Abstract
Macrosatellite DNA is composed of large repeat units, arranged in tandem over hundreds of kilobases. The macrosatellite repeat DXZ4, localized at Xq23-24, consists of 50–100 copies of a CpG-rich 3-kb monomer. Here I report that on the active X chromosome (Xa), DXZ4 is organized into constitutive heterochromatin characterized by a highly organized pattern of H3K9me3. DXZ4 is expressed from both strands and generates an antisense transcript that is processed into small RNAs that directly correlate with H3K9me3 nucleosomes. In contrast, on the inactive X chromosome (Xi) a proportion of DXZ4 is packaged into euchromatin characterized by H3K4me2 and H3K9Ac. The Xi copy of DXZ4 is bound by the chromatin insulator, CTCF, within a sequence that unidirectionally blocks enhancer–promoter communication. Immediately adjacent to the CTCF-binding site is a bidirectional promoter that, like the sequence flanking the CTCF-binding region, is completely devoid of CpG methylation on the Xi. As on the Xa, both strands are expressed, but longer antisense transcripts can be detected in addition to the processed small RNAs. The euchromatic organization of DXZ4 on the otherwise heterochromatic Xi, its binding of CTCF, and its function as a unidirectional insulator suggest that this macrosatellite has acquired a novel function unique to the process of X chromosome inactivation.
Approximately half of the human genome consists of repetitive DNA (Schmid and Deininger 1975), a small fraction of which is defined as satellite DNA (Lander et al. 2001). Satellite DNAs are tandemly repeated sequences that are a major component of constitutive heterochromatin (Miklos and John 1979). The most extensive examples of human satellite DNA include the alphoid arrays that define active centromeres (Schueler et al. 2001) and the pericentromeric classical satellite I, II, and III repeats found on most chromosomes (Lee et al. 1997). In addition to these, several members of a novel family of human tandem repeats have emerged that have tentatively been termed macrosatellites or megasatellites. These include DXZ4 on Xq23-24 (Giacalone et al. 1992), RS447 primarily on 4p15 (Kogi et al. 1997), and D4Z4 on 4q35 (Van Deutekom et al. 1993; Hewitt et al. 1994). While the sequences themselves are unrelated, these macrosatellites share several features, including high GC content and the extensive size of each repeat monomer; 3 kb for DXZ4 (Giacalone et al. 1992), 4.7 kb for RS447 (Kogi et al. 1997), and 3.3 kb for D4Z4 (Van Deutekom et al. 1993; Hewitt et al. 1994). Each array can include as many as 100 copies of a monomer, although the precise number of copies is highly polymorphic in the general population (Giacalone et al. 1992; Van Deutekom et al. 1993; Hewitt et al. 1994; Kogi et al. 1997; Gondo et al. 1998). The function of macrosatellites is unclear, although RS447 contains an open reading frame coding for a novel deubiquitinating enzyme (Saitoh et al. 2000). Although RS447 shows significant meiotic instability (Okada et al. 2002), no biological significance for low or high copy number has been determined. However, contraction in the size of the D4Z4 array is associated with facioscapulohumeral muscular dystrophy (Wijmenga et al. 1992), indicating that these sequences are not simply “junk” DNA (Ohno 1972), but are integral to genome function.
By virtue of its location on the X chromosome, DXZ4 is exposed to a biological phenomena not experienced by autosomal macrosatellites. Early during development, female embryos undergo X inactivation to balance X-linked gene dosage between the sexes (Lyon 1961). This process essentially renders one of the two X chromosomes genetically silent by the chosen inactive X chromosome (Xi) being repackaged into facultative heterochromatin (Heard and Disteche 2006). Giacalone et al. (1992) made the unexpected observation that DXZ4 CpGs on the Xi were hypomethylated. Our interest in DXZ4 came about through investigation of the Xi chromatin organization. Markers of euchromatin are generally absent from the dosage compensated regions of the Xi, with one notable exception. A single region on Xq is highlighted by histone H3 dimethylated at lysine-4 (H3K4me2) (Boggs et al. 2002). This region resides at the distal edge of an extensive macroH2A band and is inseparable from DXZ4 by FISH (Chadwick and Willard 2002). At interphase the signal is retained and appears as a single distinct foci within the territory of the hypo-H3K4me2 Xi (Boggs et al. 2002; Chadwick and Willard 2003). This pattern is shared by several other covalent histone modifications and chromatin proteins, including the CCCTC-binding factor CTCF (Chadwick and Willard 2003). CTCF is a multifunctional DNA-binding protein involved in transcription regulation, chromatin insulation, and chromatin organization (for review, see Filippova 2008 and references therein). Furthermore, CTCF has been found to be an important factor not only in the choice of the future Xi (Chao et al. 2002; Boumil et al. 2006; Donohoe et al. 2007; Xu et al. 2007) but also in regulating regions of the Xi from which genes escape X-inactivation (Filippova et al. 2005).
The present study sought to confirm that DXZ4 is the site of H3K4me2 and CTCF binding, and if this arrangement is unique to the Xi, what potential gain of function this alternate chromatin organization provides.
Results
DXZ4 on the inactive X chromosome is packaged into euchromatin; a macrosatellite organization unique to X chromosome inactivation
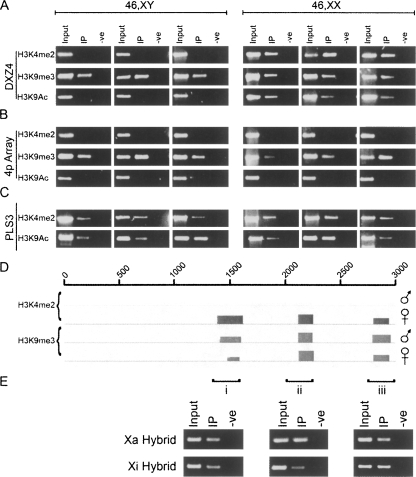
To confirm that DXZ4 is the physical site of H3K4me2 on Xq23-24, anti-H3K4me2 chromatin immunoprecipitation (ChIP) was performed on several independent male and female samples. As can be seen in Figure 1A (top row), DXZ4 is positive for H3K4me2, but only in females, indicating that the interphase Xi signal of H3K4me2 (Supplemental Fig. 1) is a Xi-specific feature of DXZ4 chromatin.
Figure 1.
DXZ4 on the inactive X chromosome adopts a Xi-specific euchromatin-like conformation. (A) Ethidium bromide (EtBr) agarose gels of H3K4me2, H3K9me3, and H3K9Ac ChIP assessed by PCR for co-immunoprecipitation of DNA from the Xq23 macrosatellite DXZ4, and (B) an unrelated macrosatellite on chromosome 4p (4p array). Results are shown for three independent primary male and female cell lines. (C) PCR analysis of the plastin 3 promoter (PLS3). Samples include the input, experimental (IP), and control (-ve). (D) Close-up of ChIP-chip data for a single 3-kb DXZ4 monomer (0–3000 bp) with H3K4me2 and H3K9me3 in a male and female sample. The coordinates at the top of the image represent nucleotides 114,785,064–114,788,023 of Human Genome Build HG17. Immediately below the line is the NimbleGen Signal Map 1.8 output for the H3K4me2 and H3K9me3 ChIP-chip. The width of the signal indicates the extent of the hybridization. (E) H3K9me3 ChIP from the Xa and Xi hybrid assessed by PCR. The three regions analyzed by PCR are indicated by i–iii and correspond to the sites of the H3K9me3 peaks defined by ChIP-chip.
