Abstract
Peptidoglycan glycosyltransferases (PGTs), enzymes that catalyze the formation of the glycan chains of the bacterial cell wall, have tremendous potential as antibiotic targets. The moenomycins, a potent family of natural product antibiotics, are the only known active site inhibitors of the PGTs and serve as blueprints for the structure-based design of new antibacterials. A 2.8 Å structure of a Staphylococcus aureus PGT with moenomycin A bound in the active site appeared recently, potentially providing insight into substrate binding; however, the protein:ligand contacts were not analyzed in detail and the implications of the structure for inhibitor design were not addressed. We report here the 2.3 Å structure of a complex of neryl-moenomycin A bound to the PGT domain of Aquifex aeolicus PBP1A. The structure allows us to examine protein:ligand contacts in detail, and implies that six conserved active site residues contact the centrally-located F-ring phosphoglycerate portion of neryl-moenomycin A. A mutational analysis shows that all six residues play important roles in enzymatic activity. We suggest that small scaffolds that maintain these key contacts will serve as effective PGT inhibitors. To test this hypothesis, we have prepared, via heterologous expression of a subset of moenomycin biosynthetic genes, a novel moenomycin intermediate that maintains these six contacts but does not contain the putative minimal pharmacophore. This compound has comparable biological activity to the previously proposed minimal pharmacophore. The results reported here may facilitate the design of antibiotics targeted against peptidoglycan glycosyltransferases.
INTRODUCTION
Peptidoglycan glycosyltransferases (PGTs) are highly conserved bacterial enzymes that catalyze the polymerization of the NAG-NAM disaccharide subunit of bacterial peptidoglycan (Figure 1) (1-5). These enzymes are regarded as attractive antibiotic targets because their structures are conserved, their functions are essential, they have no eukaryotic counterparts, and they are located on the external surface of the bacterial membrane where they are readily accessible to inhibitors (6-8). While there are not yet any antibiotics in clinical use that directly target these enzymes, the phosphoglycolipid natural product moenomycin (1, Figure 2) inhibits them at nanomolar concentrations and has potent antibiotic activity, with minimum inhibitory concentrations (MICs) 10-1000 times lower than vancomycin’s MICs against various Gram-positive microorganisms (6, 9). Although moenomycin is a potential lead, its therapeutic utility is limited by poor pharmacokinetic properties, including a long half-life and minimal oral bioavailability (9). In addition, although moenomycin strongly inhibits Gram-negative PGTs, its spectrum is restricted to Gram-positive microorganisms, apparently because it cannot penetrate the outer membrane of Gram-negative bacteria (10). It may be possible to overcome moenomycin’s limitations by altering its structure, and the recent completion of the total synthesis of moenomycin (11-13), combined with the identification of the biosynthetic genes for its production (14), make wide-ranging explorations of structural changes feasible for the first time. Nevertheless, the complexity of moenomycin is sufficiently daunting that detailed information on how it interacts with its PGT targets is required to guide effectively the synthesis of analogs. In a major accomplishment by Strynadka and coworkers, the 2.8 Å structure of a co-complex of moenomycin with S. aureus PBP2 was recently solved (MmA:saPGT complex). This complex shows that moenomycin binds in the active site cleft of the PGT domain (15). Compared with the apo-enzyme structure, large conformational changes attributed to moenomycin binding were observed in the vicinity of the active site, particularly in the small lobe of the PGT domain (15). Data interpretation focused on what moenomycin binding suggests about how the substrates bind and what catalysis involves, but the implications of this complex for antibiotic design were not addressed.
Figure 1.
The last two steps in bacterial cell wall biosynthesis. Moenomycin A is the only known antibiotic that directly targets PGTs.
Figure 2.

Chemical structures of moenomycin A and n-MmA. The different parts of moenomycin are designated A through I as labeled.
