Abstract
Silencers and enhancer-blockers (EBs) are cis-acting, negative regulatory elements (NREs) that control interactions between promoters and enhancers. Although relatively uncharacterized in terms of biological mechanisms, these elements are likely to be abundant in the genome. We developed an experimental strategy to identify silencers and EBs using transient transfection assays. A known insulator and EB from the chicken beta-globin locus, cHS4, served as a control element for these assays. We examined 47 sequences from a 1.8-Mb region of human chromosome 7 for silencer and EB activities. The majority of functional elements displayed directional and promoter-specific activities. A limited number of sequences acted in a dual manner, as both silencers and EBs. We examined genomic data, epigenetic modifications, and sequence motifs within these regions. Strong silencer elements contained a novel CT-rich motif, often in multiple copies. Deletion of the motif from three regions caused a measurable loss of silencing ability in these sequences. Moreover, five duplicate occurrences of this motif were identified in the cHS4 insulator. These motifs provided an explanation for an uncharacterized silencing activity we measured in the insulator element. Overall, we identified 15 novel NREs, which contribute new insights into the prevalence and composition of sequences that negatively regulate gene expression.
The goal of the ENCODE Consortium (The ENCODE Project Consortium 2007) toward comprehensively annotating all functional elements of the noncoding genome has fueled interest in identifying novel types of elements such as negative regulators of gene expression (negative regulatory elements, NREs). In contrast to the large body of literature on positively acting elements such as enhancers and promoters, cis-acting NREs have not been extensively studied. Despite their scarcity in the literature, these elements are likely to be abundant in the genome. Examples of NREs include silencers, which decrease expression of a gene under their regulation and enhancer-blocking (EB) elements, which prevent the action of an enhancer on a promoter when placed between the two, but not otherwise (Gaszner and Felsenfeld 2006). A further refinement of the definition applies to barrier elements, or insulators, which function as physical barriers in the DNA to block progression of closed chromatin into active regions. The definitions of these elements are based on their behaviors in experimental assays. The outcome of such assays is a measurable loss of expression from a reporter gene.
Examples of NREs are extremely limited (Ogbourne and Antalis 1998; Gaszner and Felsenfeld 2006). Only one protein is known to bind barrier elements in mammals—the zinc finger protein, CCCTC-binding factor (CTCF). Binding of this protein marks boundaries between different chromatin domains (Barski et al. 2007) and separates genes with discordant expression patterns (Xie et al. 2007). CTCF binds to the boundary element of the chicken beta-globin locus, known as hypersensitive site 4, or cHS4. This element is described as a “compound insulator” due to the presence of EB and barrier functions (Saitoh et al. 2000; Recillas-Targa et al. 2002). A single confirmed binding site for the protein CTCF is present in footprint region II of cHS4 and contributes the EB activity. This CTCF site alone is not sufficient to produce the barrier function (Burgess-Beusse et al. 2002), indicating that additional functional elements are present, yet uncharacterized.
EB and silencer assays typically require integration of the reporter gene into the chromatin of genomic DNA. This process is too laborious to use in large-scale analyses. To improve scalability, our system uses nonintegrated recombinant plasmids, which have been shown to support EB activity (Recillas-Targa et al. 1999). We evaluated silencing and EB function in genomic sequences by developing a transient transfection assay suitable for high-throughput screening. To ensure that we identified elements capable of overcoming the effects of strong enhancers, the assay utilizes an enhancer from the human beta-globin locus control region. Known as DNase I hypersensitive site II (HS2) (Talbot and Grosveld 1991), this enhancer is functional in multiple cell lines, with multiple promoters (Elnitski et al. 1997). Furthermore, the presence of HS2 provides a large window of expression to reliably measure loss-of-function effects. Our experimental strategy employed a screen for functional elements from the 1.8-Mb ENCODE region encompassing the CFTR gene. Additionally, we evaluated whether sequence conservation was a characteristic of silencers and EBs and identified candidate proteins acting at these sites through comparison to ChIP-chip and ChIP-seq data (Barski et al. 2007; The ENCODE Project Consortium 2007). We present data for 15 novel NRE regions and directly address whether silencers and EBs act in an absolute and invariant manner or if genomic context influences their function. Furthermore, these data indicate that studies of NREs can be performed on the same scale as enhancer and promoter assays.
