Abstract
The ability to sense molecular tension is crucial for a wide array of cellular processes, including the detection of auditory stimuli, control of cell shape, and internalization and transport of membranes. We show that myosin-I, a motor protein which has been implicated in powering key steps in these processes, dramatically alters its motile properties in response to tension. We measured the displacement generated by single myosin-I molecules, and we determined the actin-attachment kinetics with varying tensions using an optical trap. The rate of myosin-I detachment from actin decreases > 75-fold within 2 pN of tension, resulting in myosin-I transitioning from a low (< 0.2) to a high duty-ratio motor (> 0.9). This impressive tension sensitivity supports a role for myosin-I as a molecular force sensor.
Myosin-Is are the widely expressed, single-headed, and membrane-associated members of the myosin superfamily that participate in regulating membrane dynamics and structure in nearly all eukaryotic cells. Eight myosin-I isoforms are expressed in humans, making it the largest “unconventional” myosin family (1). One specific and well characterized molecular function of a myosin-I isoform (myo1c) is to dynamically provide tension to sensitize mechano-sensitive ion channels responsible for hearing (2-4). Myosin-Is also power the transport and deformation of membranes in the cell cortex and in apical cell projections (5-8). To perform these roles, myosin-Is have been proposed to act as tension-sensing proteins that alter their ATPase and mechanical properties in response to changes in loads imparted by their cellular cargos (3, 9).
Biochemical, structural, and single molecule experiments suggest that some myosin-I isoforms (myo1a, myo1b, myo1c) are adapted to sense tension. Specifically, it has been shown that myosin-I produces its working stroke displacement in two substeps (10). An initial displacement of the lever arm is followed by an additional ∼ 32° rotation that accompanies ADP release (11). Since ADP release kinetically limits the rate of the detachment of myosin from actin (12, 13), the extra lever arm rotation has been proposed to be a force-sensing substep, with the presence of resisting loads preventing this lever arm rotation, and thus inhibiting ADP release and actin detachment. A similar model has been proposed for gating of myosin-V motor activity during processive motility (14).
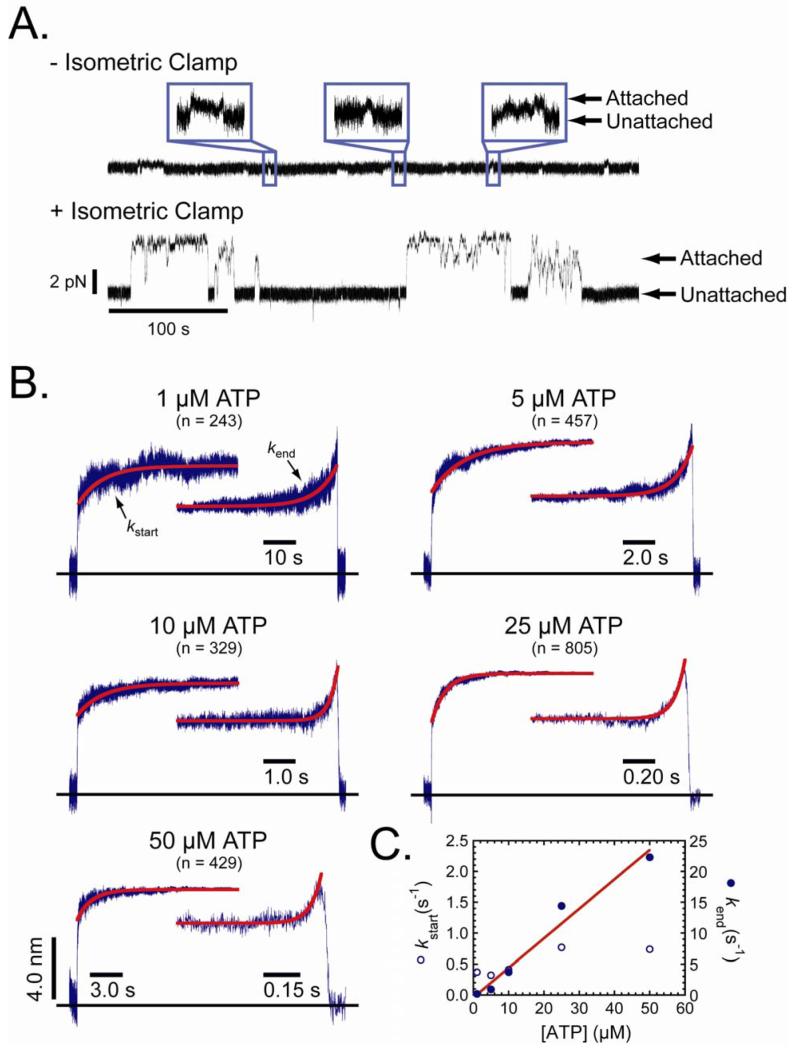
We characterized the motor activity of myosin-I by measuring single-molecule force-generating events using the three-bead configuration, in which a single actin filament, suspended between two beads held separate by optical traps, is brought close to the surface of a pedestal-bead that is sparsely coated with myosin (15). A recombinant myo1b splice isoform containing five IQ motifs and a C-terminal biotinylation tag (16) was attached to streptavidin-coated pedestal beads (17). Single-molecule actomyosin interactions at low loads were acquired using low trap stiffness (∼ 0.022 pN/nm; Fig. 1A) in the presence of 1 - 50 μM ATP. Since the myo1b working stroke is the sum of two substeps, we examined the substep sizes and kinetics of the actomyosin interactions by ensemble averaging the time courses of individual actomyo1b interactions that were synchronized at the times the interactions started or ended (Fig. 1B). Time courses of the start-time averages reveal the lifetimes of the first substep, and time courses of the end-time averages reveal the lifetimes of the second substep (18).