We recently reported the 2.1 Å structure of the PGT domain from Aquifex aeolicus PBP1A (aaPGT) (16). The aaPGT domain shows high structural similarity to the saPGT domain and is inhibited by moenomycin, making it a good model system for exploring the structural basis for moenomycin inhibition. Here we describe the 2.3 Å structure of a complex of neryl-moenomycin A (n-MmA) bound to the PGT domain of A. aeolicus PBP1A (n-MmA:aaPGT complex). From the structure we infer putative H-bond contacts and analyze the binding site interactions in detail. Below we describe these binding interactions and provide mutational data demonstrating the importance of six contacted residues in enzymatic activity. We also report that a moenomycin analog that satisfies the identified interactions, but does not contain the putative minimal pharmacophore (15, 17), is biologically active. The results described here, taken together with the previous structure, provide information that will enable the structure-based design of moenomycin analogs.
RESULTS AND DISCUSSION
Initial attempts to obtain well-ordered crystals of the A. aeolicus PGT domain with moenomycin bound were unsuccessful, possibly because the 25 carbon chain on the reducing end of the antibiotic makes crystallization challenging. Therefore, we prepared an analog of moenomycin (2, Figure 2) containing a ten carbon neryl chain in place of the longer natural chain (12). The neryl analog was found to inhibit a panel of enzymes available in our laboratory, including A. aeolicus PBP1A (Supplementary Text 1), S. aureus PBP2, and E. faecalis PBP2A, with a potency similar to the parent compound (12). Since these results suggested that the neryl analog contains all the structural features required for PGT binding, we explored strategies for obtaining co-complexes and found that n-MmA soaks readily into the aaPGT crystals reported previously (16).
Structure description
We solved the structure of the co-complex by molecular replacement and refined it to 2.3 Å resolution. Refinement statistics are shown in Table 1. In the structure, 92.3% of the residues are in the most favored region of the Ramachandran plot. As described previously, PGT domains are largely alpha helical. They resemble lysozyme in several respects, including the presence of a large and a small lobe separated by a deep cleft that comprises the active site (15, 16). This cleft is where moenomycin binds. Except for the A-ring of the ligand, which appears to adopt more than one conformation, and the neryl chain, for which the electron density is weak, the final structure of the ligand fits well to the electron density, with an average temperature factor of 89 Å2. The average for the protein is 54 Å2. The A-ring was excluded from the model during refinement.
Table 1.
Data processing and structure refinement statistics
| n-MmA:aaPGT | |
|---|---|
| Data collection | |
| Space group | I222 |
| Unit cell (Å) | a = 54.9 |
| b = 100.4 | |
| c = 104.0 | |
| Resolution (Å) | 30-2.31 |
| Observations | 77660 |
| Unique reflections | 12846 |
| Completeness (%) | 99.1 (94.8)1 |
| Rsymm2(%) | 5.9 (39.6)1 |
| Refinement statistics | |
| Resolution (Å) | 30-2.31 |
| Rwork/Rfree3 | 0.225/0.260 |
| Non-H protein atoms | 1422 |
| Water molecules | 48 |
| Ligand atoms | 87 |
| <B> (Å2) | |
| Protein atoms | 54 |
| Water molecules | 58 |
| Ligand | 89 |
| Ramachandran plot (%) (core/allow/general) | 92.3/7.1/0.6 |
Highest resolution shell (2.39-2.31 Å).
Rsymm = Σ |Ih - <Ih>|/ Σ Ih, where <Ih> is the average intensity over symmetry equivalents.
Rwork and Rfree = Σ | | Fo | - | Fc | |/ Σ | Fo | for the working set and test set (6%) of reflections.
No significant conformational changes in the aaPGT domain occurred upon n-MmA binding (Figure 3b; see Supplementary Text 2). In contrast, large conformational changes, including an ordering of helix 2 and of the coil between helix 3 and helix 4 in the small lobe, were observed in the saPGT domain upon binding of moenomycin (Figure 3b) (15). Although these changes were attributed to moenomycin binding, it is worth noting that the altered regions of the saPGT complex resemble the conformation observed in both the free and the bound aaPGT structures. Therefore, the aaPGT apo-enzyme is already preorganized into a conformation competent to bind moenomycin. This implies that restructuring of the PGT active site (induced fit) is not a necessary consequence of moenomycin binding. We note that the two saPGT structures were crystallized in two different space groups and that the small lobe of the protein in the apo-enzyme structure makes extensive packing interactions. It is possible that the apo-saPGT domain crystallized in a conformation that is not competent to bind moenomycin in order to accommodate these packing interactions.