Results
Experimental design
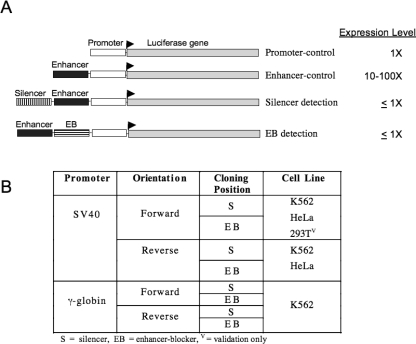
Four plasmids were necessary to determine whether silencing or EB activity was present in a test region (Fig. 1). Two control plasmids were used to define the upper and lower limits of luciferase signal in the assay. The promoter-control produced the baseline level of luciferase activity, normalized to 1×. The enhancer-control revealed the upper threshold of expression, up to 100× over the promoter alone, contributed by the HS2 enhancer. The enhancer-control expression vector provided the backbone into which the candidate silencer and EB elements were cloned. A cloning position upstream of the enhancer ascertained negative activity called “silencing.” In contrast, when positioned between the enhancer and promoter, an element that inhibited gene expression was designated as an EB. The HS2 enhancer was included in all silencer and EB plasmids to confirm an ability to overcome strong activating events and as a measure of the interference between promoter and enhancer interactions, respectively. Confirmation of silencer or EB status required an expression signal that was equivalent to, or below, that produced by the promoter alone (1×), despite the presence of the enhancer (Fig. 1A). Such a response was consistent with a complete loss of enhancer activity. Several cloning vectors and conditions were tested to assess whether the promoter, cell line, and orientation of the cloned sequences affected the phenotype (Fig. 1B).
Figure 1.
Experimental design. (A) A series of four plasmids was utilized in the experimental assay system. Functional components of the luciferase-reporter expression vectors are indicated: (Promoter-control) the minimal promoter-only control plasmid; (Enhancer-control) the control plasmid containing the enhancer. The insertion sites for silencer elements and enhancer-blocking (EB) sites are designated relative to the enhancer location. Expression levels are high for enhancer-driven expression (10–100×) and low for full repressive activity (1×). (B) The experimental options included the choice of promoter, cloning orientation, the function (assessed by virtue of the cloning position), and cell line.
Silencing conferred by the chicken HS4 insulator
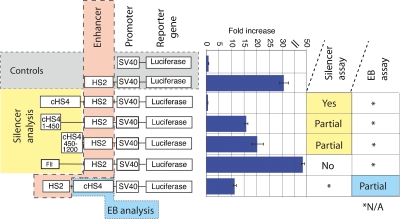
As a known insulator element, the 1.2-kb region from the chicken beta-globin locus was examined for a role in the silencing and EB functions assayed in this newly developed system. When placed upstream of the enhancer, the cHS4 insulator element completely silenced expression, causing a 30-fold reduction in signal (Fig. 2). Deletion of sequences at positions 1–450 bp or 450–1200 bp of the 1.2-kb fragment partially diminished luciferase activity by twofold or 1.5-fold, respectively. This result indicated that both halves of the element were necessary for silencing function. In contrast, a fragment containing the verified CTCF binding site, known as footprint II or FII, was unable to silence expression, indicating that CTCF alone was not sufficient for this activity. The 1.2-kb element in the EB position reduced gene expression by 2.8-fold. This result was consistent with the level of EB activity reported by Bell et al. (1999) (2.2-fold) in a stable transfection assay.
Figure 2.
Expression results obtained with the chicken HS4 compound insulator. Plasmids are depicted to the left of their expression data generated in transiently transfected K562 cells. The control plasmids contain the promoter-only or the promoter with the HS2 enhancer. The 1.2-kb region of the cHS4 insulator was subcloned upstream of HS2 as a test of silencing. Deletions of the 3′ or 5′ ends of the cHS4 insulator, containing bases 1–450 or 450–1200 of the element, were also assessed for silencing. The FII region contains the 46-bp footprint region that binds CTCF. Positions of the fragments are shown relative to the full-length cHS4 sequence. The plasmid to test cHS4 in the EB position is the final plasmid in the image. The sizes of the DNA elements are for illustration only and are not drawn to scale.
Assessing the frequency of silencers and EBs in genomic DNA
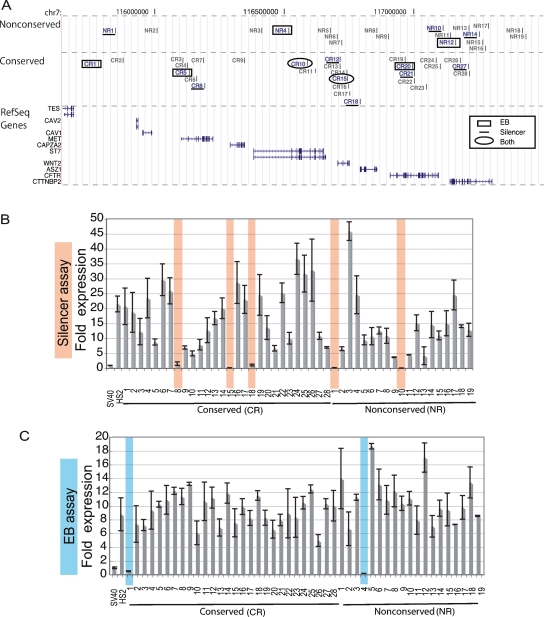
The 1.8-Mb locus encompassing the CFTR gene was chosen for testing. This region is designated as the greater CFTR locus by the ENCODE Consortium. In addition to CFTR, the locus contains nine genes including the proto-oncogene, MET, and the developmental gene, WNT2. Of the 47 target regions, two thirds were selected for testing because they included conserved noncoding sequences (Margulies et al. 2003), whereas one third did not (Fig. 3A). The conserved regions (CR) all had a signature of selective constraint as determined by the ENCODE Multi-Species Analysis group: 78% were defined under the “strict” classification, requiring assignment by three analysis programs using three alignment methods, whereas 22% met the condition for the “moderate” classification being called constrained by two programs and at least two alignment methods (Margulies et al. 2007). The average length of the sequences was 400 bp. The nonconserved regions (NR), averaging 600 bp, contained limited sequence similarity over no more than 12% of their total length. The majority of the 47 elements (60%) were located in introns, with no consistent distance to the nearest promoters. The choice of the CFTR locus was designed to complement studies of the ENCODE Consortium, which aimed to define all functional elements in 1% of the human genome.
