Abstract
Ca+-activated Cl− channel (CLCA) proteins are encoded by a family of highly related and clustered genes in mammals that are markedly upregulated in inflammation and have been shown to affect chloride transport. Here we describe the cellular processing and regulatory sequences underlying murine (m) CLCA4 proteins. The 125-kDa mCLCA4 gene product is cleaved to 90- and 40-kDa fragments, and the NH2- and COOH-terminal fragments are secreted, where they are found in cell media and associated with the plasma membrane. The 125-kDa full-length protein is only found in the endoplasmic reticulum (ER), and specific luminal diarginine retention and dileucine forward trafficking signals contained within the CLCA4 sequence regulate export from the ER and proteolytic processing. Mutation of the dileucine luminal sequences resulted in ER trapping of the immaturely glycosylated 125-kDa peptide, indicating that proteolytic cleavage occurs following recognition of the trafficking motifs. Moreover, the mutated dileucine and diarginine signal sequences directed processing of a secreted form of enhanced green fluorescent protein in a manner consistent with the effects on mCLCA4.
Keywords: chloride channel, asthma, protein trafficking, epithelium, endoplasmic reticulum retention/trafficking signal
ca2+-activated cl− channel (CLCA) proteins are highly upregulated in human asthma and animal models of mucosal inflammation (15, 17, 23). Since their initial cloning in bovine epithelium (1) and endothelium (4), numerous mammalian CLCA genes have been described with distinct, tissue-specific expression patterns (2, 13, 18). Originally described as integral membrane proteins that may be chloride channels, CLCAs have recently been shown to undergo extensive processing, including proteolytic cleavage and secretion (3, 6, 14), suggesting a more complex role than the regulation of chloride permeability and mucous secretion. In the mouse, the highly homologous murine (m) CLCA genes are tightly clustered on chromosome 3 (H3) in a manner that may enable coordinated transcriptional regulation, and the marked alteration in expression of CLCA genes in several disease states has provoked further interest in their function (12, 20, 23). mCLCA4, which is most closely related to mCLCA2 and mCLCA1 [the orthologs of human (h) CLCA3], is expressed in epithelial and smooth muscle cells (2) and contains conserved family features, including predicted proteolytic cleavage and serine phosphorylation sites, although neither the processing nor phosphorylation of mCLCA4 has been demonstrated experimentally.
Posttranslational processing in the endoplasmic reticulum (ER) is critical for the proper trafficking of proteins to their cellular targets, including the plasma membrane, secretory vesicles, and other organelles (21). Proper assembly and processing occurs through the folding and glycosylation of the native peptide and the recognition of specific motifs by chaperone proteins (5). With respect to proteins that traverse the secretory pathway, specific glycosylations occur that are recognized by carbohydrate binding proteins that serve to sort and direct the peptides to the appropriate destination (10). Specific motifs regulating ER retention and forward trafficking have recently been demonstrated in the integral membrane sodium-dependent glutamate subtype glial transporter (GLT-1), consisting of dileucine (LL) pairs NH2 terminal to a diarginine (RxR) sequence (11). In the GLT-1 transporter, three LL pairs occur as part of a pattern of heptad leucines closely followed by a highly conserved RxR sequence that is conserved in glutamate transporters and found in several channel or receptor proteins (11). The former sequences regulate the forward transport of the full-length GLT-1 translation product in monomeric and multimeric forms for endosomal transport to the plasma membrane, and the latter RxR sequence serves as an ER retention motif (11).
We have used heterologous expression of tagged mCLCA4 proteins to determine the intracellular processing and cellular localization of the full-length and processed peptides. Tagged mCLCA4 proteins specific for the NH2- and COOH-terminal fragments were created that were processed in the same way as native proteins. We report that the full-length mCLCA4 proteins are cleaved into 90- and 40-kDa fragments within the ER compartment, that both peptides are secreted and associate with the plasma membrane, and that internal LL and RxR sequences serve as ER trafficking signals in mCLCA4 proteins.