Figure 3.
Co-complex structure of n-MmA:aaPGT. a) The overall co-complex structure of n-MmA:aaPGT in a ribbon representation. The protein structure is shown in gray with the five conserved motifs that typify PGTs colored in red, yellow, blue, green and purple, and the flexible loop region colored in pink with a dashed line representing the missing residues. n-MmA is shown in stick and ball (carbon, yellow; oxygen, red; nitrogen, blue; phosphate, orange). b) A comparison of the conformations of helix 3, 4 and the coil between apo-enzyme (light blue) and complex structure (brown) of aaPGT (left) and saPGT (right). c) A close-up view of the electron density maps (pink: 1σ 2fo-fc for the protein; blue: 1σ omit 2fo-fc map calculated using the final protein structure annealed at 3000K in the absence of the ligand) at the n-MmA binding site (the structure is rotated from that in Figure 3a for a better presentation). The protein is shown in sticks (carbon, white; oxygen, red; nitrogen, blue) with its map colored in pink; n-MmA is also shown in sticks with its map colored in blue. The figures are made in Pymol.
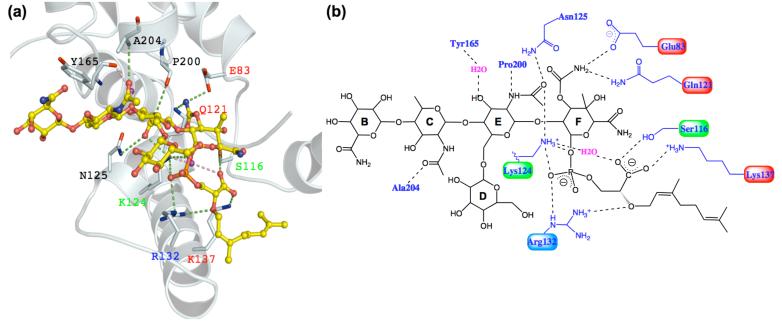
The active site cleft in the n-MmA:aaPGT complex is well-ordered and the conformations of the side chains can be clearly discerned from the electron density map (Figure 3c). Figure 4 illustrates the apparent contacts between n-MmA and the aaPGT domain, represented by interactions under 3.2 Å. The majority of the contacts involve polar interactions between conserved side chains of the PGT domain and the F-ring phosphoglycerate portion of n-MmA, with a smaller number of interactions to the E-ring, a single backbone amide interaction to the C-ring, and no interactions to the other two sugars. Notably, four of the contacts to the F-ring phosphoglycerate portion of the ligand are from invariant or highly conserved residues that help define the five signature motifs of peptidoglycan glycosyltransferases (1), with two additional contacts from other conserved residues in the active site. For example, the invariant side chains of the glutamate residue in motif 1 (E83) and the second glutamine residue in motif 2 (Q121) both contact the carbamate at C3 of the F-ring. The side chains of a conserved serine in motif 2 (S116) and an invariant lysine in motif 3 (K137) contact the carboxylate of the anomeric phosphoglycerate of the F-ring. The phosphate portion of the phosphoglycerate is contacted by the conserved side chains of a lysine (K124) and an arginine (R132) that lie between conserved motifs 2 and 3 in the small lobe of the PGT domain at the floor of the active site cleft. These and a handful of other polar contacts form an extensive hydrogen-bonding network that anchors moenomycin in the active site cleft. There is good shape complementarity between the extended ligand and the cleft itself (Figure 3a).
Figure 4.
Contacts between n-MmA and aaPGT. a) A close-up view of the n-MmA binding site. The protein backbone is shown in white ribbon; residues having contacts with moenomycin are shown in sticks; their labels are colored according to the degree of conservation in PGTs (invariant, red; highly conserved, green; strong preference, blue; not conserved, black). The dashed lines represent polar interactions under 3.2 Å. b) A schematic view of the key contacts between n-MmA and aaPGT. The figures are made in Pymol and Chemdraw.