Figure 3.
Silencer and EB functions assayed from chromosome 7 sequences. (A) An illustration depicting 47 subcloned elements appears above RefSeq gene annotations from the UCSC Human Genome Browser. The elements are divided into groups as conserved or nonconserved regions (CR or NR, respectively). The numbering scheme increases from left to right across the genomic region. (B) Transfection results for the silencer assay in K562 cells. Controls are the promoter-only (SV40) or the enhancer plasmid (HS2). All of the candidate regions are cloned into the enhancer plasmid, in the forward orientation, upstream of HS2. (C) Transfection results for the EB assay in K562 cells. All candidate regions are cloned into the enhancer plasmid, in the forward orientation, between HS2 and SV40. Silencers and EBs with strong negative effects are shaded light orange and blue, respectively. The results shown are the means from three replicates ± the standard deviation in the error bars. All silencers and EBs were retested in triplicate and resequenced to confirm the results.
In total, 47 regions were evaluated for a negative regulatory effect as measured by a complete loss of enhancer activity. Figure 3A summarizes the data obtained after all experimental analyses were conducted. Figure 3B,C illustrates the individual assays necessary for assignment of an NRE phenotype, in which the absence of enhancer-driven expression indicated a complete silencing or EB effect. For example, when cloned in the forward orientation with the SV40 promoter, five regions conferred complete silencing activity in K562 cells (i.e., 1× expression; CR8, CR15, CR18, NR1, and NR10). A distinct set of elements functioned as EBs by reducing expression to the 1× level in K562 cells, in the forward orientation with the SV40 promoter (CR1 and NR4; Fig. 3C). As indicated in Figure 3A, NREs mainly acted uniquely as silencers or EBs, and rarely as both.
Sequence conservation was not required for NRE function. Five of the nonconserved elements were able to stifle gene expression (26%) versus 35% of CR regions. Plasmids with unaffected expression levels illustrated that not all sequences could disrupt the enhancer–promoter interaction, even when placed strategically between them.
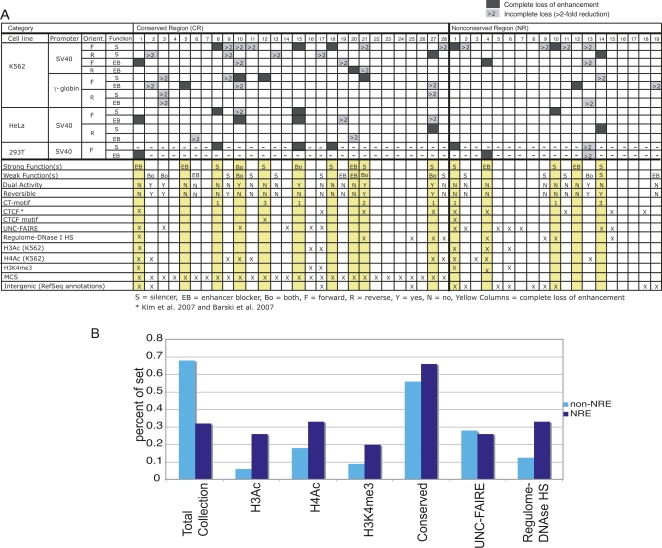
Full silencing or EB activity was observed for 15 regions, designated as NREs, under at least one experimental condition (Fig. 4A, dark gray boxes and yellow columns). In addition, a partial functional effect was obtained from 29% (or 14) of the regions under at least one condition (Fig. 4A, columns with light gray boxes). The remaining 38% of the clones showed no effect under any experimental condition we tested. Two of the NREs had dual activity causing a complete loss of enhancer function as both silencers and EBs (CR10 and CR15, labeled Bo for “Both”). These elements suggested a possible redundancy in the mechanism used by some silencers and EBs. Twelve of 15 NRE elements were orientation-dependent in their activity. Reversibility was observed more frequently when we considered the weaker phenotypes in the analysis. For instance, six regions with dual activity as silencers and EBs were obtained in this weak group (Fig. 4A, including CR10). Of the dual functioning regions, two elements (CR10 and CR15) showed an orientation-dependent effect. The five remaining weak elements (CR2, CR3, CR21, CR27, and NR13) showed some function in both orientations.
