of top hits to each genome with the requirement that the top alignment exceeded the second best alignment by at least 2 BLAT score points.
Clustering of GS20 and Illumina sequences
To search for identical sequences within datasets, all against all alignments were performed using BLAT with output format out = blast8 (similar to BLAST tab-formatted output). For GS20 sequences, two sequences aligned in the same orientation, and where the difference between the start positions of the two aligned sequences was less than 5 bp they were considered identical. For Illumina sequences, two sequences aligned with the same start position, and with an alignment length of >29 bp were accepted. Clustering of self-alignments was performed using the MCL algorithm (Enright et al. 2002).
Annotation of mammalian sequences
The reference genome coordinates of aligned ancient mammalian DNA sequences were used to query the corresponding genome annotation tables from the UCSC genome browser using Galaxy (Giardine et al. 2005).
Sequencing and phylogenetic analysis of ruminant DNAs
PCR primers were designed to amplify fragments ∼400 bp corresponding to 44 regions of the cow reference genome (bosTau2) covering alignments to ancient incisor sequences >50 bp in nonrepetitive regions. PCR amplification was performed in duplicate on genomic DNA from cow, deer, and sheep, and PCR products were sequenced on an ABI3730 sequencer.
Ruminant sequences corresponding to the ancient incisor sequence were concatenated and aligned using ClustalW (Larkin et al. 2007). Tree topologies were determined using TREE-PUZZLE and default parameters except the Tamura–Nei model of nucleotide substitutions (Schmidt et al. 2002). The same topology was also obtained by maximum likelihood and maximum parsimony methods using the PHYLIP package. Branch lengths and lineage-specific substitutions for the resulting topology were re-estimated using baseml under the REV model excluding gapped sites (Yang 2007).
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory (contract DE-AC52-07NA27344), Lawrence Berkeley National Laboratory (contract DE-AC0205CH11231), and Los Alamos National Laboratory (contract DE-AC02-06NA25396) and was supported by the National Institutes of Health (grant HG004123) and SSHRC Canada (grant 410-2005-0841).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076091.108.
References
- Briggs A.W., Stenzel U., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Stenzel U., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Meyer M., Krause J., Ronan M.T., Lachmann M., Krause J., Ronan M.T., Lachmann M., Ronan M.T., Lachmann M., Lachmann M., et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derevianko A.P., Shunkov M.V., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Shunkov M.V., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Kulik N.A., Postnov A.V., Anoikin A.A., Postnov A.V., Anoikin A.A., Anoikin A.A. Paleoenvironment and paleolithic human occupation of Gorny Altai (Subsistence and adaptation in the vicinity of Denisova Cave) Russian Academy of Sciences; Novosibirsk: 2003. [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A., Van Dongen S., Ouzounis C.A., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Zhang Y., Blankenberg D., Albert I., Taylor J., Blankenberg D., Albert I., Taylor J., Albert I., Taylor J., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M.T., Binladen J., Miller W., Wiuf C., Willerslev E., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Binladen J., Miller W., Wiuf C., Willerslev E., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Miller W., Wiuf C., Willerslev E., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Wiuf C., Willerslev E., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Willerslev E., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Poinar H., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Carlson J.E., Leebens-Mack J.H., Schuster S.C., Leebens-Mack J.H., Schuster S.C., Schuster S.C. Recharacterization of ancient DNA miscoding lesions: Insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 2007a;35:1–10. doi: 10.1093/nar/gkl483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M.T., Tomsho L.P., Rendulic S., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Tomsho L.P., Rendulic S., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Rendulic S., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Drautz D.I., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Sher A., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Tikhonov A., Dalen L., Kuznetsova T., Kosintsev P., Dalen L., Kuznetsova T., Kosintsev P., Kuznetsova T., Kosintsev P., Kosintsev P., et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007b;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- Green R.E., Krause J., Ptak S.E., Briggs A.W., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Krause J., Ptak S.E., Briggs A.W., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Ptak S.E., Briggs A.W., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Briggs A.W., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Du L., Egholm M., Rothberg J.M., Paunovic M., Egholm M., Rothberg J.M., Paunovic M., Rothberg J.M., Paunovic M., Paunovic M., et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Hawkins T.L., Detter J.C., Richardson P.M., Detter J.C., Richardson P.M., Richardson P.M. Whole genome amplification—Applications and advances. Curr. Opin. Biotechnol. 2002;13:65–67. doi: 10.1016/s0958-1669(02)00286-0. [DOI] [PubMed] [Google Scholar]