MATERIALS AND METHODS
Cell culture and heterologous expression.
HEK293T and Chinese hamster ovary (CHO) cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, 1% penicillin/streptomycin (Invitrogen), and 1% l-glutamine (Invitrogen). Cells were transfected with mCLCA4 constructs cloned into the mammalian expression vector pIRES2-green fluorescent protein reporter vector (EGFP; Clontech Laboratories, Mountain View, CA), using Lipofectamine 2000 (Invitrogen) and the manufacturer's protocol (for transient transfection of adherent cells using a half dosage of Lipofectamine 2000). For immunocytochemistry experiments, cells were initially transfected with myc-tagged and other constructs. Later (24 h), transfected cells were plated on slide chambers coated with 17 μg/ml poly-d-lysine in the same growth medium. Following plating (24 h), transfected cells were washed three times with PBS and then fixed with 2% paraformaldehyde. After fixation, cells were washed with PBS, blocked with 10% nonfat dried milk in Tris-buffered saline (TBS), and permeabilized with 0.05% Triton X-100. For Myc-tagged mCLCA4 constructs, mCLCA4 expression was determined by sequential incubation with 4A6 anti-Myc antibody (1:100; Upstate, Lake Placid, NY) and Texas Red-conjugated [or fluorescein isothiocyanate (FITC) conjugated] horse anti-mouse IgG secondary antibody (1:80; Vector Laboratories, Burlingame, CA). Slides were visualized with an Olympus Fluoview (FV5-PSU) confocal microscope. The hCLCA2ss-EGFP-4L/4A construct was made by fusing EGFP with the putative trafficking sequences. Expression was detected using an EGFP polyclonal antibody (1:8; Chemican, Temecula, CA), followed by a FITC-conjugated goat anti-rabbit IgG secondary antibody (1:80; Vector). Cells were imaged using a Zeiss Meta confocal microscope.
mCLCA4 expression.
All insertions were performed by PCR-based, site-directed mutagenesis (ExSite; Strategene, La Jolla, CA). Myc insertions were expressed, and proteins were separated by gel electrophoresis to ensure proper processing of mCLCA4 to 90- and 40-kDa fragments. NH2-terminal specific tagged mutants were produced by insertion of the Myc tag between Ser21 and Ser22 (mCLCA4-21myc). COOH-terminal specific mutants were produced by inserting a Myc tag between Asn713 and Asp714 (mCLCA4-713myc). Both NH2- and COOH-terminal Myc insertion mutants were found to be processed normally. Amino acid substitutions were produced by site-directed mutagenesis of mClCA4 as previously described (16). All mutants were confirmed by DNA sequencing. To test the activity of mCLCA4 trafficking sequences, 120-bp sequences containing the putative LL and RxR trafficking signals (amino acids 565–605), or the respective mutants (2R/2A or 4L/4A), were inserted in the COOH terminus of EGFP cDNA containing the NH2-terminal hCLCA2 (secretion) signal sequence (hCLCA2ss), by PCR amplification of the pcDNA3.1Zeo vector (3) using a primer containing the putative trafficking sequences. The three DNA constructs (hCLCA2ss-EGFP-WT, 2R/2A, or 4L/4A-pcDNA3.1Zeo) were transfected into HEK293T cells as above. EGFP fluorescence was detected at 24 and 48 h, and cells were imaged using confocal (Zeiss Meta) or widefield epifluorescence (Nikon TE300) microscopes. Cell lysates were collected using the same amount of RIPA buffer, and 15 μg protein were used for immunoblot in each condition. After transfection (72 h), the media was collected for immunoprecipitation and immunoblot as described below.
Protein preparation and immunoblotting.