Typically satellite sequences and repeats are organized into constitutive heterochromatin characterized by H3K9me3 (Martens et al. 2005). ChIP analysis was repeated with anti-H3K9me3 and readily detected at DXZ4 in both male and female samples (Fig. 1A, middle row). To further support the Xi-specific euchromatin organization of DXZ4, ChIP was performed with anti-H3K9Ac. As with H3K4me2, IP of DXZ4 with anti-H3K9Ac was female specific (Fig. 1A, bottom row).
To determine if the contrasting chromatin organization of the Xa and Xi alleles of DXZ4 is unique to the X chromosome and X inactivation, the chromatin organization of the autosomal macrosatellite RS447 (Kogi et al. 1997) on chromosome 4p was investigated. The choice of the 4p-array was based upon its having a similar copy number, genomic organization, and CpG content to DXZ4 (Kogi et al. 1997). Investigation of the 4p-array in the same samples clearly shows the presence of H3K9me3, but the lack of H3K4me2 and H3K9Ac in males and females, in contrast to what was observed at DXZ4 (Figure 1, cf. A and B). To rule out that the absence of H3K4me2 and H3K9Ac signals in the male samples was not due to poor ChIP, the promoter of the transcriptionally active plastin 3 gene was assessed and shown to be positive in all samples (Fig. 1C).
In order to investigate at higher resolution the pattern of H3K4me2 and H3K9me3 at DXZ4, ChIP was performed combined with custom microarray hybridization. Figure 1D shows data for a single complete 3-kb DXZ4 monomer. Peaks obtained for the 1.5-Mb genomic interval flanking DXZ4, and a close-up of the DXZ4 array are shown in the Supplemental material (Supplemental Figs. 2, 3). ChIP-chip identified three specific regions of DXZ4 associated with H3K4me2, each peak spanned 100–241 bp. Consistent with the ChIP-PCR analysis described above, H3K4me2 signals were female specific. DNA immunoprecipitated with anti-H3K9me3 hybridized to exactly the same three regions of DXZ4 as H3K4me2 for the males and female samples, each spanning 120–150 bp (Fig. 1D). The most likely interpretation of the H3K9me3 data is that DXZ4 in males and on the Xa in females is organized into constitutive heterochromatin characterized by H3K9me3. However, it remains a possibility that some of the 50–100 Xi copies of DXZ4 are also organized into H3K9me3 heterochromatin. To evaluate this possibility, H3K9me3 ChIP was performed on somatic cell hybrids containing either a Xa or Xi. Figure 1E clearly indicates that H3K9me3 is detected at DXZ4 on the Xa and Xi. How the H3K9me3 defined monomers spatially relate to the H3K4me2 monomers within the context of the full macrosatellite array remains to be determined.
Long DXZ4 transcripts originate from the sense strand only on the Xa and both strands from the Xi
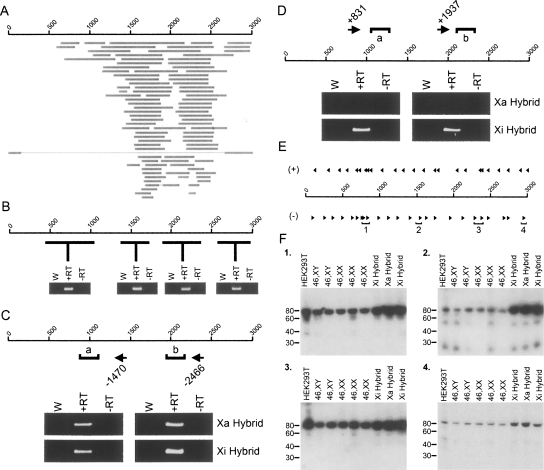
A comparison of DXZ4 DNA sequence against entries in the expressed sequence tags (ESTs) database revealed numerous matches suggesting that DXZ4 is expressed (Fig. 2A). Furthermore, comparative genome-wide microarray hybridization with nuclear and cytoplasmic RNA indicated that DXZ4 RNA is almost exclusively nuclear (Kapranov et al. 2007). Transcription of DXZ4 was confirmed by RT-PCR (Fig. 2B). Strand-specific RT-PCR revealed that DXZ4 is transcribed from the sense strand only in males, but both sense and antisense strands in females (Supplemental Fig. 4). By use of RNA isolated from somatic cell hybrids containing either a Xa or Xi, it was possible to demonstrate that sense transcripts originate from both the Xa and Xi (Fig. 2C), whereas an antisense transcript originates specifically from the Xi (Fig. 2D).
Figure 2.
A long DXZ4 sense transcript and antisense small RNAs are common to the Xa and Xi arrays, whereas an antisense transcript is specific to the Xi. (A) Location of human ESTs with >94% sequence match to DXZ4. The scale of 1–3000 represents the 3-kb coordinates of a single DXZ4 monomer in bp. Horizontal gray bars represent individual EST matches. (B) A single 3-kb DXZ4 monomer is represented by a 1–3000 bp scale. Below the scale are the results of a selection of RT-PCR analyses on random-hexamer primed cDNA (i.e. strand-independent). The approximate region of DXZ4 PCR amplified is represented by the horizontal black lines. Each RT-PCR shows water control (W), with reverse transcriptase (+RT) and a no reverse transcriptase control (−RT). (C) Strand-specific RT-PCR analysis of DXZ4 RNA in Xa and Xi hybrids for the sense transcript. The 1–3000 bp scale represents a single 3-kb DXZ4 monomer. The location and starting coordinate of antisense or sense priming oligonucleotides are indicated by the left- and right-pointing arrows, respectively. PCR for each strand-specific RT reaction “a” (980–1221) and “b” (2106–2358) are indicated. Ethidium bromide gel images of water control (W), with reverse transcriptase (+RT) or without (−RT) are shown. (D) Strand-specific RT-PCR analysis of DXZ4 RNA in Xa and Xi hybrids for the antisense transcript. (E) The approximate location of 19–21 bp oligonucleotides hybridized to Northern blots are shown above and below DXZ4 for sense (+) and antisense (−) transcript detection, respectively. Small RNA hybridizing oligonucleotides are highlighted by regions 1–4. (F) Northern blots for oligonucleotides from regions 1–4. The size in bp is shown to the left. An example of an ethidium bromide stained polyacrylamide gel showing the small RNAs can be seen in Supplemental Figure 5A.
Small antisense RNA molecules originate from four specific regions of DXZ4
The detection of DXZ4 transcripts from both strands suggests that double-stranded DXZ4 RNA could form and be processed into small RNAs (Zaratiegui et al. 2007). Small RNA molecules were isolated from several male and female cell lines as well as Xi and Xa containing hybrids. A total of 50 labeled oligonucleotides (25 each specific to either sense or antisense DXZ4) (Fig. 2E), were hybridized to Northern blots of the small RNAs. Antisense-specific oligonucleotides from four specific regions of DXZ4 detected an ∼85-nucleotide (nt) RNA (Fig. 2F). (Positive and negative examples for sense and antisense oligonucleotides can be seen in Supplemental Fig. 5B.) Some oligonucleotides detected an additional series of smaller RNA species of ∼50 nt and 25 nt along with the 85-nt RNA. These are unlikely to be degradation products, as not all oligonucleotides from the same DXZ4 interval (i.e., region-1) could detect them. Rather, it is more likely that these represent further processed versions of the 85-nt RNA; some of the oligonucleotide probes overlap with the end product smaller species and hence can detect them. Two other intriguing observations can be made from the Northern data. First, the small antisense RNAs are detected from both the Xa and Xi. Second, the site of origin for three of the four small RNAs corresponds precisely to the H3K9me3 and H3K4me2 peaks detected by ChIP-chip (Fig. 1D).