Figure 4.
Overview of silencer and EB sites and their genomic features. (A) Summary of all transfection results organized by conserved and nonconserved categories. Dark gray boxes indicate strong phenotypes for silencing or EB, producing expression levels at 1× or below. (Light gray boxes) Modest silencing or EB phenotypes (at least twofold below the enhancer-control); (white boxes) no decrease in expression or a value less than twofold below the enhancer. Columns below the transfection results indicate characteristics of each element, including activity levels, reversibility, motif content, and genomic features. (B) Analysis of genomic attributes in strong silencers and EBs. The data are graphed to show the percent of NREs and non-NREs carrying each feature.
NREs demonstrated consistency across cell lines and specificity for promoters. For instance, CR15 was a silencer in K562, HeLa, and 293T. Out of the 15 NREs, 10 regions recapitulated results from K562 cells in HeLa or 293T cells (Fig. 4A). Some NREs functioned with multiple promoters whereas others switched or lost their function when the promoter changed. For example, the CR15 element completely silenced expression in K562 cells with the SV40 promoter, but had no effect with the gamma-globin promoter. Additionally, assays with the gamma-globin promoter uniquely identified three strong silencers and two strong EBs (in K562 cells, forward orientation). These strong functions were weak or nonexistent with the SV40 promoter. Only the element CR10 had a strong phenotype with both the SV40 and gamma-globin promoters. However, CR10 functioned in two different ways, acting as a silencer of the interaction between the gamma-globin promoter and HS2 or as an EB when placed between HS2 and the SV40 promoter. The reverse was seen for the weakly functioning CR2 element, where SV40 and gamma-globin promoters enabled silencer or EB functions, respectively. Some regions maintained consistent function despite changes in promoter composition. For instance, CR27 functioned as a weak EB with either promoter.
Considering weak effects, seven elements displayed dual activity as silencers and EBs. However the vast majority of elements conferred position-specific activity by acting in a mutually exclusive manner as a silencer or EB (22 of the 29 sites demonstrating activity). Furthermore, the majority of elements (22 of 29) exhibited a directional effect.
The 47 regions were divided into a set of 15 strong-acting NREs, and the remaining 32 weak or nonactive regions were designated non-NREs. Although the measurements indicated a spectrum of repressive levels, this classification eliminated false positives from the NRE data set. Comparison of the two data sets to ENCODE data indicated the presence of distinct differences between the NREs and non-NREs (Fig. 4B). In ChIP-chip data collected from K562 cells (Koch et al. 2007) the 15 NRE regions showed a larger signal for histone acetylation at H3 and H4 in K562 cells than did the 32 non-NREs. Furthermore, the tri-methylation signal, H3K4me3, was larger in NREs than non-NREs. Other indicators of functional regions such as UNC-FAIRE locations (Giresi et al. 2007) and Regulome DNase I hypersensitive sites (Dorschner et al. 2004) occurred as frequently or more frequently in the NREs than non-NREs (tabulated in Fig. 4A). Nevertheless, the number of observations was too small to accurately quantitate the statistical significance of these differences.
Motif analyses of silencer elements
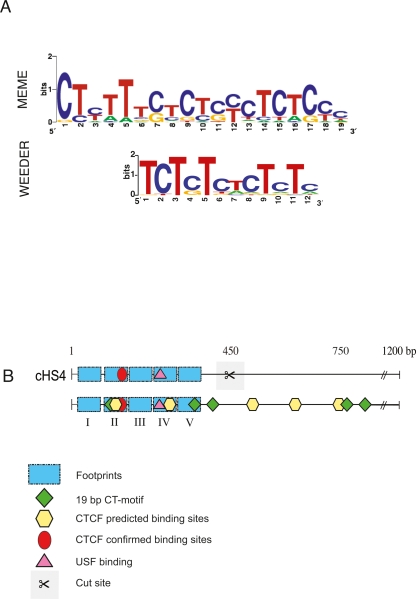
Given the abundant appearance of silencers and EBs in our assay, we searched for the presence of novel DNA motifs using the programs MEME (Multiple Em for Motif Elicitation) (Bailey et al. 2006) and Weeder (Pavesi et al. 2004). MEME uses a gapless, local, multiple sequence alignment to search for statistically significant motifs in the input set compared to a random background set. In contrast, Weeder uses a consensus-based method that allows substitutions to enumerate enriched sequences compared to a random background set. The strong silencer group (Fig. 4A) was selected for analysis because it contained a functionally comparable collection of NREs. A novel 19-bp pattern was identified that had an expectation value (E-value) of 1.9 × 10−2, indicating a very low likelihood of finding the same motif from a random set of sequences (Fig. 5A and tabulated in Fig. 4A). The motif was present in eight out of 10 strong SV40 silencers. The CT-rich motif was quite simple, yet captured variability at individual positions in the silencer sequences. An example of a simple, yet potent, functional motif includes CCNCNCCCN, bound by KLF1 (erythroid Krüppel-like factor, formerly known as EKLF) (Pilon et al. 2006). The Weeder analysis identified a 12-bp CT-rich motif in the same data set. The 12-bp motif had a nearly identical nucleotide-level signature as the MEME motif (Fig. 5A).