Figure 1.
Single and ensemble-averaged actomyo1b interactions. (A) Representative time traces showing force during unitary actomyo1b attachments acquired ± engagement of the isometric clamp in the presence of 50 μM ATP. Note the dramatic increase in attachment durations during high-force attachments as a result of engagement of the isometric clamp. Insets show 4 s of three different attachment events acquired without the isometric clamp. Arrows indicate attached and detached times. (B) Ensemble-averaged interactions at five different ATP concentrations. Single attachments were synchronized at the times the attachments started or ended and were averaged as described (17). The number of unitary interactions in each average is indicated. The time scale for all start-time averages is the same, and the time scales for the end-time averages vary as indicated. The red lines are fits of the start-time (kstart) and end-time (kend) averages to a single exponential rate function. (C) ATP concentration dependence of (○) kstart and (●) kend rates. The red line is a linear fit of the kend rates, yielding a slope of 0.48 μM-1s-1 (correlation coefficient = 0.985).
The time courses of the start-time averages have rapid initial 5.1 ± 0.43 nm substeps of actin displacement followed by slower 3.3 ± 0.35 nm increases to the final displacement. If the second 3.3 nm substep is the result of a 32° lever-arm rotation (11), we calculate the effective myo1b lever-arm length to be 6.0 ± 0.63 nm. This length is shorter than expected given the regulatory domain (i.e., lever-arm region) contains five calmodulin-binding IQ-motifs, which would have a length of ∼ 20 nm if it was a rigid lever-arm (19). The short effective lever-arm of myo1b is likely due to weak calmodulin binding to a subset of the IQ-motifs resulting in a flexible regulatory domain (16). The regulatory domain is alternatively spliced in vivo (20), so it is possible that the compliance of this region is transcriptionally regulated so as to modulate the mechanical properties of this motor (16).
The initial 5.1 nm working-stroke substep occurred within the experimental response-time and most likely corresponds to the displacement that accompanies phosphate release (10). The time courses of the 3.3 nm increase in the start-time averages reveal the lifetimes of the 5.1 nm substep at the different ATP concentrations. These time courses, at all ATP concentrations, were well fit by a single exponential function with rates (kstart = 0.37 s-1 - 0.77 s-1) slower than reported for ADP release from actomyo1b in solution (1.8 s-1, (13)). Small loads on the actomyo1b interactions likely lead to these slower rates (see below).