Identification of a bidirectional promoter in DXZ4
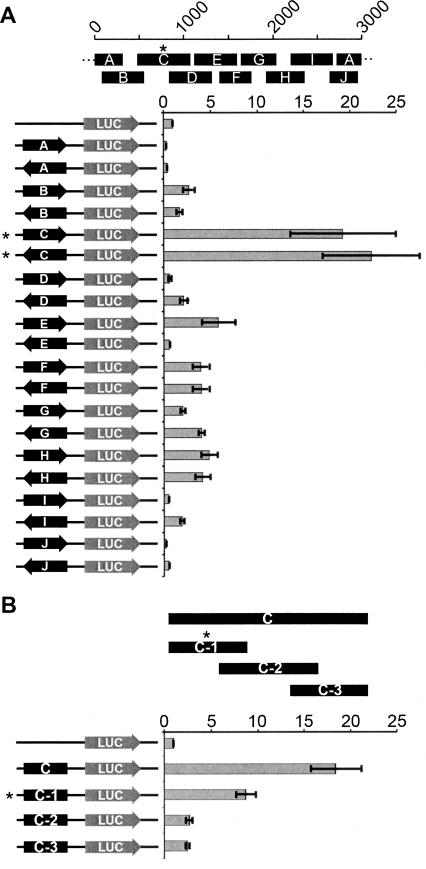
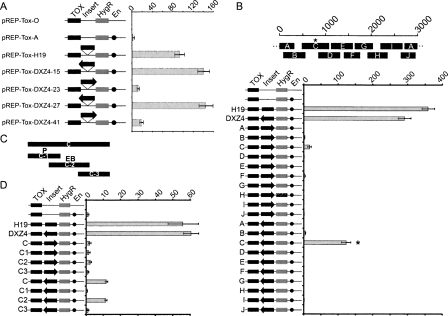
To understand what sequences were driving the expression of the larger transcripts and the small RNAs, DXZ4 was investigated for intrinsic promoter activity. Ten overlapping DXZ4 fragments encompassing a full monomer were cloned in either orientation upstream of a luciferase reporter gene. Strong promoter activity was detected in one fragment, regardless of orientation (Fig. 3A). To narrow down the region containing promoter activity, the fragment was divided into three overlapping subfragments, and most activity was retained in fragment C-1 (Fig. 3B). Independent ChIP-chip data immunoprecipitated DNA from fragment C-1 using antibodies specific to the general transcription factor TAF1 (Kim et al. 2005), supporting the hypothesis that fragment C-1 acts as a promoter.
Figure 3.
Identification of a bidirectional promoter in DXZ4. (A) A single 3-kb DXZ4 monomer is represented by 0–3000 bp scale. The approximate location of subfragments A–J are shown below the monomer. DXZ4 subfragments cloned upstream of the Luciferase reporter gene in pGL3-Basic are shown to the left. The orientation of the fragment is represented by the arrow. The level of luciferase activity detected relative to that of the empty vector (assigned a value of 1) is indicated (0–25×). (B) DXZ4 subfragment C is represented by the black solid bar, below which are shown the relative locations of the three subregions. Below are shown the five different pGL3-Basic constructs tested for luciferase activity. All data (A,B) represent the mean and standard error for four independent experiments. The asterisk highlights the subregion fragment that retained the most promoter activity.
The insulator protein CTCF binds DXZ4 in vitro and specifically associates with DXZ4 on the inactive X chromosome in vivo
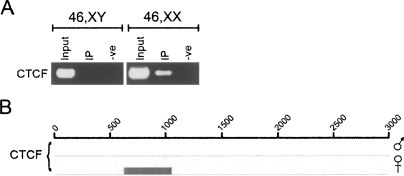
As is the case with H3K4me2, the insulator protein CTCF is underrepresented at the Xi, with the exception of a single intense signal (Supplemental Fig. 1; Chadwick and Willard, 2003). ChIP was performed to determine if CTCF is directly associated with DXZ4, as opposed to sequences immediately adjacent to the macrosatellite. The result clearly indicated that anti-CTCF antibodies could co-immunoprecipitate DXZ4 DNA, but only in females (Fig. 4A). The most likely interpretation is that CTCF associates specifically with DXZ4 on the Xi. In order to determine more specifically what sequence within DXZ4 is bound by CTCF, two complementary approaches were used: ChIP-chip and electrophoresis mobility shift assays (EMSA).
Figure 4.
CTCF binds to a single region of DXZ4 on the Xi only. (A) ChIP with CTCF co-immunoprecipitates DXZ4 DNA in females only. The samples shown are one example of four independent male and female cell line ChIP experiments that showed the same result. (B) Close-up of ChIP-chip data for a single 3-kb DXZ4 monomer (0–3000 bp) with CTCF in a male and female sample. The coordinates at the top of the image represent nucleotides 114,785,064–114,788,023 of Human Genome Build HG17. Immediately below the line is the NimbleGen Signal Map 1.8 output for the CTCF ChIP-chip. The width of the signal indicates the extent of the hybridization.
CTCF ChIP was performed and hybridized to the same microarray as described above for H3K4me2 and H3K9me3. Consistent with recent genome-wide analysis of CTCF binding (Kim et al. 2007), numerous CTCF-binding sites were identified throughout the region flanking DXZ4 (Supplemental Fig. 2), many of which overlapped (data not shown). Within the DXZ4 array a single hybridizing peak per monomer (Supplemental Fig. 3) in both female but not the male sample. Data showing the single hybridizing female specific peak within a single monomer are shown in Figure 4B. Two interesting correlations can be made from these data. First, the region bound by CTCF corresponds to the site of origin of the small antisense RNA that does not overlap with the H3K4me2 and H3K9me3 peaks (Fig. 2F, group-1). Second, the region bound by CTCF is immediately adjacent to the bidirectional promoter within DXZ4 (Fig. 3B).
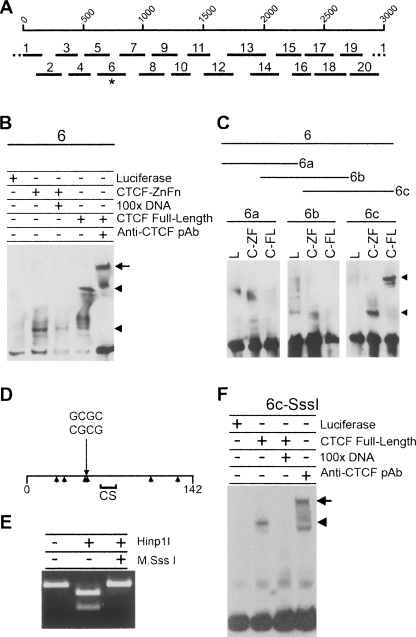
For EMSA analyses, the 3-kb DXZ4 sequence was subdivided into 20 overlapping fragments that were incubated with the in vitro translated 11 zinc finger DNA-binding domain of CTCF (CTCF-ZnFn) (Klenova et al. 1993; Filippova et al. 1996). Only fragment-6 (F6) was bound by CTCF-ZnFn (Fig. 5A; Supplemental Fig. 6), and the specificity of the binding was confirmed by the significantly reduced shift in the presence of a 100-fold excess of unlabeled F6 (Fig. 5B). Furthermore, full-length in vitro translated CTCF bound to F6, and the shift in migration was further impeded in the presence of anti-CTCF polyclonal antisera (Fig. 5B). In order to refine the CTCF-binding site, F6 was divided into three overlapping fragments, of which only the 142-bp F6c was positive by EMSA (Fig. 5C). This could be further reduced to 84 bp without impairing CTCF binding (Supplemental Fig. 7). These data are consistent with the localization of CTCF observed by ChIP as the F6c sequence falls within the binding peak observed in the DXZ4 microarray analysis (Fig. 6B). Cytosine methylation at the dinucleotide CpG prevents binding of CTCF to CpG containing target sites (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000b; Filippova et al. 2001). F6c contains six CpG sites (Fig. 5D), yet complete CpG methylation of F6c in vitro (Fig. 5E) did not interfere with CTCF binding (Fig. 5F). The most likely reason for this is that the CTCF-binding motif within F6c does not contain a CpG. Comparison of the DNA sequence of F6c with the genome-wide human consensus sequence for CTCF binding (Kim et al. 2007), revealed a good match (AGTTCCTCT TGATGGCAGTA) that does not contain a CpG. This indicates that flanking CpG methylation does not prevent CTCF binding in vitro. The DNA sequence of F6c highlighting the minimal 84-bp EMSA fragment, Hinp1I recognition sequence, and putative CTCF binding site is shown in Supplemental Figure 8.