Figure 5.
Bioinformatic analyses of silencer sequences. (A) The sequence motif(s) identified in eight strong silencer regions from chromosome 7 and the cHS4 insulator element. The upper motif is detected using the MEME program and the lower motif is from the Weeder program. (B) Motifs detected in the 1.2-kb cHS4 insulator sequence. The upper panel represents functional annotations prior to our analysis. The lower panel implicates novel functional regions as a result of our analysis. Blue rectangles depict the five published footprint regions, green diamonds illustrate positions of the 19-bp CT-motifs, yellow hexagons correspond to newly predicted CTCF binding sites, the red circle denotes one characterized CTCF site, and the pink triangle represents a verified USF binding site. CTCF sites that intersect the 19-bp CT-motif (yellow and green icons, respectively) are within footprint region FII and at the position marked at 750 bp. The scissors indicate the site of the truncation analysis presented in Figure 2.
To evaluate the motif predictions, the 32 non-NRE sequences that were unable to function as strong silencers or EBs were examined in the same way as the silencer set. Neither a significant motif of equivalent size (10 bp or greater) nor any E-values with a likelihood <1 was produced. Additionally, the set of seven strong EBs did not show significant enrichment for any motif.
The inclusion of the cHS4 insulator sequence in the analysis of strong silencers decreased the E-value of the 19-bp CT-motif to 1.9 × 10−6. Through this approach five copies of the CT-motif were identified in the cHS4 region (Fig. 5B, green diamonds, lower panel). Two of these motifs coincided with the footprint regions of cHS4 (Fig. 5B, blue rectangles, lower panel), implicating function through in vivo evidence. One of these motifs overlapped the verified CTCF site in footprint FII (red circle). One motif outside the footprint regions again overlapped a CTCF site we predicted at position 750 bp. In all, we predicted four novel CTCF binding sites by their sequence identity to a CTCF binding-motif database (http://www.essex.ac.uk/bs/molonc/spa.htm) (yellow hexagons). Notably, these predicted motifs provided an explanation for the partial silencing activity detected in our deletion analysis of cHS4 (Fig. 2).
Experimental assessment of the CT-motif
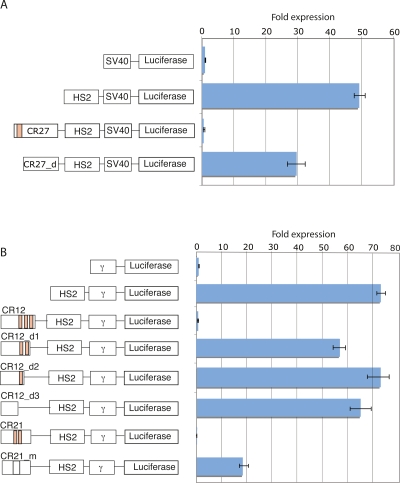
The presence of the CT-motif in multiple silencer regions provided an opportunity to test the functional consequences of deleting or altering these sequences. Three of the strong silencer regions contained two or more copies of the 19-bp CT-motif (tabulated in Fig. 4A). Multiple occurrences implied the possibility of additive or redundant contributions from each motif. We examined motifs from three regions: CR12, 21, and 27. A deletion of the CR27 region (CR27_d) removed the terminal end of the sequence and did not change the spacing of any of the remaining elements in the construct. Deletion of 47 bases containing the motif reduced the silencing function from the previous 80-fold repression to a smaller 1.6-fold repression (Fig. 6A). CR12 and CR21 silenced expression with the gamma-globin promoter. A deletion analysis removed three copies of the CT-motif sequentially from CR12 (Fig. 6B). Removal of the outermost motif (CR12_d1) reinstated 80% of the expression level. The remaining 20% of the silencing was counteracted after removal of the second CT-motif (CR12_d2). The third motif (CR12_d3) did not contribute to silencing on its own, conferring expression that was equivalent to the second deletion. Region CR21 contained tandem copies of the CT-motif in an internal location. A 41-bp sequence from a nonsilencer element was used to replace these motifs collectively. A 19.5-fold increase in luciferase expression resulted after replacement of the CT-motif (CR21_m) (Fig. 6B).
Figure 6.
Deletion of the 19-bp CT-motif from three silencer elements. The components of each plasmid are shown to the left of the expression data. (A) A series of plasmids utilizing the SV40 promoter, HS2 enhancer, and CR27 silencer are shown. The 19-bp CT-motif is shown in the pink box. (B) Transient transfection results showing the effect of deletions or replacement of the CT-motif in gamma-globin silencing vectors. Three copies of the 19-bp CT-motif are sequentially removed from region CR12 by a deletion analysis. The internal location of two motifs in CR21 (light orange boxes) was replaced simultaneously by a neutral sequence from a nonsilencing element (white box).