The time courses of end-time averages have the same initial and total displacements observed in the start-time averages (Fig. 1B), and they reveal the lifetimes of attachment after the 3.3 nm substep. The time courses were well fit by single-exponential functions with rates (kend) that are linearly related to the ATP concentration with a slope of 0.48 μM-1s-1 (Fig. 1C), which is similar to solution measurements of the apparent second-order rate constant for ATP binding (0.22 μM-1s-1, (13)). A two-step model for myo1b detachment from actin with the rates obtained from the ensemble averaged interactions describes the distributions of actomyo1b attachment lifetimes (Fig. S2). These data confirm that the myo1b working-stroke occurs in two substeps (10) with lifetimes consistent with solution kinetic measurements (13) of the rates of ADP release (lifetime of 5.1 nm substep) and ATP binding (lifetime of 3.3 nm substep).
The force-dependence of actomyo1b attachment lifetimes was measured using a feedback system that applies a dynamic load to the actomyo1b to keep the actin near its isometric position while myosin undergoes its working stroke (21). With the feedback system, the resisting force applied to the actomyo1b attachment was determined primarily by the size of the myo1b powerstroke, the stiffness of the myo1b lever arm, and the position at which myosin binds to the actin filament.
We observed dramatic increases in actomyo1b attachment durations in the presence of 50 μM ATP upon engagement of the isometric clamp, with many durations exceeding 1 min (Fig. 2A). The attachment durations increased with increasing force until ∼ 1.5 pN, after which they appeared to be force independent. We assumed a model for the rate of actomyo1b detachment that includes force-dependent and force-independent pathways:
| (1) |
where kg is a force-dependent rate constant and ki is a force-independent rate constant for actomyo1b dissociation. The force dependence of the detachment rate (kdet) can be calculated as (22):
| (2) |
where kg0 is the rate of kg in the absence of force, ddet is the distance parameter (i.e., the distance to the transition state), F is force, k is the Boltzmann constant and T is the temperature. Because the attachment durations at each force are expected to be exponentially distributed, we used bootstrap Monte Carlo to simulate data to use in Maximum Likelihood Estimations (MLEs) (17). From the MLEs we determined the values and confidence limits of ddet, kg0, and ki that best describe the distribution of attachment durations. The best fit value of kgo = 1.6 s-1 (+0.5/-0.35 s-1) is consistent with the rate of ADP release measured via biochemical methods (1.8 s-1, (13)), which limits kdet at 50 μM ATP in the absence of force. The distance parameter ddet = 12 nm (+1.6/-3.0 nm), which is a measure of strain sensitivity, is strikingly large and distinguishes myo1b as an extraordinarily strain-dependent motor at loads < 2 pN. This sensitivity is very different from other characterized myosins, including strain-sensitive myosin-VI (23). Unlike myo1b, strain-sensitivity of myosin-VI is only seen at low forces (< 2 pN) in the presence ADP.