Figure 5.
CTCF binds to DXZ4 DNA in vitro independent of flanking CpG methylation. (A) A single 3-kb DXZ4 monomer is represented by 1–3000 bp scale, under which are shown the 20 different fragments used in the original EMSA analysis (see Supplemental Fig. 2). The asterisk indicates fragment-6, the mobility of which is retarded in the presence of CTCF protein. (B) Confirmation of CTCF specificity by 100× excess competition with unlabeled 6-DNA and supershift (upper arrowhead and arrow, respectively). (C) Refining CTCF to 6c only in the presence of the CTCF zinc fingers region (C-ZF) and full-length CTCF (C-FL) as indicated by the arrowheads, but not by luciferase (L). (D) Map of fragment 6c. The 142-bp fragment 6c is represented by the horizontal line, with the location of the Hinp1I site (GCGC) indicated. Arrowheads below the line indicate location of CpG sequences. The region denoted CS indicates a region with a good match to the CTCF genome-wide consensus sequence (Kim et al. 2007). (E) In vitro methylation of fragment 6c by M.SssI CpG methyltransferase blocks digestion by the methyl-sensitive Hinp1I restriction endonuclease. (F) CTCF binding to CpG methylated fragment 6c (Me-6c).
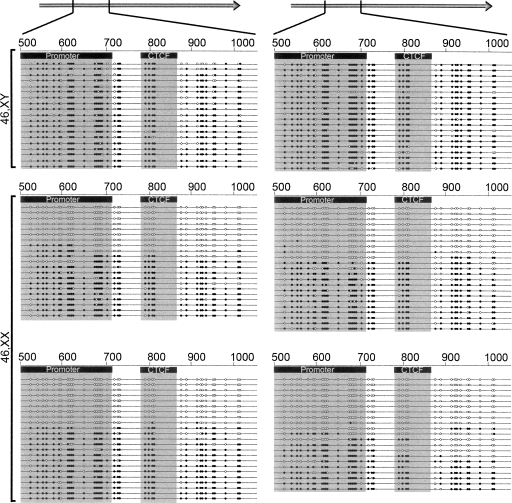
Figure 6.
CpG sequences at the promoter and CTCF-binding regions of DXZ4 are methylated on the active X but unmethylated on the inactive X chromosome. Gray arrows at the top of the image represent a single DXZ4 monomer, with the region under analysis highlighted (498–1033 bp). Below shows the CpG methylation status of the region for two independent male and four independent female cell lines. The location of the minimal promoter region and the smallest CTCF EMSA fragment are indicated by the black boxes and the gray shadow. Each line represents the sequence of an individual clone. The location of CpG sequences is indicated by the circles on the lines, with open circles indicating unmethylated and black filled circles as methylated CpGs.
CpG dinucleotides across the promoter and flanking the CTCF-binding site of DXZ4 are methylated on the Xa and unmethylated on the Xi
DXZ4 CpG dinucleotides on the Xi are hypomethylated, whereas those on the Xa are hypermethylated (Giacalone et al. 1992). CTCF associates specifically with the DXZ4 array on the Xi (Fig. 4), and in vivo plays a crucial role in maintaining the flanking DNA methylation-free (Schoenherr et al. 2003; Fedoriw et al. 2004; Pant et al. 2004; Filippova et al. 2005; Engel et al. 2006), which for DXZ4 would include the adjacent promoter (Fig. 3). Therefore it seemed appropriate to determine the extent of CpG methylation across this region. In males, most CpGs at the promoter, CTCF-binding region, and a few hundred base pairs distal are methylated (Fig. 6, top two panels). In contrast, the same interval was completely unmethylated in approximately half of all female clones (Fig. 6, bottom four panels), confirming that DXZ4 is differentially methylated on the Xa and Xi (Giacalone et al. 1992).
The CTCF-binding region of DXZ4 unidirectionally interferes with promoter–enhancer communication
Multiple functions have been attributed to CTCF (for recent review, see Filippova 2008)), including acting as a chromatin insulator (Bell et al. 1999; Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000b). To examine DXZ4 for insulator activity, DXZ4 monomers were cloned into an episomal insulator assay (Mukhopadhyay et al. 2004) and assessed for the ability to block promoter–enhancer communication. In the assay, DXZ4 does demonstrate characteristics of an insulator (Fig. 7A). However, the activity is almost entirely dependent upon the orientation of DXZ4, similar to other cis acting enhancer-blocking elements used in this assay (Kanduri et al. 2000a). To identify the specific sequences from which this activity originates, DXZ4 was divided into 10 overlapping fragments and assayed in both orientations. A single region of DXZ4 could reproducibly block enhancer–promoter communication effectively in one orientation (Fig. 7B). Encouragingly, this sequence corresponds precisely with the region of DXZ4 bound by CTCF in vivo and in vitro (Figs. 4, 5). However, this DXZ4 fragment also contains the bidirectional promoter activity (Fig. 3A). To ensure that the unidirectional enhancer–promoter blocking activity is not an artifact of promoter activity, the same three subfragments used to refine the promoter (Fig. 3B) were used in both orientations in the insulator assay (Fig. 7C). Reassuringly, the promoter–enhancer blocking activity was physically separated from the promoter activity (Fig. 7D).
Figure 7.
DXZ4 unidirectionally interferes with promoter–enhancer communication in vitro, an attribute confined to DNA encompassing the CTCF binding site. (A) Schematic representation of the different constructs are labeled and shown on the left. The location of the diphtheria toxin-A gene (TOX), hygromycin-B resistance gene (HygR), insert (Insert), and SV40 enhancer (En) are indicated for the rectangles and circles. Insert identity is indicated to the left of each construct. The orientation of DXZ4 inserts are represented by the black arrows. The graph to the right indicates the number of colonies obtained for each construct after 2–3 wk of selection for hygromycin-B. Data represents the mean and standard error for four independent experiments. (B) A single 3-kb DXZ4 monomer is represented by the 1–3000 bp scale. The approximate location of subfragments A–J are shown below the monomer. Data obtained as for A. The asterisk highlights the subfragment that retained most of the insulator activity. (C) Representation of the subfragments of C (C1–C3) that were cloned in either orientation into pREP-ToxA. The P indicates the fragment containing the highest promoter activity, whereas the EB indicates the fragment containing enhancer-blocking activity determined below. (D) Data obtained for the C subfragments determined as for A.