After 48 h transfection, cells were washed with ice-cold PBS and lysed in (150 μl) RIPA buffer (20 mM Tris buffer at pH 7.5 containing 10% glycerol, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton 100-X, 5 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM sodium vanadate, 0.1% SDS, 0.5% sodium deoxycholate, 1.46 μM pepstatin A, 10.5 μM leupeptin, 960 μM benzamidine, 1.53 μM aprotinin, and 570 μM phenylmethylsulfonyl fluoride). Samples were centrifuged (34,000 rpm for 30 min at 4°C) to remove insoluble cellular debris, and the supernatants were boiled in denaturing sample buffer (200 mM Tris buffer at pH 6.8 containing 8% SDS, 40% sucrose, 0.4% bromphenol blue, and 100 mM dithiothreitol) and proteins resolved by SDS-PAGE on a 10% gradient gel. Gels were electroblotted to polyvinylidene difluoride membranes, and the membranes were blocked in TBS containing 0.1% Triton X-100 and 5% nonfat dried milk. Subsequently, the membranes were immunostained with the anti-Myc antibody 4A6 (1:1,000; Upstate), anti-EGFP antibody (1:1,000; Chemicon), or anti-α-tubulin antibody (1:10,000; Sigma, St. Louis, MO), washed extensively, and then exposed to a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:3,000; Bio-Rad, Hercules, CA). Immunoblots were visualized by enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL) on a Kodak Image Station 440 (NEN Life Science Products, Boston, MA). For immunoprecipitation, culture media was collected 48 and 72 h after transfection and precleared with protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for 30 min. Precleared media were then incubated overnight with an anti-Myc antibody (4A6) (1:100), followed by incubation with protein A/G-agarose beads. Protein A/G-agarose beads were washed extensively in RIPA buffer and boiled in an equal volume of 2× SDS loading buffer, followed by Western blotting as described above.
ER-Golgi transport inhibition.
Posttransfection (24 h) of mCLCA4-21-Myc, HEK293T cells were washed and treated with 2 μg/ml brefeldin A (BFA; 0.02% ethanol in DMEM); control wells were washed and exposed to the same medium without BFA. BFA-treated cells were either exposed to the drug for 24 h (BFA condition), or the inhibition was reversed by wash after 4 h and exposure to 0.02% ethanol in DMEM for an additional 20 h (wash). After 24 h, the medium was collected for immunoprecipitation; cells were lysed for protein extraction, and equal concentrations of protein/sample were used for Western blotting. All experiments were performed in triplicate.
Subcellular fractionation.
After transfection (48 h), cells were washed with cold PBS and scraped in 1 ml of homogenization buffer containing 250 mM sucrose, 10 mM Tris at pH 8.0, 50 mM NaF, 1.9 mM benzamidine, 1.1 mM phenylmethylsulfonyl fluoride, and 1 mM EDTA, with 2.92 μM pepstatin A, 21 μM leupeptin, and 3 μM aprotinin. Cells were homogenized and centrifuged at 600 g for 10 min to remove nuclei, and the supernatants were transferred to a new tube and subsequently centrifuged at 100,000 g for 60 min to pellet membrane fractions. Supernatants were adjusted to 100 mM NaCl and 1% Triton X-100; this fraction was designated S2 and was immunoblotted as described above. The pellets were washed in homogenization buffer and centrifuged at 100,000 g for 30 min, and the pellets were solubilized in 150 μl of homogenization buffer containing 100 mM NaCl and 1% Triton X-100; this fraction was designated P2 and was immunoblotted as described above.
In vitro deglycosylation.
Deglycosylation assays were carried out with peptide N-glycosidase F (PNGase) and endoglycosidase H (Endo H) using proprietary provided buffers (New England Biolabs, Ipswich, MA). Briefly, cells were lysed as described above and centrifuged at 13,000 rpm for 25 min, and the supernatants were mixed with denaturation buffer (2–60 μl cell lysate). Samples were boiled for 10 min, divided into thirds, and treated with either G7/NP-40 buffer and PNGase, G5 buffer and Endo H, or with no enzymes. After incubation, all samples were boiled for 5 min, subjected to SDS-PAGE, and immunoblotted, as described above.