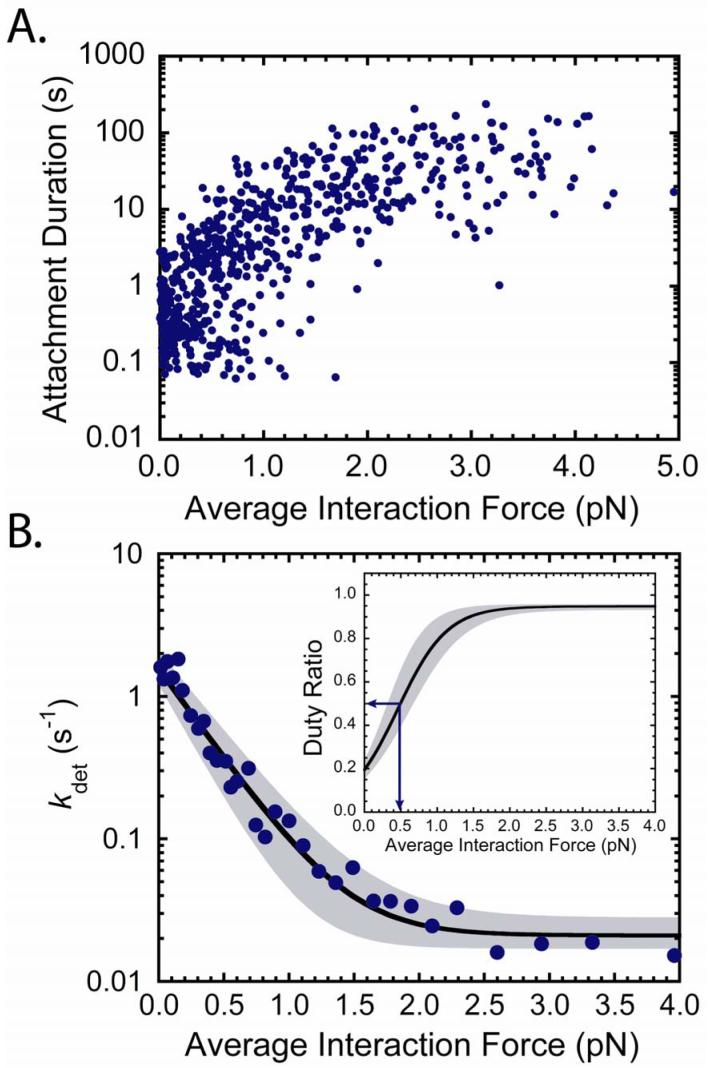
Figure 2.
Force dependence of actomyo1b attachment durations. (A) Attachment durations (n = 638 from 12 experiments) plotted as a function of the average force during the attachment in the presence of 50 μM ATP. (B) Force dependence of the actin-detachment rate (kdet) as calculated using the parameters from the fit of the data in (A) to eq. 2 as determined by MLE. The best fit parameters are kg0 = 1.6 s-1 (+0.5/-0.35 s-1), ddet = 12 nm (+1.6/-3.0 nm), and ki = 0.021 s-1 (+0.007/-0.004 s-1). Points are the inverse averages of 20 consecutive attachments durations from the plot in (A). (Inset) Predicted myo1b duty-ratio as a function of force. The shaded areas show the 97% confidence limit based on the uncertainties listed above. Arrows show the force at which myo1b becomes a high duty-ratio motor (∼ 0.5 pN).
The strain sensitivity of myo1b is most clearly illustrated in a plot of kdet versus force (Fig. 2B), where it can be seen that kdet decreases > 75-fold with < 2 pN of resisting force. Note that at the low forces experienced by myo1b in the absence of the isometric clamp (< 0.2 pN; Fig. 1B), a 1.5-fold decrease in kdet is predicted over the unloaded rate (kg0), which is consistent with the values of kstart measured from the start-time averages.
The fraction of the total ATPase cycle in which myo1b is bound to actin in a force-bearing state is termed the duty ratio, and the force dependence of the duty ratio can be calculated as,
| (3) |
where kdet is defined above and katt is the rate of entry into the strong binding states. katt can not be determined directly from the force time-courses, but can be estimated from the rate of phosphate release (katt = 0.38 s-1; Fig. S3). Myo1b transforms from a low duty ratio motor (< 0.2) to a high duty ratio motor (> 0.5) when working against as little as 0.5 pN of force, and it approaches the duty ratio of processive myosins (> 0.9) at forces as low as 1.5 pN (Fig. 2B, inset).