Discussion
DXZ4 on the Xa: Heterochromatin within euchromatin
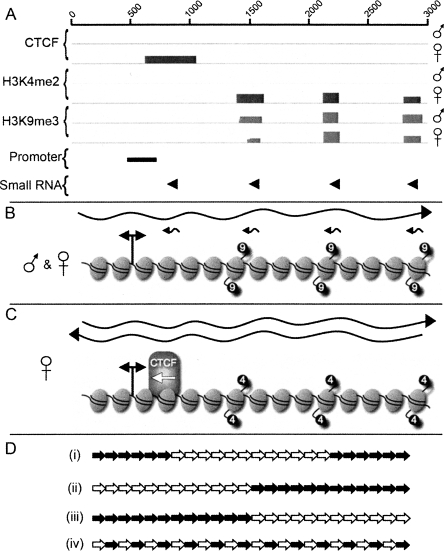
Consistent with the chromatin organization of major and minor satellites in mouse (Martens et al. 2005), DXZ4 chromatin on the Xa is characterized by the heterochromatic histone modification H3K9me3. ChIP-chip data reveal that H3K9me3 nucleosomes are not randomly distributed throughout the 3-kb DXZ4 monomer but instead are highly localized to three sites. Intriguingly, these three sites directly correspond with three of the four small RNA sites of origin. Figure 8A provides a summary of data obtained for DXZ4 chromatin features, promoter, and small RNA sites of origin. From this information, integrated models of heterochromatic DXZ4 (Fig. 8B) and euchromatic DXZ4 (Fig. 8C) are derived. The model in Figure 8B suggests a mechanism of heterochromatin assembly at DXZ4 similar to that observed in small RNA mediated heterochromatin formation at tandem repeats in Schizosaccharomyces pombe (Grewal and Elgin 2007). In S. pombe, centromeric tandem repeats are expressed from both strands, resulting in double-stranded RNAs (Hall et al. 2002; Volpe et al. 2002) that are subsequently processed into small RNAs (Reinhart and Bartel 2002). These small RNAs associate with an RNAi-induced transcriptional silencing complex (RITS) (Verdel et al. 2004), which is then thought to recruit the H3 lysine-9 histone methyltransferase (HMTase) Clr4 and nucleate the formation of heterochromatin (Hall et al. 2002; Volpe et al. 2002). Both strands of DXZ4 are expressed from a bidirectional promoter. The sense strand is detected as a long stable RNA, whereas the antisense strand is processed into small RNAs of 85 nt and smaller, that align with the H3K9me3 nucleosomes (Fig. 8B). The evidence thus far suggests that similar to the S. pombe pathway, the DXZ4 small RNAs are directing an HMTase to produce the H3K9me3 nucleosomes, potentially via a RITS complex. Indeed, RNAi components are necessary for some heterochromatin formation in humans (Fukagawa et al. 2004; Kim et al. 2006), silencing of LINE retrotransposon elements (Yang and Kazazian Jr. 2006), and packaging of the CTG repeats at the myotonic dystrophy locus into heterochromatin (Cho et al. 2005).
Figure 8.
Summary of the chromatin organization and transcription of DXZ4 on the active and inactive X chromosomes and a schematic model of how this organization might be arranged at Xq23. (A) Combined summary of ChIP-chip, luciferase, and small RNA Northern data for a single 3-kb monomer. (B) Summary illustration of DXZ4 chromatin and transcripts in males and at the Xa. The wavy right-pointing black arrow represents the >3-kb sense strand transcript. Small left-pointing arrows represent small antisense RNAs. The elliptical shapes represent nucleosomes. A total of 15 nucleosomes are arbitrarily assigned to a single DXZ4 monomer based on ∼200-bp per nucleosome. The nucleosomes corresponding to the peaks of H3K9me3 signal are indicated by the “9”-labeled black circles. The bidirectional promoter is indicated by the double-headed arrow. (C) Summary illustration of DXZ4 chromatin and transcripts as determined for the Xi. The left- and right-facing wavy arrows represent the sense and antisense DXZ4 transcripts. The nucleosomes corresponding to the peaks of H3K4me2 signal are indicated by the “4”-labeled black circles. The position of CTCF binding is indicated by the CTCF labeled shape, and the left-pointing white arrow represents the direction of promoter–enhancer interference activity. (D) Schematic representation of possible Xi arrangement of H3K4me2 defined monomers (white arrows) and H3K9me3 monomers (black arrows). Each arrow represents a full 3-kb monomer. Only 24 of a potential 50–100 monomers were used for illustrative purposes.
Several questions remain to be addressed about the relationship between the DXZ4 small RNAs and the heterochromatin organization. First, why is only one strand of DXZ4 processed into small RNAs and not both? Second, why are only specific regions of the complementary strand processed into small RNA? And finally, why are all four small RNA sites not marked by H3K9me3? The small RNA site of origin that is not characterized by an H3K9me3 nucleosome corresponds exactly to the CTCF-binding site on the Xi (Fig. 8C). It is possible that this site is occupied by an alternative DNA-binding protein/complex that blocks access to CTCF and displaces the nucleosome, hence accounting for the lack of H3K9me3.
DXZ4 on the Xi: Euchromatin within heterochromatin
DXZ4 on the Xi stands out as a euchromatic island embedded within the extensive surrounding facultative heterochromatin (Boggs et al. 2002; Chadwick and Willard 2002, 2003). Like on the Xa, both strands of DXZ4 are expressed. However, the antisense transcript that is likely processed into small RNAs on the Xa is stabilized sufficiently here that it can be detected as longer RNA species (Fig. 8C). Nevertheless, small RNAs are still being produced from DXZ4 on the Xi. The most likely explanation for this is that not all of the Xi array is packaged into euchromatin. Some proportion of the Xi array is characterized by H3K9me3 and therefore is likely to share features of the Xa constitutive heterochromatin organization. If the hypothesis outlined above for DXZ4 on the Xa is correct, then these small RNAs would be involved in driving formation of the H3K9me3 portion of the Xi array. Figure 8D proposes four possible arrangements of DXZ4 monomers packaged into euchromatin relative to those packaged into heterochromatin. The first example (Fig. 8D, i) suggests that all euchromatic monomers are found toward the center of the array, flanked by heterochromatin. This shares significant similarity to the organization of alpha satellite chromatin at human centromeres (Schueler and Sullivan 2006). Alternatively, DXZ4 could be packaged into a homogenous euchromatin domain adjacent to a heterochromatin domain (Fig. 8D, ii and iii). Finally, DXZ4 could be a mix of euchromatin and heterochromatin (Fig. 8D, iv). Either outcome is likely to influence the flanking chromatin differently, as will the ratio of euchromatic monomers to heterochromatic. The precise arrangement and impact on flanking chromatin is under investigation.
CTCF plays an important role in multiple epigenetic phenomena (Filippova 2008). The binding of CTCF to DXZ4 on the Xi only implores the following question—what is its purpose in this situation? While methylation of CpG sites within a CTCF target sequence blocks the ability of CTCF to bind (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000b; Filippova et al. 2001), the putative CTCF-binding site within DXZ4 does not contain any CpG sequences, and surrounding DNA methylation does not interfere with binding in vitro. Nevertheless, CTCF is only associated with the Xi and its binding site, and surrounding DNA is completely unmethylated. How, therefore, is CTCF prevented from recognizing and binding to DXZ4 on the Xa considering that CpG methylation is not a factor? One possibility is that some other DNA-binding protein/complex already occupies the site blocking access to CTCF. Alternatively, the heterochromatic packaging of DXZ4 may be sufficient to conceal the binding motif. Finally, the exact overlap of a small RNA with the predicted CTCF-binding motif, and the lack of an H3K9me3 nucleosome, suggests that the RNA may have some role in obstructing CTCF binding.