Comparison to CTCF binding sites and other genomic data
The connection between CTCF and EB function is well established in the literature. We sought to use available data about the location of CTCF binding in the genome to assess the possibility of CTCF involvement in our silencer and EB phenotypes. Experimental evidence showed that the CTCF protein could bind several of the silencer sequences. For example, the CR27 element, which acted as a strong silencer and weak EB, was bound by CTCF in ChIP-chip assays (Fig. 4A; Kim et al. 2007). Complementary evidence of CTCF occupancy at the silencers and EBs came from precisely mapped CTCF binding sites retrieved by the ChIP-seq technique (Barski et al. 2007). Four NREs have this type of CTCF evidence. A few CTCF sites localized to positions with no activity or weak activity in our assay (CR17, NR11, NR15, and NR18). The remaining silencers and EBs showed no evidence for CTCF binding from any surveys of the community-wide, high-throughput ChIP data. In two of the strong silencer sequences, CR12 and NR1, the CTCF binding-motif coincided with the 19-bp CT-motif. Furthermore, CT-motifs in NR1, CR27, and CR21 also colocalized with the CTCF evidence.
Additional considerations
As a test of the ability of randomly selected regions to act as NREs, we randomly cloned 17 regions averaging 500 bp in length from Escherichia coli. Two of the E. coli clones acted as strong silencers. No functional information was available for these sequences; however, one of them contained a match to the 19-bp CT-motif. A search of the E. coli genomic sequence identified 16 contigs that contained at least three copies of the CT-motif, 52 occurrences in total. Although the function of this motif will differ in E. coli and human sequences, it is clearly discernable in the prokaryotic genome. Thus a random selection of DNA sequences captured one of them. Three other randomly cloned E. coli sequences functioned as EBs; however, when pooled together with the set of seven strong EB sequences from this analysis, did not identify an enriched motif. We do not yet know what human proteins are capable of binding these bacterial sequences and therefore cannot discount them as being false positives.
We further examined the expression data generated from the human sequences for additional functions such as enhancer activity, when no repressive activity was present. However, due to the presence of the HS2 enhancer in all of the plasmids, the assay did not show strong evidence for enhancer function. Very few elements increased expression more than twofold above the enhancer-control plasmid (as indicated in Fig. 3B,C). We concluded that the presence of the HS2 enhancer might define the upper limit of expression achievable in these cells, precluding increased luciferase signals. Conversely, these cloned sequences might not interact additively with the HS2 enhancer.
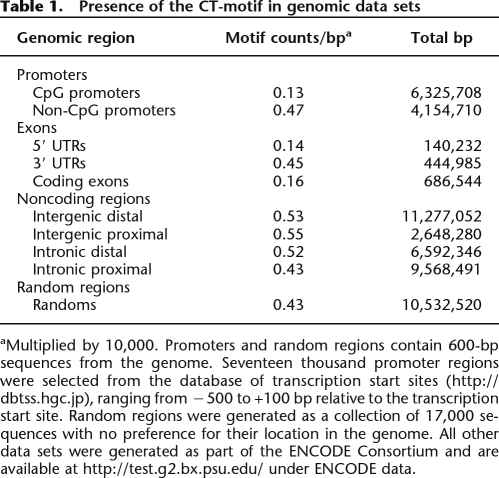
As a test of the location of the CT-motif in the genome, we examined several data sets representing genomic regions. A collection of 17,000 promoter regions was divided into CpG islands and non-CpG islands. Furthermore, exonic regions were separated into coding, 5′ untranslated regions (UTRs) and 3′ UTRs. Noncoding regions representing intergenic and intronic elements were categorized based on distance to the nearest exon, either proximal or distal (i.e., < or >5 kb, respectively). In comparison, a set of 17,000 random sequences was generated with no knowledge of the original positions in the genome. CT-motifs were mapped in each data set and the total counts were normalized to the number of nucleotides in that data set. No significant enrichment was recorded for any of the categories examined. However, CpG island promoters showed a threefold depletion of the CT-motif compared to non-CpG islands (Table 1). Furthermore, 5′ UTRs and coding regions also had reduced levels of the motif.
Table 1.
Presence of the CT-motif in genomic data sets
aMultiplied by 10,000. Promoters and random regions contain 600-bp sequences from the genome. Seventeen thousand promoter regions were selected from the database of transcription start sites (http://dbtss.hgc.jp), ranging from −500 to +100 bp relative to the transcription start site. Random regions were generated as a collection of 17,000 sequences with no preference for their location in the genome. All other data sets were generated as part of the ENCODE Consortium and are available at http://test.g2.bx.psu.edu/ under ENCODE data.