- Hodges E., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., Rodesch M.J., Albert T.J., Hannon G.J., Albert T.J., Hannon G.J., Hannon G.J., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hofreiter M., Serre D., Poinar H.N., Kuch M., Paabo S., Serre D., Poinar H.N., Kuch M., Paabo S., Poinar H.N., Kuch M., Paabo S., Kuch M., Paabo S., Paabo S. Ancient DNA. Nature Rev. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J., Lalueza-Fox C., Orlando L., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Lalueza-Fox C., Orlando L., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Orlando L., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., Hanni C., Fortea J., de la Rasilla M., Fortea J., de la Rasilla M., de la Rasilla M., et al. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr. Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Stanke M., Smith K.E., Siepel A., Smith K.E., Siepel A., Siepel A., et al. The UCSC genome browser database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalueza-Fox C., Rompler H., Caramelli D., Staubert C., Catalano G., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Rompler H., Caramelli D., Staubert C., Catalano G., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Caramelli D., Staubert C., Catalano G., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Staubert C., Catalano G., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Catalano G., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Hughes D., Rohland N., Pilli E., Longo L., Condemi S., Rohland N., Pilli E., Longo L., Condemi S., Pilli E., Longo L., Condemi S., Longo L., Condemi S., Condemi S., et al. Science. Vol. 318. 2007. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals; pp. 1453–1455. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Valentin F., Wallace I.M., Wilm A., Lopez R., Wallace I.M., Wilm A., Lopez R., Wilm A., Lopez R., Lopez R., et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J.P., Wain-Hobson S., Vartanian J.P., Wain-Hobson S., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J.P., Hofreiter M., Smith D., Priest J.R., Rohland N., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Hofreiter M., Smith D., Priest J.R., Rohland N., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Smith D., Priest J.R., Rohland N., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Priest J.R., Rohland N., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Rohland N., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Rabeder G., Krause J., Detter J.C., Paabo S., Rubin E.M., Krause J., Detter J.C., Paabo S., Rubin E.M., Detter J.C., Paabo S., Rubin E.M., Paabo S., Rubin E.M., Rubin E.M. Genomic sequencing of Pleistocene cave bears. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- Noonan J.P., Coop G., Kudaravalli S., Smith D., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Coop G., Kudaravalli S., Smith D., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Kudaravalli S., Smith D., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Smith D., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Chen F., Platt D., Paabo S., Pritchard J.K., Platt D., Paabo S., Pritchard J.K., Paabo S., Pritchard J.K., Pritchard J.K., et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar H.N., Schwarz C., Qi J., Shapiro B., MacPhee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Schwarz C., Qi J., Shapiro B., MacPhee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Qi J., Shapiro B., MacPhee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Shapiro B., MacPhee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., MacPhee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., Huson D.H., Tomsho L.P., Auch A., Tomsho L.P., Auch A., Auch A., et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Polz M.F., Cavanaugh C.M., Cavanaugh C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Wu L., Huang H., McDonel P.E., Palumbo A.V., Tiedje J.M., Zhou J., Wu L., Huang H., McDonel P.E., Palumbo A.V., Tiedje J.M., Zhou J., Huang H., McDonel P.E., Palumbo A.V., Tiedje J.M., Zhou J., McDonel P.E., Palumbo A.V., Tiedje J.M., Zhou J., Palumbo A.V., Tiedje J.M., Zhou J., Tiedje J.M., Zhou J., Zhou J. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompler H., Rohland N., Lalueza-Fox C., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Rohland N., Lalueza-Fox C., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Lalueza-Fox C., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M., Bertranpetit J., Schoneberg T., Hofreiter M., Schoneberg T., Hofreiter M., Hofreiter M. Nuclear gene indicates coat-color polymorphism in mammoths. Science. 2006;313:62. doi: 10.1126/science.1128994. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Williams R., Peisajovich S.G., Miller O.J., Magdassi S., Tawfik D.S., Griffiths A.D., Peisajovich S.G., Miller O.J., Magdassi S., Tawfik D.S., Griffiths A.D., Miller O.J., Magdassi S., Tawfik D.S., Griffiths A.D., Magdassi S., Tawfik D.S., Griffiths A.D., Tawfik D.S., Griffiths A.D., Griffiths A.D. Amplification of complex gene libraries by emulsion PCR. Nat. Methods. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang D.Y., Eng B., Waye J.S., Dudar J.C., Saunders S.R., Eng B., Waye J.S., Dudar J.C., Saunders S.R., Waye J.S., Dudar J.C., Saunders S.R., Dudar J.C., Saunders S.R., Saunders S.R. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Yang D.Y., Cannon A., Saunders S.R., Cannon A., Saunders S.R., Saunders S.R. DNA species identification of archaeological salmon bone from the Pacific Northwest Coast of North America. J. Archaeol. Sci. 2004;31:13. [Google Scholar]