The lack of DNA methylation in the vicinity of CTCF on the Xi is consistent with a critical role for CTCF in maintaining and preventing the spread of CpG methylation (Schoenherr et al. 2003; Fedoriw et al. 2004; Pant et al. 2004; Filippova et al. 2005; Engel et al. 2006). Therefore, CTCF likely contributes to the hypomethylation of DXZ4 at the Xi (Giacalone et al. 1992) and the adjacent methylation-free promoter. How the methylation status of the promoter region relates to transcription is unclear and warrants further investigation. Obviously, DXZ4 is expressed from both the Xa and Xi, yet the promoter region is heavily methylated on the Xa. Perhaps the methylation status influences the fate of the RNA.
The CTCF-binding region of DXZ4, is capable of unidirectionally interfering with enhancer–promoter communication in vitro analogous to DNA sequence upstream of the mouse H19 gene (Kanduri et al. 2000a). Whether this reflects its activity in vivo remains an important question to be addressed, as does how the arrangement of CTCF bound euchromatin monomers that could all possess this activity influences the flanking Xq chromatin (Fig. 8D). Proximal to DXZ4 is a major band of macroH2A and polycomb characterized heterochromatin (Chadwick and Willard 2002; Chadwick 2007), whereas the distal chromatin is most often characterized by constitutive-like heterochromatin signatures, including H3K9me3 and heterochromatin protein-1 (Chadwick and Willard 2003, 2004; Chadwick 2007). Therefore, it is conceivable that the euchromatic Xi allele of DXZ4 is somehow involved in maintaining this chromatin organization, a hypothesis that is currently under investigation. The binding of CTCF adjacent to the DXZ4 promoter region and an independently identified TAF1 site (Kim et al. 2005) suggests that CTCF may be involved in recruiting RNA polymerase II to DXZ4 (Chernukhin et al. 2007). Research into what role CTCF has upon maintaining DXZ4 euchromatin and the stability of the antisense transcript is ongoing.
This report confirms that the single intense foci of CTCF and H3K4me2 observed within the territory of the Xi at interphase (Boggs et al. 2002; Chadwick and Willard 2003) are physically associated with DXZ4 DNA, and that this arrangement is specific to the DXZ4 array on the Xi. DXZ4 on the Xa and some fraction of the Xi array adopts a constitutive heterochromatin organization, and the data presented here are consistent with RNAi-mediated heterochromatin formation. The differential organization of macrosatellite chromatin is unique to the X chromosome and the process of X-inactivation, and is to our knowledge, the first example of a novel macrosatellite gain of function.
Methods
Cell lines
Human diploid female cell lines used include hTERT-RPE1 (Clontech Laboratories, No. C4000-1), hTERT-HME1 (Clontech Laboratories, No. C4002-1), IMR90 (ATCC, CCL-186), and WI38 (ATCC, CCL-75). Human diploid male cell lines used include hTERT-BJ1 (Clontech Laboratories, No. C4001-1), CCD-1139Sk (ATCC, CRL-2708), and CCD-1140Sk (ATCC, CRL-2714). Additional cell lines include JEG-3 (ATCC, HTB-36) and HEK-293 (ATCC, CL-1573). Cells were maintained according to ATCC or Clontech recommendations. Somatic cell hybrids containing either an active X chromosome (t60-12) or inactive X chromosomes (t48-1a-1Daz4A and t86-B1maz1b-3a) were maintained as described (Brown and Willard 1989).
Electrophoresis mobility shift assay
EMSA were performed on fragments of DXZ4 generated by PCR. The DXZ4 coordinates of PCR fragments along with the DNA sequence of oligonucleotides used can be found in Supplemental Table 1. PCR products were end-labeled with digoxigenin-11-ddUTP (DIG) using recombinant Terminal Transferase (Roche Applied Science). Luciferase, CTCF zinc finger region, or full-length CTCF (Quitschke et al. 2000) were synthesized in vitro using the Quick-coupled cell-free T7 and SP6 transcription/translation system (TNT, Promega). The CTCF zinc finger template was generated by RT-PCR amplification of the 11 zinc finger region of CTCF (CTCF-ZnFn-Fwd, 5′-ATGCTGCTATCAGAGGT TAATGC-3′; CTCF-ZnFn-Rev, 5′-CTAATTGTCGTCCAGATCTG GTTC-3′). The 1.1 kb product was TA cloned into pCR2.1 (Invitrogen) prior to sequence verification, excision with HindIII and XbaI (NEB), and cloning into pGEM3 (Promega). Binding was performed at room temperature for 30 min using 1 μL of corresponding TNT protein with DIG-DNA in 1× phosphate buffered saline supplemented with 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% NP40, and 10% glycerol. Reactions were separated on 6% polyacrylamide gels in 0.5× TBE buffer at 80 V and 4°C. DNA was transferred and fixed to nylon membrane and DIG detected (Roche). Supershifts were performed using the full-length CTCF protein only, with polyclonal rabbit anti-CTCF (Upstate Biotech, 07-729). In vitro CpG methylation of DNA was achieved using M.SssI CpG methyltransferase (NEB) and methylation assessed using the methyl-sensitive restriction endonuclease Hinp1I (NEB).
Chromatin immunoprecipitation
ChIP was performed essentially as described (Chadwick 2007) except chromatin was sheared using a Bioruptor (Diagenode) with the following conditions: cell density of ∼1 × 107 cells/mL, sonicated at 4°C using 12 cycles of 30 sec on, 30 sec off at maximum power. ChIP used 1–10 μL of rabbit polyclonal antisera (Upstate Biotech: anti-H3K4me2, 07-030; anti-CTCF, 06-917; anti-H3K9Ac, 06-942; anti-H3K9me3, 07-523) or 10 μL of rabbit serum as a negative control (Calbiochem).
Microarray hybridization
ChIP DNA was prepared for labeling and microarray hybridization exactly as described (www.nimblegen.com). The custom microarray design and analysis was as described (NimbleGen Systems) (Chadwick 2007).
Insulator assay
Full-length DXZ4 monomers were isolated from the BAC clone 2272M5 (Research Genetics) by digestion with XhoI. Single 3-kb monomers were cloned into the XhoI site of pREP-ToxA (Mukhopadhyay et al. 2004). The orientation of inserts was determined by restriction digest mapping and sequencing. DXZ4 subregions A–J were prepared by PCR and TA cloned into pCR2.1 (Invitrogen). The DXZ4 coordinates of PCR fragments along with the DNA sequence of oligonucleotides used can be found in Supplemental Table 2. DNA sequence integrity and orientation was confirmed by sequencing prior to transfer of a representative forward and reverse insert of each into pREP-ToxA by excision from pCR2.1 with KpnI-XhoI and cloning into the corresponding sites of pREP-ToxA. Subregion H contains an internal XhoI site. For this fragment, pREP-ToxA was first cut with XhoI, blunt ended, and then cut with KpnI. The forward and reverse H fragments were transferred from pCR2.1 by excision with KpnI-EcoRV. The assay was performed in JEG3 cells as described (Yu et al. 2004).