Statistical analysis.
The density of proteins in images of Western blots was quantitated using ImageJ. Statistical comparisons were made by Student's t-test or one-way ANOVA (Student-Newman-Keul's) using SigmaStat, with between-group P values <0.05 considered statistically significant.
RESULTS
mCLCA4 proteins undergo proteolytic cleavage and membrane association.
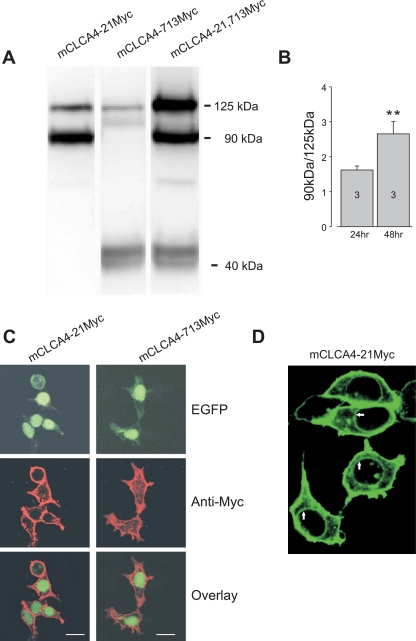
To determine the degree to which the 909-amino-acid, full-length mCLCA4 translation product undergoes intracellular processing and the fate of the processed peptides, NH2- and COOH-terminal Myc insertion mutants were created, expressed in HEK293T, and analyzed by Western blotting and immunocytochemistry. Expression of the NH2-terminal Myc construct (mCLCA4-21Myc) and analysis of cell lysates demonstrated immunoreactive bands of ∼125 and 90 kDa (Fig. 1A), whereas expression of the COOH-terminal insertion clone (mCLCA4-713) resulted in immunoreactive bands of 125 and 40 kDa, and NH2- and COOH-fragment Myc insertion clones (mCLCA4-21, 713Myc) produced all three bands (Fig. 1A). In time course experiments in which the proportion of 90-kDa relative to 125-kDa protein (90- to 125-kDa ratio) was examined 24 and 48 h following transfection, the proportion of 90-kDa protein increased progressively with time (Fig. 1B; see also Fig. 3A). These results are consistent with the endocytic cleavage of the 125-kDa full-length peptide to 90- and 40-kDa NH2- and COOH-terminal peptides, respectively.
Fig. 1.
Proteolytic processing and cellular localization of murine (m) Ca2+-activated Cl− channel (CLCA) 4. A: Western blot of lysates from HEK293T cells expressing NH2- and COOH-terminal Myc-tagged mCLCA4 cDNA. mCLCA4 proteins migrate as a 125-kDa full-length protein that is cleaved to a 90-kDa NH2-terminal and a 40-kDa COOH-terminal fragments. Note that the apparent ratio of 125- and 90-kDa proteins in the blot at right is distorted by the double labeling of the full-length peptide only. B: ratio of 90- to 125-kDa proteins (mClCA4-21Myc) in 3 separate Western blot experiments as in A after 24 and 48 h. The relative amount of proteolytic product was significantly increased at 48 h compared with 24 h (**P < 0.05; see also Fig. 3A) C: HEK293T cells were transfected with a mCLCA4-IRES-green fluorescent protein reporter vector (EGFP) plasmid and immunostained, revealing a prominent plasma membrane expression pattern for both NH2- and COOH-terminal-specific Myc-tagged proteins. D: HEK293T cells were transfected with mCLCA4-21Myc-IRES-EGFP plasmid, followed by anti-myc immunostaining. Nuclear rim staining (arrows) is consistent with endoplasmic reticulum (ER) localization of the full-length protein.
Fig. 3.