The contacts to the F-ring from the conserved E and Q residues in the upper part of the binding cleft can also be identified in the MmA:saPGT structure (E114 and Q152 in S. aureus PBP2). However, there are significant differences in contact residues in the lower portion of the cleft, formed by the small lobe, which is where the phosphoglycerate lipid portion of moenomycin binds. The phosphoglycerate lipid portion of moenomycin A is proposed to mimic the diphospholipid leaving group of the donor substrate (18), and both the phosphate and carboxylate moieties are thought to play critical roles in the biological activity of moenomycin (17). In the saPGT complex, the only residue within H-bonding distance to the carboxylate moiety is E171; however, carboxylate-carboxylate interactions are typically not stabilizing. In addition, the corresponding residue in the n-MmA:aaPGT complex, E140, is more than 6 Å from the carboxylate. Thus, the carboxylate-carboxylate interaction in the MmA-saPGT structure is unlikely to explain the proposed critical role of the carboxylate moiety. With regard to the phosphate moiety of moenomycin, some positively charged residues contact it in the MmA:saPGT complex, but there are fewer than in the n-MmA:aaPGT complex; furthermore, not all the contacting residues in the two complexes correspond. The presence of different contacts to the phosphate of moenomycin in the two structures may reflect the large number of positively charged residues in close proximity that are capable of forming electrostatic interactions. Regardless of the explanation, these differences in the complexes should be considered by those attempting the structure-based design of analogs.
Activity of the mutated proteins
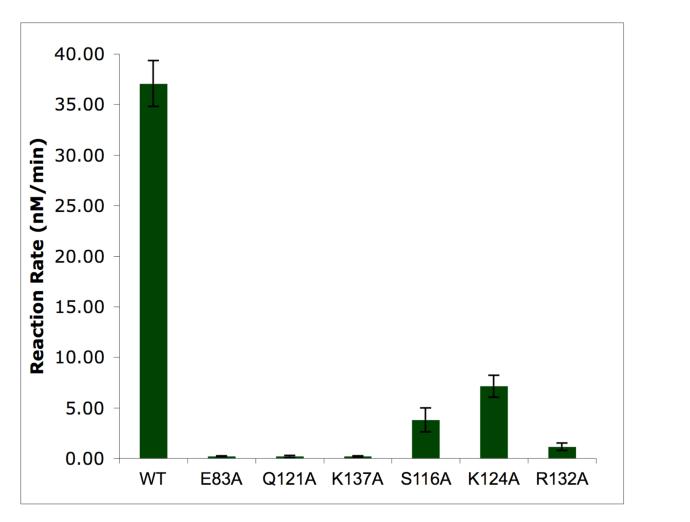
Based on the structure of the n-MmA:aaPGT complex we have identified six residues that make hydrogen bonds to the F-ring phosphoglycerate portion of n-MmA. An alignment of 200 hundred PGTs from both Gram-positive and Gram-negative bacteria shows that three of these six residues, E83, Q121, and K137, are invariant in all PGTs; two others, S116 and K124, can only be replaced by threonine or arginine, respectively; the remaining residue, R132, is also conserved. The high conservation of these six residues in all PGTs, combined with their central location in the active site cleft, suggests that they play critical roles in catalytic activity. Although E83A was previously shown to be inactive (16, 19, 20), confirming its importance, none of the other five mutant proteins has been examined. Therefore, we measured the catalytic activity of each mutant protein under initial rate conditions using an established assay that measures the formation of peptidoglycan polymer (21). The results are summarized in Figure 5. Activity was undetectable for E83A, Q121A, and K137A, and barely detectable for R132A (∼3% of the wild-type enzyme). The activities of S116A and K124A were reduced by 10- and 5-fold, respectively, compared with the wild-type enzyme. The inactive mutant proteins were also evaluated at 30-fold higher enzyme concentrations and these reactions were analyzed using an SDS-PAGE assay that separates peptidoglycan products to single disaccharide resolution (19). No turnover was observed for E83A, Q121A, or K137A even under these forcing conditions (data not shown). We have concluded that most of the residues that anchor moenomycin in the active site are essential for good enzymatic activity, which is consistent with their high degree of conservation. Moenomycin evidently evolved to recognize these conserved residues, and their importance in enzymatic activity may slow the development of resistance due to mutations in the target.