Reverse transcription PCR
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen). First strand cDNA was prepared using random hexamers and M-MLV Reverse transcriptase (Invitrogen). PCR was performed using combinations of primers described in Supplemental Tables 1–3. Strand-specific cDNA was prepared using 1 μg of total RNA that was first DNase I treated using 1 unit of amplification grade DNase I (Invitrogen) for 60 min at 37°C prior to addition of EDTA to 2.5 mM and heat inactivation for 10 min at 68°C. DNase I–treated RNA was mixed with 1.5 pmol of forward or reverse DXZ4 oligo or water as a control for nonspecific priming by any residual small DNA fragments not removed by DNase I treatment. The mix was incubated for 5 min at 75°C prior to dropping to 42°C. The mix was then adjusted to 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 1 M Betaine, and 20 units of RNase-Out (Invitrogen) with or without (for –RT control) 200 units of M-MLV reverse transcriptase (Invitrogen). The reaction was incubated for 60 min at 42°C before heat inactivation.
Luciferase reporter assays
The inserts for the forward and reverse DXZ4 subregions A–J in pCR2.1 (described above), were excised with KpnI-XbaI and cloned into KpnI-NheI–digested pGL3-basic (Promega). DXZ4 subregions C-1, C-2, and C-3 were prepared by PCR, TA cloned (Invitrogen), and sequence verified. Inserts were transferred to pGL3-Basic as described above. DXZ4-pGL3-Basic constructs were transfected into HEK-293 cells using lipofectamine 2000 (Invitrogen) and cell extracts prepared and assayed for luciferase activity 48 h later using the Luciferase Reporter Gene Assay kit (Roche). Luciferase activity was measured on a GloMax-96 Luminometer (Promega) and signals normalized to total protein as determined using the Bradford Assay (Bio-Rad).
Small RNA Northern analysis
Small RNA was isolated using the PureLink miRNA isolation kit (Invitrogen), and separated on 15% Novex TBE-Urea gels (Invitrogen). RNA was transferred to Hybond N+ (Amersham) and hybridization performed for 2 h at 37°C in ExpressHyb (Clontech). Blots were washed at room temperature three times in 2× SSC 0.1% SDS for 10 min each, followed by twice in 0.2× SSC 0.1% SDS for 10 min each before exposure to autoradiographic film overnight at −80°C. Oligo probes were end labeled with T4 poly nucleotide kinase (NEB) using >7000 Ci/mmol adenosine 5′-triphosphate gamma-32P (MP Biomedicals) for 60 min at 37°C before heat inactivation for 1 min at 95°C, quenching on ice, and addition to hybridization solution. Nucleic acid size was measured by comparison to 10-bp ladder (Invitrogen) that was separated alongside RNA samples and hybridized with end labeled 10-bp ladder.
Bisulfite modification of genomic DNA, cloning, and sequencing
Genomic DNA was bisulfite modified essentially as described (Grunau et al. 2001), incorporating adaptations described by others (Huang et al. 2006). Primers specific to bisulfite-modified DNA were used to amplify nucleotides 498–1033 of DXZ4 (BiS-Fwd, CCAAACAAACTACCCAAAACC; BiS-Rev, GAAGGTAG GTTAGTAAGAAGG). PCR products were cleaned using the Qiaquick PCR purification kit (Qiagen) before TA cloning into pCR2.1 (Invitrogen) and DNA sequencing.
Acknowledgments
I thank Wolfgang Quitschke for provision of the human full-length CTCF cDNA cloned into pGEM-7Zf(−); Hunt Willard for provision of the Xa/Xi somatic cell hybrids; Rolf Ohlsson for provision of the pREP-ToxO, A, and H19 constructs; and Susan Murphy for technical advice on bisulfite DNA modification. I thank Lisa Chadwick and Kristin Scott for critically evaluating the manuscript and the following individuals for helpful discussions and suggestions: Paul Wade, Carolyn Brown, John Greally, and Beth Sullivan. I also thank the anonymous reviewers for their useful suggestions. This work was supported by a grant from the National Institutes of Health (GM073120, B.P.C.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075713.107.
References
- Bell A.C., Felsenfeld G., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D., Heard E., Spector D.L., Chinault A.C., Allis C.D., Spector D.L., Chinault A.C., Allis C.D., Chinault A.C., Allis C.D., Allis C.D. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Boumil R.M., Ogawa Y., Sun B.K., Huynh K.D., Lee J.T., Ogawa Y., Sun B.K., Huynh K.D., Lee J.T., Sun B.K., Huynh K.D., Lee J.T., Huynh K.D., Lee J.T., Lee J.T. Differential methylation of Xite and CTCF sites in Tsix mirrors the pattern of X-inactivation choice in mice. Mol. Cell. Biol. 2006;26:2109–2117. doi: 10.1128/MCB.26.6.2109-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., Willard H.F., Willard H.F. Noninactivation of a selectable human X-linked gene that complements a murine temperature-sensitive cell cycle defect. Am. J. Hum. Genet. 1989;45:592–598. [PMC free article] [PubMed] [Google Scholar]
- Chadwick B.P. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F., Willard H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F., Willard H.F. Chromatin of the Barr body: Histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum. Mol. Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F., Willard H.F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W., Huynh K.D., Spencer R.J., Davidow L.S., Lee J.T., Huynh K.D., Spencer R.J., Davidow L.S., Lee J.T., Spencer R.J., Davidow L.S., Lee J.T., Davidow L.S., Lee J.T., Lee J.T. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- Chernukhin I., Shamsuddin S., Kang S.Y., Bergstrom R., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Shamsuddin S., Kang S.Y., Bergstrom R., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Kang S.Y., Bergstrom R., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Bergstrom R., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Mukhopadhyay R., Docquier F., Farrar D., Docquier F., Farrar D., Farrar D., et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J., Analau E., Filippova G.N., Tapscott S.J., Filippova G.N., Tapscott S.J., Tapscott S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Donohoe M.E., Zhang L.F., Xu N., Shi Y., Lee J.T., Zhang L.F., Xu N., Shi Y., Lee J.T., Xu N., Shi Y., Lee J.T., Shi Y., Lee J.T., Lee J.T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Engel N., Thorvaldsen J.L., Bartolomei M.S., Thorvaldsen J.L., Bartolomei M.S., Bartolomei M.S. CTCF-binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- Fedoriw A.M., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S., Svoboda P., Schultz R.M., Bartolomei M.S., Schultz R.M., Bartolomei M.S., Bartolomei M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Filippova G.N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Neiman P.E., Collins S.J., Lobanenkov V.V., Collins S.J., Lobanenkov V.V., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Klesert T.R., Lobanenkov V.V., Tapscott S.J., Lobanenkov V.V., Tapscott S.J., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Tsuchiya K.D., Disteche C.M., Disteche C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Takami Y., Nakayama T., Oshimura M., Nakayama T., Oshimura M., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Giacalone J., Friedes J., Francke U., Friedes J., Francke U., Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1992;1:137–143. doi: 10.1038/ng0592-137. [DOI] [PubMed] [Google Scholar]