Discussion
These data show that silencer and EB functions are present at high frequencies in the 1.8-Mb region encompassing the CFTR gene. We have succeeded in detecting these negative regulatory elements using a transient transfection assay, which can easily scale to accommodate an even greater throughput in the future. Using this assay, we showed that strong silencer elements contained a 19-bp CT-motif. Deletion of the 19-bp motifs from three of the silencers significantly reduced the ability of the elements to silence gene expression, though it did not eliminate the silencer function in all cases. Clearly, multiple sites can work in combination to confer the fully silenced phenotype. Results from the cHS4 deletion series also supported this conclusion, where the removal of either half of the 1.2-kb cHS4 insulator sequence showed residual, albeit partial, silencing. Conversely, the characterized CTCF binding site in the cHS4 FII footprint region contributed none of the silencing when tested alone. The identification of five copies of the 19-bp CT-motif in the cHS4 insulator, coupled with our deletion data, provided definitive evidence that sequences outside the cHS4 core contribute to its silencing activity. We also predicted the presence of additional copies of CTCF motifs outside the core region. Our data support the idea that the individual components of the full-length insulator element act together to create a powerful silencer. As a compound insulator element, cHS4 represents the best-characterized negative regulatory element to date; yet new features continue to emerge regarding its functional composition.
Although predictive approaches have not been developed for NREs, functional assays can provide a large set of confirmed examples. We have developed a highly efficient assay to determine if a silencer or EB is present in DNA. Changing the parameters of the assay revealed the specialization of the function: The orientation of the sequence, the identity of the promoter, and the cellular environment all influenced the transcriptional output from the plasmid in unique ways. These responses indicate that silencer function can be modified to respond to changing conditions in the cell. Furthermore, directionality is a known feature of insulator elements (Gerasimova et al. 1995). The dual nature of some elements, acting as both silencers and EBs, is illustrated dramatically through our assay system. If they also function as barrier elements, these sequences would make exciting candidates for use in gene therapy vectors. This analysis also clearly proves that sequences recognized by CTCF inhibit gene expression in some situations but are not active under all experimental conditions. As is typical of large-scale enhancer analyses, we use promoters in an artificial combination with cis-acting regulatory elements, for economy of scale. In the case of enhancers, artificial combinations have proven effective and quite often recapitulate the exact developmental expression patterns seen in the embryo (Pennacchio et al. 2006). Here, we present data on 15 novel NRE regions. Although the approach could be scaled to examine a larger portion of the human genome, analyses of 1% of the genome may provide enough insight into critical NRE attributes to enable predictive approaches. This experimental assay would then support validation efforts.
The motif identified by the MEME analysis does not closely match the recently published CTCF consensus from Kim et al. (2007) or Xie et al. (2007). Nevertheless, our motif incorporates an optional CCCTC pattern, for which CTCF was named. Overall, the 19-bp pattern seems extremely simple, generalizing to an enriched CT-motif. Similar patterns have been recorded previously for silencer elements (Ogbourne and Antalis 1998). Additional supporting evidence from ChIP data corroborates that CTCF binds some of our verified silencer and EB regions from chromosome 7. However, the differences between cell lines make it impossible to confirm CTCF occupancy in our cell lines using data obtained in CD4+ cells (Barski et al. 2007). Furthermore, the sites with no evidence for CTCF binding may elicit their silencing effect through a protein that recruits CTCF or through a protein other than CTCF. Persuasive evidence for the existence of additional proteins that function as silencers and EBs comes from other species. For instance, GAGA and the paired scs/scs′ elements also perform these activities in Drosophila (Kellum and Schedl 1992; Ohtsuki and Levine 1998).
If CTCF is functioning at NRE sites, then the high frequency of silencers and EBs identified in our assay is entirely plausible, given the prevalence of CTCF binding sites recently identified in the human genome. The functional variability of individual elements indicates that not all sites convey a uniform response. This conclusion is reinforced by experimental data showing that CTCF can act as either a silencer or an activator (Reik and Murrell 2000), suggesting a modulatory role. We noted that the genomic landscape of the NRE elements resembles that of active regions, containing histone H3 and H4 acetylation and H3K4tri-methylation. Furthermore, the CT-motifs appear more frequently in promoters that require modulation across different cell types (tissue restricted) than in promoters that have CpG islands and may be widely expressed. Our assay provides one approach to studying silencer and EB functions of both ubiquitously acting and condition-specific elements. We are currently testing these same expression vectors in stable transfection assays to ascertain the role of chromatin in these silencer and EB regions.
Methods
DNA expression constructs
Candidate regions were cloned into plasmid DNA using the Invitrogen Gateway System. Eight cloning vectors containing either the SV40 or the human (G)gamma-globin promoter were designed using the PGL3-basic expression plasmid of Promega. Gateway recombination sites were created upstream or downstream of the core human beta-globin HS2 enhancer, in either the forward or reverse configuration.