Figure 5.
Activity of the six aaPGT mutant proteins compared to the wild type enzyme. Error bars are based on reactions carried out in triplicate.
The minimal pharmacophore of moenomycin
The crystal structure shows that the majority of directional contacts observed between the protein and the ligand involve the EF-ring phosphoglycerate portion of moenomycin (Figure 4b), which suggests that this part of the molecule is the most critical portion for inhibition (22). A disaccharide degradation product of moenomycin containing the EF rings was reported to have in vitro inhibitory activity but no biological activity, and the CEF trisaccharide analog 3 (Figure 6a), has been identified as the minimal biologically active pharmacophore (22). The C2-N-acetyl group of the C-ring makes a single polar contact to the backbone amide of A204 in the aaPGT crystal structure. Unless this contact is critical, the role of the C-ring in biological activity is not obvious from the structural analysis. To evaluate the proposed importance of the C-ring, we prepared the DEF trisaccharide analog 4 (Figure 6b) by heterologous expression of a subset of moenomycin biosynthetic genes in S. lividans TK24 (17). This compound was purified from cell extracts and characterized by 1H NMR and high-resolution mass spectrometry (Supplementary Methods). It was then evaluated for PGT inhibitory activity in vitro as well as biological activity.
Figure 6.

Chemical structures for the CEF (3) and DEF (4) trisaccharides. The sugars are labeled as in moenomycin A shown in Figure 2.
Compound 4 was found to inhibit S. aureus PBP2 with an IC50 value of 870 ± 110 nM, approximately ten fold higher than that of moenomycin itself. Against S. aureus, compound 4 was found to have an MIC value of 8 μg/mL, which is close to the reported value of 1.3 μg/mL for the CEF trisaccharide (22). We have concluded from these results that the C-ring per se is not required for biological activity; instead, the minimal pharmacophore appears to include the EF-ring phosphoglycerate portion, which makes extensive contacts to the enzyme, and either the C or the D ring. The D-ring protrudes into the solvent in both structures and the structural basis for its contribution to activity is not clear. In the saPGT complex, there is one H-bonding interaction that could potentially play a stabilizing role. However, it is worth noting that the D-ring glycosidic bond torsion angles are different in the two structures, reflecting the flexibility of the 1,6 linkage, but also making it impossible for similar protein:ligand contacts to form in the two structures. X-ray crystallographic information on DEF analog bound to a PGT domain will be required to understand the role of D-ring better. Nevertheless, the identification of this new trisaccharide pharmacophore confirms the central importance of the EF-ring phosphoglycerate in binding and implies that it will be possible to produce PGT-active antibiotics from relatively small scaffolds as long as they maintain the conserved network of polar contacts to key active site residues.
Conclusion
Structures of two different PGT domains, one from a Gram-positive mesophile and one from a Gram-negative thermophile, were recently reported and confirm that these enzymes are structurally highly homologous (15, 16). They also share key mechanistic similarities (16, 23, 24). Therefore, it is reasonable to expect that it will be possible to develop broad-spectrum antibiotics against these enzymes. The moenomycins, which are potent inhibitors of the PGTs, provide a blueprint to guide the design of such antibiotics.
An analysis of the complex structure of n-MmA:aaPGT reveals a well-defined network of conserved polar contacts to the EF-ring phosphoglycerate portion of moenomycin. We have shown that each of the residues involved in this network of contacts plays an important role in catalytic activity, which may help explain the observation that resistance has apparently not developed to moenomycin despite three decades of use as an animal growth promoter. The crystal structure, in revealing the central importance of the EF-phosphoglycerate portion of the molecule, also suggests that prospects for the design of novel moenomycin analogs are promising. The availability of the biosynthetic genes (14), combined with chemistry to manipulate moenomycin intermediates (11-13), makes it feasible to explore a variety of di- and trisaccharide derivatives containing substitutions in order to obtain compounds that may have more favorable properties than the parent molecule. The crystal structure provides information on where substitutions to the EF-ring are sterically allowed, and the ability to obtain co-complexes rapidly by soaking methods should facilitate iterative improvements in design.