- Gondo Y., Okada T., Matsuyama N., Saitoh Y., Yanagisawa Y., Ikeda J.E., Okada T., Matsuyama N., Saitoh Y., Yanagisawa Y., Ikeda J.E., Matsuyama N., Saitoh Y., Yanagisawa Y., Ikeda J.E., Saitoh Y., Yanagisawa Y., Ikeda J.E., Yanagisawa Y., Ikeda J.E., Ikeda J.E. Human megasatellite DNA RS447: Copy-number polymorphisms and interspecies conservation. Genomics. 1998;54:39–49. doi: 10.1006/geno.1998.5545. [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Elgin S.C., Elgin S.C. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C., Clark S.J., Rosenthal A., Clark S.J., Rosenthal A., Rosenthal A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I., Noma K., Ayoub N., Cohen A., Grewal S.I., Ayoub N., Cohen A., Grewal S.I., Cohen A., Grewal S.I., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Ingram R.S., Levorse J.M., Tilghman S.M., Levorse J.M., Tilghman S.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Heard E., Disteche C.M., Disteche C.M. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes & Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., Francis F., Sharpe P.T., Hofker M., Sharpe P.T., Hofker M., Hofker M., et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- Huang Z., Wen Y., Shandilya R., Marks J.R., Berchuck A., Murphy S.K., Wen Y., Shandilya R., Marks J.R., Berchuck A., Murphy S.K., Shandilya R., Marks J.R., Berchuck A., Murphy S.K., Marks J.R., Berchuck A., Murphy S.K., Berchuck A., Murphy S.K., Murphy S.K. High throughput detection of M6P/IGF2R intronic hypermethylation and LOH in ovarian cancer. Nucleic Acids Res. 2006;34:555–563. doi: 10.1093/nar/gkj468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C., Holmgren C., Pilartz M., Franklin G., Kanduri M., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Holmgren C., Pilartz M., Franklin G., Kanduri M., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Pilartz M., Franklin G., Kanduri M., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Franklin G., Kanduri M., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Kanduri M., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Liu L., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Ginjala V., Ulleras E., Mattsson R., Ohlsson R., Ulleras E., Mattsson R., Ohlsson R., Mattsson R., Ohlsson R., Ohlsson R. The 5′ flank of mouse H19 in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 2000a;10:449–457. doi: 10.1016/s0960-9822(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Wolffe A., Ohlsson R., Lobanenkov V.V., Ohlsson R., Lobanenkov V.V., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000b;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Hertel J., Hackermuller J., Hofacker I.L., Hackermuller J., Hofacker I.L., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J., Villeneuve L.M., Morris K.V., Rossi J.J., Morris K.V., Rossi J.J., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Zhang M.Q., Lobanenkov V.V., Ren B., Lobanenkov V.V., Ren B., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Goodwin G.H., Neiman P.E., Lobanenkov V.V., Neiman P.E., Lobanenkov V.V., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogi M., Fukushige S., Lefevre C., Hadano S., Ikeda J.E., Fukushige S., Lefevre C., Hadano S., Ikeda J.E., Lefevre C., Hadano S., Ikeda J.E., Hadano S., Ikeda J.E., Ikeda J.E. A novel tandem repeat sequence located on human chromosome 4p: Isolation and characterization. Genomics. 1997;42:278–283. doi: 10.1006/geno.1997.4746. [DOI] [PubMed] [Google Scholar]
- Lander E.S.L.M., Linton B., Birren C., Nusbaum M.C., Zody J., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Linton B., Birren C., Nusbaum M.C., Zody J., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Birren C., Nusbaum M.C., Zody J., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Nusbaum M.C., Zody J., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Zody J., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Baldwin K., Devon K., Dewar M., Doyle W., FitzHugh R., Devon K., Dewar M., Doyle W., FitzHugh R., Dewar M., Doyle W., FitzHugh R., Doyle W., FitzHugh R., FitzHugh R., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C., Fisher R.B., Ferguson-Smith M.A., Lin C.C., Ferguson-Smith M.A., Lin C.C., Lin C.C. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Opravil S., Radolf M., Steinlein P., Jenuwein T., Radolf M., Steinlein P., Jenuwein T., Steinlein P., Jenuwein T., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G.L., John B., John B. Heterochromatin and satellite DNA in man: Properties and prospects. Am. J. Hum. Genet. 1979;31:264–280. [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Yu W., Whitehead J., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Yu W., Whitehead J., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Whitehead J., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Kanduri M., Ginjala V., Vostrov A., Ginjala V., Vostrov A., Vostrov A., et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. So much “junk” DNA in our genome. Brookhaven Symp. Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- Okada T., Gondo Y., Goto J., Kanazawa I., Hadano S., Ikeda J.E., Gondo Y., Goto J., Kanazawa I., Hadano S., Ikeda J.E., Goto J., Kanazawa I., Hadano S., Ikeda J.E., Kanazawa I., Hadano S., Ikeda J.E., Hadano S., Ikeda J.E., Ikeda J.E. Unstable transmission of the RS447 human megasatellite tandem repetitive sequence that contains the USP17 deubiquitinating enzyme gene. Hum. Genet. 2002;110:302–313. doi: 10.1007/s00439-002-0698-2. [DOI] [PubMed] [Google Scholar]
- Pant V., Kurukuti S., Pugacheva E., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Kurukuti S., Pugacheva E., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Pugacheva E., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R., Klenova E., Lobanenkov V., Ohlsson R., Lobanenkov V., Ohlsson R., Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke W.W., Taheny M.J., Fochtmann L.J., Vostrov A.A., Taheny M.J., Fochtmann L.J., Vostrov A.A., Fochtmann L.J., Vostrov A.A., Vostrov A.A. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 2000;28:3370–3378. doi: 10.1093/nar/28.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Bartel D.P., Bartel D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Saitoh Y., Miyamoto N., Okada T., Gondo Y., Showguchi-Miyata J., Hadano S., Ikeda J.E., Miyamoto N., Okada T., Gondo Y., Showguchi-Miyata J., Hadano S., Ikeda J.E., Okada T., Gondo Y., Showguchi-Miyata J., Hadano S., Ikeda J.E., Gondo Y., Showguchi-Miyata J., Hadano S., Ikeda J.E., Showguchi-Miyata J., Hadano S., Ikeda J.E., Hadano S., Ikeda J.E., Ikeda J.E. The RS447 human megasatellite tandem repetitive sequence encodes a novel deubiquitinating enzyme with a functional promoter. Genomics. 2000;67:291–300. doi: 10.1006/geno.2000.6261. [DOI] [PubMed] [Google Scholar]
- Schmid C.W., Deininger P.L., Deininger P.L. Sequence organization of the human genome. Cell. 1975;6:345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J., Levorse J.M., Tilghman S.M., Levorse J.M., Tilghman S.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Sullivan B.A., Sullivan B.A. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 2006;7:301–313. doi: 10.1146/annurev.genom.7.080505.115613. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F., Rudd M.K., Gustashaw K., Willard H.F., Gustashaw K., Willard H.F., Willard H.F. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., van Ommen G.J., Hofker M.H., Frants R.R., Hofker M.H., Frants R.R., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D., Sugiyama T., Gygi S., Grewal S.I., Moazed D., Gygi S., Grewal S.I., Moazed D., Grewal S.I., Moazed D., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Teng G., Grewal S.I., Martienssen R.A., Grewal S.I., Martienssen R.A., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Gruter A.M., Hofker M.H., Moerer P., Williamson R., Hofker M.H., Moerer P., Williamson R., Moerer P., Williamson R., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Xu N., Donohoe M.E., Silva S.S., Lee J.T., Donohoe M.E., Silva S.S., Lee J.T., Silva S.S., Lee J.T., Lee J.T. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Yang N., Kazazian H.H., Kazazian H.H. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Tavoosidana G., Mukhopadhyay R., Kanduri C., Mukhopadhyay R., Kanduri C., Kanduri C., et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A., Irvine D.V., Martienssen R.A., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]