Transfection of cells and measurement of expression
To assess transient expression, 4 × 105 K562 cells and 6 × 104 HeLa or 293T cells were transfected with 0.4 μg of test DNA and 4.0 or 40 ng of pRL-Tk Renilla plasmid for lipofection (using the reagent TFX-50) or electroporation (using the Amaxa 96-well nucleofector II). Both approaches used Renilla luciferase as a control for transfection efficiency. Luciferase expression was measured in a 96-well plate format with detection of fluorescence using the dual luciferase “Stop and Glo” procedure from Promega. Measurements were recorded on a Berthold plate-reader luminometer. The average expression level from three replicate transfections was normalized to the Renilla luciferase cotransfection control. This value was further normalized to the average expression level from three normalized replicates of the promoter-only plasmid to yield a “fold” enhancement measurement (Elnitski et al. 2001). The standard deviation on the averages is plotted as the value of the error bars. Upon producing a silencing phenotype, each construct was resequenced to confirm the integrity of the plasmid.
Acknowledgments
We thank Ann Dean, David Bodine, and Elliott Margulies for suggestions, comments, and materials; and Ross Hardison for support in the earliest stages of this project. Anonymous reviewers provided helpful suggestions toward the final presentation of materials. L.E. is supported by the Intramural Research Program of the National Human Genome Research Institute, US National Institutes of Health.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073817.107.
References
- Bailey T.L., Williams N., Misleh C., Li W.W., Williams N., Misleh C., Li W.W., Misleh C., Li W.W., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Wang Z., Wei G., Chepelev I., Zhao K., Wei G., Chepelev I., Zhao K., Chepelev I., Zhao K., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Recillas-Targa F., Simpson M., West A., Felsenfeld G., Simpson M., West A., Felsenfeld G., West A., Felsenfeld G., Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. 2002;99 (Suppl. 4):16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Mack J., Hall R., Goldy J., Sabo P.J., Hall R., Goldy J., Sabo P.J., Goldy J., Sabo P.J., Sabo P.J. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Miller W., Hardison R., Miller W., Hardison R., Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the β-globin locus control region. Role of basic helix-loop-helix proteins. J. Biol. Chem. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Li J., Noguchi C.T., Miller W., Hardison R., Li J., Noguchi C.T., Miller W., Hardison R., Noguchi C.T., Miller W., Hardison R., Miller W., Hardison R., Hardison R. A negative cis-element regulates the level of enhancement by hypersensitive site 2 of the β-globin locus control region. J. Biol. Chem. 2001;276:6289–6298. doi: 10.1074/jbc.M009624200. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M., Felsenfeld G., Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Gdula D.A., Gerasimov D.V., Simonova O., Corces V.G., Gdula D.A., Gerasimov D.V., Simonova O., Corces V.G., Gerasimov D.V., Simonova O., Corces V.G., Simonova O., Corces V.G., Corces V.G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D., McDaniell R.M., Iyer V.R., Lieb J.D., Iyer V.R., Lieb J.D., Lieb J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P., Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Zhang M.Q., Lobanenkov V.V., Ren B., Lobanenkov V.V., Ren B., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C.M., Andrews R.M., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Andrews R.M., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Beare D.M., Fowler J.C., Couttet P., Fowler J.C., Couttet P., Couttet P. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., NISC Comparative Sequencing Program. Haussler D., Green E.D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbourne S., Antalis T.M., Antalis T.M. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem. J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes & Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesole G., Mereghetti P., Mauri G., Pesole G., Mauri G., Pesole G., Pesole G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Dubchak I., Holt A., Lewis K.D., Holt A., Lewis K.D., Lewis K.D. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pilon A.M., Nilson D.G., Zhou D., Sangerman J., Townes T.M., Bodine D.M., Gallagher P.G., Nilson D.G., Zhou D., Sangerman J., Townes T.M., Bodine D.M., Gallagher P.G., Zhou D., Sangerman J., Townes T.M., Bodine D.M., Gallagher P.G., Sangerman J., Townes T.M., Bodine D.M., Gallagher P.G., Townes T.M., Bodine D.M., Gallagher P.G., Bodine D.M., Gallagher P.G., Gallagher P.G. Alterations in expression and chromatin configuration of the alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol. Cell. Biol. 2006;26:4368–4377. doi: 10.1128/MCB.02216-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F., Bell A.C., Felsenfeld G., Bell A.C., Felsenfeld G., Felsenfeld G. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc. Natl. Acad. Sci. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F., Pikaart M.J., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G., Pikaart M.J., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G., Litt M.D., West A.G., Gaszner M., Felsenfeld G., West A.G., Gaszner M., Felsenfeld G., Gaszner M., Felsenfeld G., Felsenfeld G. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl. Acad. Sci. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Murrell A., Murrell A. Genomic imprinting. Silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Bell A.C., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G., Bell A.C., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G., West A.G., Simpson M., Pikaart M., Felsenfeld G., Simpson M., Pikaart M., Felsenfeld G., Pikaart M., Felsenfeld G., Felsenfeld G. Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot D., Grosveld F., Grosveld F. The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 1991;10:1391–1398. doi: 10.1002/j.1460-2075.1991.tb07659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Lindblad-Toh K., Kellis M., Lander E.S., Kellis M., Lander E.S., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]