Finally, we note that the co-complex structure reported here makes possible other structure-based design efforts because it implies that non-carbohydrate based scaffolds that establish the same type of polar interaction network described for the n-MmA:aaPGT complex should also function as inhibitors.
METHODS
Materials
Radiolabeled heptaprenyl-Lipid II was synthesized as described by Ye et al (25). Moenomycin A was extracted and purified from the feed stock flavomycin and purified as described (12). The QuikChange site-directed mutagenesis kit was obtained from Stratagene. All other reagents and buffer components were purchased from Sigma-Aldrich.
Chemical synthesis of n-MmA
n-MmA was synthesized as described by Adachi et al (12) with some modifications (Supplementary Methods).
Site-directed mutagenesis of ΔPBP1A[N29-K243]
The parent plasmid, ΔPBP1A[N29-K243], was constructed as reported (16). The QuikChange Site-Directed mutagenesis kit was used to make the mutant proteins S116A, Q121A, K124A, R132A and K137A from ΔPBP1A[N29-K243], using the primer pairs given in Supplementary Table 1. Wild-type and mutant proteins were expressed as N-terminal thioredoxin fusions and purified as described previously (16, 19).
Activity assays of the mutated proteins
Typical conditions for aaPGT activity assays are as follows unless otherwise noted: 50 mM HEPES, pH 7.5, 20% DMSO, 10 mM CaCl2, 4 μM C14-labeled Lipid II and 60 nM enzyme. 2 μM enzyme was added for reactions carried out under forcing conditions. All activity assays of PGT and its mutant proteins were prepared on ice and initiated with a rapid temperature ramp to 55°C in a PCR cycler and stopped by putting on ice. A paper chromatography assay and an SDS-PAGE assay were then used to characterize the initial reaction rate and product distribution respectively (19, 21). For initial rate calculations, substrate conversions were kept under 10%. MIC values (μg/mL) were obtained using a standard microdilution assay. The MIC is defined as the lowest antibiotic concentration that resulted in no visible growth after incubation at 37°C for 22h.
Crystallization and structural determination
Crystals of protein PBP1A[51-243] were obtained as described previously (16). n-MmA was soaked into PGT crystals at a concentration of 1.5 mM in a stabilizing solution containing 100 mM HEPES, pH 7.5, 15% polyethylene glycol 6000 for 8h. The crystals then were cryoprotected with 25% glycerol and flash frozen in liquid nitrogen.
Complete data sets for the co-complex crystals were collected at NE-CAT ID-24 of the Advanced Photon Source (Argonne National Laboratories, Chicago). Data were indexed and scaled with HKL2000 (26). The data collection statistics are shown in Table 1. The structure of apo-aaPGT (PDB 2OQO) was used as a starting point for the refinement of the n-MmA:aaPGT model. First, all solvent and other ligand molecules were removed from the apo-aaPGT model. Then the model was refined against the n-MmA:aaPGT diffraction data using CNS 1.2 (27), first by employing rigid body refinement followed by simulated annealing and temperature factor refinement. The protein model in the complex was then completed by interactive rounds of manual fitting in COOT 0.3 (28) and refinement in CNS. Inspection of 2fo-fc and fo-fc electron density maps confirmed the presence of n-MmA. When modeling n-MmA into the density, we dissected n-MmA to 9 groups (A to I) and added individual groups one by one to the model. Restraints for ideal geometry were applied to each group of n-MmA in the refinement. Finally, a limited number of ordered water molecules were modeled by the criteria of reasonable thermal factors and formation of hydrogen bonds with protein residues or neighboring water molecules. Detailed statistics pertaining to the final model are given in Table 1.
Supplementary Material
ACKNOWLEDGEMENTS
This work is based upon research conducted at the Northeastern Collaborative Access Team beam lines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38. This work was supported by the National Institutes of Health RO1 GM076710 and RO1 GM066174.
Supporting Information Available: This material is available free of charge via the Internet.
Accession Codes
The coordinates and structure factors have been deposited in the Protein Data Bank with PDB ID code 3D3H.