Mutation of putative phosphorylation site slows proteolytic cleavage. A: mutant mCLCA4 cDNAs in which the S and T residues in the R592ARSPT597 sequence (residues 592–596) were mutated to AA or DD were expressed, and lysates were blotted. The immunoblot shows the consistent finding in all experiments of a decrease in the amount of proteolytic cleavage in the AA mutant (8 experiments). B: proteins were obtained and processed as in A. Mean ratios of the density of the 90- and 125-kDa bands in 8 experiments. The relative amount of 90-kDa proteolytic product decreased significantly in the AA mutant (**P < 0.005). All other comparisons were not significant. C: decreased processing of the native peptide was observed in both HEK293T and Chinese hamster ovary (CHO) cell lines. D: subcellular fractionation revealed that most mCLCA4 protein was membrane associated, but the AA mutant resulted in an increase in the amount of 125-kDa protein. S, supernatant; P, pellet. E: immunoprecipitation indicated that both mutants are secreted in the extracellular space.
The cellular localization of the Myc-tagged mCLCA4 proteins was examined by immunocytochemistry in cells transfected with bicistronic constructs that included EGFP. As shown in Fig. 1C, both NH2- and COOH-terminal fragments are associated with the plasma membrane. This pattern of localization cannot be explained by immunodetection of the parent 125-kDa fragment, since the full-length peptide is only found in the ER. As shown in Fig. 1D, prominent nuclear rim staining was observed along with plasma membrane localization, consistent with ER localization of the full-length fragment and similar to findings for hCLCA1 and hCLCA2 (3, 7).
Secretion of the 90- and 40-kDa COOH-terminal mCLCA4 peptides.
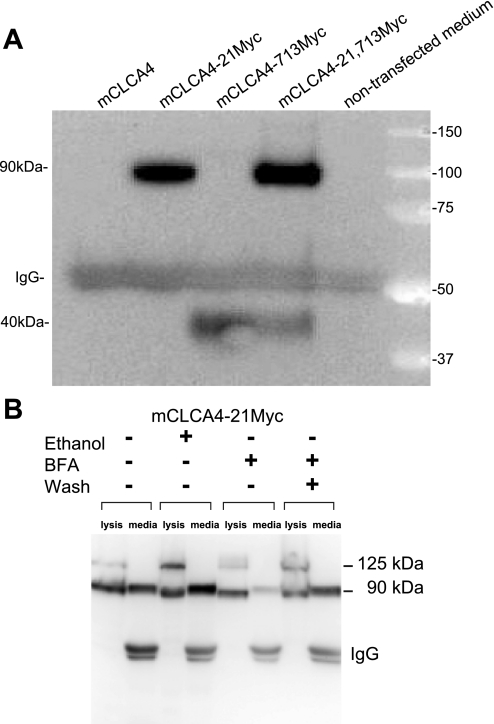
Because previous reports indicated that NH2- and COOH-terminal fragments from some CLCAs are secreted (3, 6, 8), we attempted to immunoprecipitate Myc-tagged mCLCA proteins from media surrounding transfected cells. As shown in Fig. 2A, Western blots of the immunoprecipitates revealed the presence of the 90-kDa NH2- and 40-kDa COOH-terminal peptides in the cell media. Moreover, exposure of cells to the ER-Golgi transport inhibitor BFA (2 μg/ml for 24 h) markedly reduced the amount of protein present in the media, whereas washout of BFA (4 h) reversed this effect (Fig. 2B). Thus mCLCA4 is processed into two peptides, both of which are secreted and associate with the plasma membrane.
Fig. 2.
The 90-kDa NH2-terminal fragment and COOH-terminal fragment are secreted in the extracellular space. A: media from HEK293T cells transfected with Myc-tagged cDNAs were immunoprecipitated with a Myc antibody and immunoblotted with the same antibody. Note detection of both the 90-kDa NH2- and 40-kDa COOH-terminal peptides in the cell media. B: experiments were performed as in A. Continuous exposure to 2 μg/ml brefeldin A (BFA) for 24 h markedly inhibited secretion of the 90-kDa fragment, whereas replacement of BFA after 4 h with DMEM and further incubation for 20 h (wash) reversed the inhibition. Lysates are shown next to immunoprecipitates for identification of the proteins.