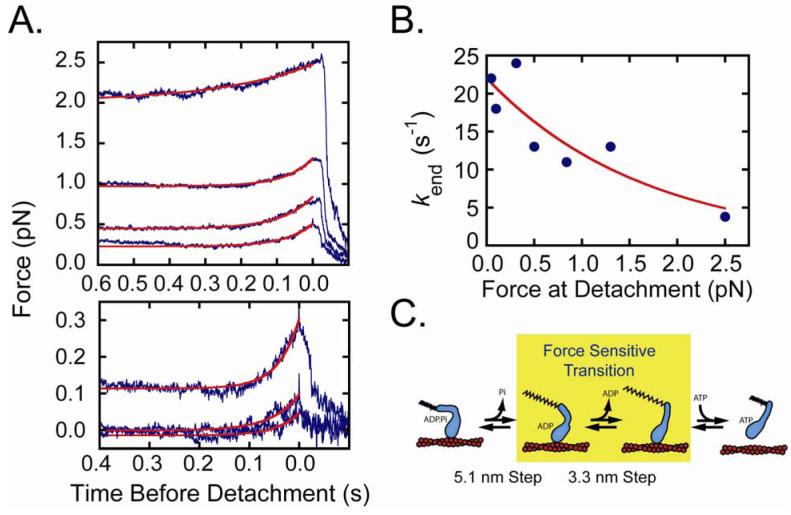
We investigated the effect of force on the lifetimes and force amplitudes of the working-stroke substeps. Interactions acquired in the presence of 50 μM ATP were sorted into bins based on the force immediately before detachment, and individual interactions were synchronized at their end-times and ensemble averaged. Transient increases in force due to substeps were observed in the 500 ms before detachment in all force bins (Fig. 3A). Single exponential fits of the time-courses yielded rates that decreased with increasing force (Fig. 3B). The force dependence of the rates was fit to the equation:
| (4) |
where kend0 is the rate of the time course in the absence of force and dend is the distance parameter for the substep (Fig. 3B). The best-fit rate of kend0 (22 ± 2.5 s -1) is consistent with the rate of 50 μM ATP binding in the absence of resisting loads (kend = 24 s-1; Fig. 1C), and the value dend = 2.5 ± 0.83 nm is much smaller than ddet (Fig. 2B). Therefore, the ATP binding step is not the force-dependent step that limits the rate of actomyo1b detachment. ADP release is the most likely candidate for the force-dependent transition (Fig. 3C). Interestingly, ddet is significantly larger than the size of the substep that correlates with ADP release (3.3 ± 0.35 nm; Fig. 1B), indicating that the force-sensitive transition-state is not on a coordinate that is in-line with a rigid lever arm rotation (24).
Figure 3.
Effect of force on working-stroke substep lifetimes. (A) Single interactions acquired with the isometric clamp in the presence of 50 μM ATP were sorted into bins based on the force immediately before detachment, synchronized to the interaction end-time, and averaged. From the bottom, the force bins in pN are: (0 - 0.125), (0.125 - 0.25), (0.25 - 0.50), (0.50 - 0.75), (0.74 - 1.0), (1.0 - 2.0), (2.0 - 4.0). For clarity, the interaction averages in the lowest force bins are shown on an expanded scale. The red lines are fits of the end-time averages to a single exponential rate function (kend). The rates of the end-time averages are faster than the feedback response time of the isometric clamp, resulting in the lower force amplitudes for the faster substep time courses. (B) Force dependence of kend rates obtained from the fits in (A). The red line is the best fit of the data to eq. 4 with kend0 = 22 ± 2.5 s-1 and dend = 2.5 ± 0.83 nm. (C) Model for myo1b (blue) bound to actin (red) undergoing a working-stroke. The 5.1 nm substep, 3.3 nm substep, and force-sensitive transition are identified. The rate of the ADP release step as a function of force is defined as kdet (eq. 2), and the rate of ATP binding and subsequent actomyo1b detachment is defined as kend (eq. 4). The extended spring signifies tension on the actomyo1b complex.
The presence of a substep in the end-time averages in all force bins indicates that actomyo1b detachment did not occur prior to ADP release, even at forces where the detachment rate is force insensitive (> 1.5 pN; Fig. 2). The force-independent detachment rate (ki in scheme 1) is likely the result of accelerated detachment due to force fluctuations (i.e., noise) in the system. Decreases in force before the ADP-release substep are observed in the force-binned start- and end-time averages (Fig. S4). Thus, when force transiently drops due to noise, there is an exponentially higher probability of ADP release (Fig. 2B), which is followed by rapid ATP binding and detachment.