References
- 1.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 3.Offant J, Michoux F, Dermiaux A, Biton J, Bourne Y. Functional characterization of the glycosyltransferase domain of penicillin-binding protein 1a from Thermotoga maritima. Biochim. Biophys. Acta. 2006;1764:1036–1042. doi: 10.1016/j.bbapap.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Terrak M, Nguyen-Disteche M. Kinetic characterization of the monofunctional glycosyltransferase from Staphylococcus aureus. J. Bacteriol. 2006;188:2528–2532. doi: 10.1128/JB.188.7.2528-2532.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 6.Halliday J, McKeveney D, Muldoon C, Rajaratnam P, Meutermans W. Targeting the forgotten transglycosylases. Biochem. Pharmacol. 2006;71:957–967. doi: 10.1016/j.bcp.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr. Opin. Chem. Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Wright GD. Biochemistry. A new target for antibiotic development. Science. 2007;315:1373–1374. doi: 10.1126/science.1140374. [DOI] [PubMed] [Google Scholar]
- 9.Goldman RC, Gange D. Inhibition of transglycosylation involved in bacterial peptidoglycan synthesis. Curr. Med. Chem. 2000;7:801–820. doi: 10.2174/0929867003374651. [DOI] [PubMed] [Google Scholar]
- 10.El-Abadla N, Lampilas M, Hennig L, Findeisen M, Welzel P, Muller D, Markus A, van Heijenoort J. Moenomycin A: The role of the methyl group in the moenuronamide unit and a general discussion of structure-activity relationships. Tetrahedron. 1999;55:699–722. [Google Scholar]
- 11.Taylor JG, Li X, Oberthur M, Zhu W, Kahne DE. The total synthesis of moenomycin A. J. Am. Chem. Soc. 2006;128:15084–15085. doi: 10.1021/ja065907x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi M, Zhang Y, Leimkuhler C, Sun B, LaTour JV, Kahne DE. Degradation and reconstruction of moenomycin A and derivatives: dissecting the function of the isoprenoid chain. J. Am. Chem. Soc. 2006;128:14012–14013. doi: 10.1021/ja065905c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welzel P. A long research story culminates in the first total synthesis of moenomycin A. Angew. Chem. Int. Ed. Engl. 2007;46:4825–4829. doi: 10.1002/anie.200700765. [DOI] [PubMed] [Google Scholar]
- 14.Ostash B, Saghatelian A, Walker S. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem. Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5348–5353. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welzel P, Kunisch F, Kruggel F, Stein H, Scherkenbeck J, Hiltmann A, Duddeck H, Muller D, Maggio JE, Fehlhaber HW, Seibert G, Vanheijenoort Y, Vanheijenoort J. Moenomycin-a - Minimum Structural Requirements for Biological-Activity. Tetrahedron. 1987;43:585–598. [Google Scholar]
- 18.Ritzeler O, Hennig L, Findeisen M, Welzel P, Muller D, Markus A, Lemoine G, Lampilas M, vanHeijenoort J. Synthesis of a trisaccharide analogue of moenomycin A(12) implications of new moenomycin structure-activity relationships. Tetrahedron. 1997;53:1675–1694. [Google Scholar]
- 19.Barrett D, Wang TS, Yuan Y, Zhang Y, Kahne D, Walker S. Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J. Biol. Chem. 2007;282:31964–31971. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala JA, Ghuysen JM, Nguyen-Disteche M. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol. Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Walker D, Sun B, Hu Y, Walker S, Kahne D. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5658–5663. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stembera K, Vogel S, Buchynskyy A, Ayala JA, Welzel P. A surface plasmon resonance analysis of the interaction between the antibiotic moenomycin A and penicillin-binding protein 1b. Chembiochem. 2002;3:559–565. doi: 10.1002/1439-7633(20020603)3:6<559::AID-CBIC559>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Barrett D, Leimkuhler C, Chen L, Walker D, Kahne D, Walker S. Kinetic characterization of the glycosyltransferase module of Staphylococcus aureus PBP2. J. Bacteriol. 2005;187:2215–2217. doi: 10.1128/JB.187.6.2215-2217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. Better substrates for bacterial transglycosylases. J. Am. Chem. Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Methods in Enzymology. Academic Press; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.