In conclusion, our results show that myosin-I responds to small resisting loads (< 2 pN) by dramatically increasing the actin-attachment lifetime more than 75-fold. This impressive tension sensitivity supports models that identify myosin-I as the adaptation motor in mechanosensory hair cells (2, 3). More generally, the load-dependent kinetics support a model in which myosin-Is function to generate and sustain tension for extended time periods, rather than to rapidly transport cargos (Fig. S5). This new understanding of myosin-I mechanics allows a more rigorous assignment of this motor’s molecular roles in controlling organelle morphology (8) and dynamics (5) in the wide-variety of cells types in which it is expressed.
Supplementary Material
Acknowledgments
We thank T. Lin for technical assistance and Y.E. Goldman for helpful discussions. Supported by NIH grants AR051174 (HS and EMO) and GM057247 (EMO).
References
- 1.Berg JS, Powell BC, Cheney RE. Mol Biol Cell. 2001 Apr;12:780. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt JR, et al. Cell. 2002 Feb 8;108:371. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 3.Batters C, Wallace MI, Coluccio LM, Molloy JE. Philos Trans R Soc Lond B Biol Sci. 2004 Dec 29;359:1895. doi: 10.1098/rstb.2004.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillespie PG, Cyr JL. Annu Rev Physiol. 2004;66:521. doi: 10.1146/annurev.physiol.66.032102.112842. [DOI] [PubMed] [Google Scholar]
- 5.Bose A, et al. Nature. 2002 Dec 19-26;420:821. [Google Scholar]
- 6.McConnell RE, Tyska MJ. J Cell Biol. 2007 May 21;177:671. doi: 10.1083/jcb.200701144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Mol Biol Cell. 2006 Nov;17:4856. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salas-Cortes L, et al. J Cell Sci. 2005 Oct 15;118:4823. doi: 10.1242/jcs.02607. [DOI] [PubMed] [Google Scholar]
- 9.Geeves MA, Perreault-Micale C, Coluccio LM. J Biol Chem. 2000 Jul 14;275:21624. doi: 10.1074/jbc.M000342200. [DOI] [PubMed] [Google Scholar]
- 10.Veigel C, et al. Nature. 1999 Apr 8;398:530. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 11.Jontes JD, Wilson-Kubalek EM, Milligan RA. Nature. 1995 Dec 14;378:751. doi: 10.1038/378751a0. [DOI] [PubMed] [Google Scholar]
- 12.Siemankowski RF, Wiseman MO, White HD. Proc Natl Acad Sci U S A. 1985 Feb;82:658. doi: 10.1073/pnas.82.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JH, Lin T, Hokanson DE, Ostap EM. Biochemistry. 2006 Sep 26;45:11589. doi: 10.1021/bi0611917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veigel C, Schmitz S, Wang F, Sellers JR. Nat Cell Biol. 2005 Sep;7:861. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 15.Finer JT, Simmons RM, Spudich JA. Nature. 1994 Mar 10;368:113. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 16.Lin T, Tang N, Ostap EM. J Biol Chem. 2005 Dec 16;280:41562. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 17.See methods in supplementary information.
- 18.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Nat Cell Biol. 2003 Nov;5:980. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 19.Warshaw DM, et al. J Biol Chem. 2000 Nov 24;275:37167. doi: 10.1074/jbc.M006438200. [DOI] [PubMed] [Google Scholar]
- 20.Ruppert C, Kroschewski R, Bahler M. J Cell Biol. 1993 Mar;120:1393. doi: 10.1083/jcb.120.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagi Y, Homsher EE, Goldman YE, Shuman H. Biophys J. 2006 Feb 15;90:1295. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell GI. Science. 1978 May 12;200:618. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 23.Altman D, Sweeney HL, Spudich JA. Cell. 2004 Mar 5;116:737. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 24.Tsygankov D, Fisher ME. Proc Natl Acad Sci U S A. 2007 Dec 4;104:19321. doi: 10.1073/pnas.0709911104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.