mCLCA4 is not phosphorylated at RARSPT but mutation alters proteolytic cleavage.
mCLCA1, mCLCA2, mCLCA4, bovine (b) CLCA1, and bCLCA2 proteins contain a predicted phosphorylation hotspot (R592ARSPT597) near the COOH terminus of the NH2-terminal peptide (2). To further understand the significance of this motif, we mutated S595 and T597 residues to A (mCLCA4-21MycAA) or D (mCLCA4-21MycDD). Our original goal was to determine whether these residues undergo phosphorylation by serine/threonine kinases. However, transfection of cells with these constructs and Western blotting of proteins failed to demonstrate a shift in electrophoretic mobility of the 90-kDa fragment; these studies included extensive serum deprivation alone or additional exposure to forskolin, phorbol ester, methacholine, ionomycin, or several protein kinase inhibitors (data not shown) and were paired with control experiments in which phosphorylation produced the expected mobility shift in similar-size proteins (data not shown). In the experiments, mCLCA4-21MycAA mutant clones displayed a consistent proteolytic phenotype, however. Western blotting indicated that the mCLCA4-21MycAA mutant underwent less proteolytic cleavage than either wild-type or mCLCA4-21MycDD proteins, resulting in a significant shift in the ratio of the 90- to 125-kDa proteins (Fig. 3, A and B). This finding was observed over a time course of 12–72 h (data not shown) and in both HEK293T and CHO cells (Fig. 3C). The apparent shift in the degree of proteolytic cleavage of the parent 125-kDa translation product was not due to a shift in the localization of the fragments. Subcellular fractionation of cellular lysate indicated that both proteins were almost exclusively associated with cellular membranes and that a relative decrease in production of the 90-kDa fragment was observed within this membrane pellet (Fig. 3D). Similarly, immunoprecipitation of the 90-kDa fragment from media of transfected cells demonstrated that the 90-kDa fragment of the mCLCA4-21MycAA mutant was secreted in the media (Fig. 3E), indicating that a marked change in distribution or folding did not occur.
LL and RxR sequences regulate ER retention and export of mCLCA4.
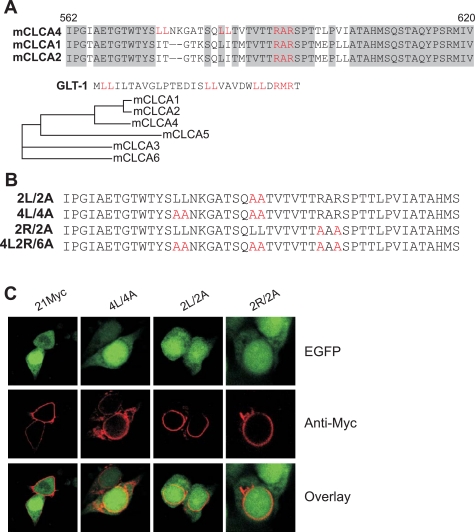
The alteration in proteolytic processing observed in the mCLCA4-21MycAA mutant suggested the possibility that the RARSPT sequence might be involved in the regulation of ER export, resulting in less efficient export of the parent 125-kDa full-length protein and an attendant decrease in processing to the 90-kDa NH2-terminal fragment. Examination of the neighboring sequence indicated the presence of two upstream LLs that have been shown to regulate ER retention and export in combination with an RxR motif (11), which is contained within the targeted RARSPT sequence. The RAR sequence is shared between mCLCA1, mCLCA2, and mCLCA4 but is not preserved in other mCLCAs, whereas the LL sequences are unique to mCLCA4. LI sequences are found in mCLCA1 and mCLCA2 (Fig. 4A), however, and may play a similar role. In other proteins, cytoplasmic LLs appear to serve as an ER export or forward trafficking signal through the binding of chaperones, whereas the RxR is an ER retention signal (11). To determine the relevance of these sequences, we created additional mutants that eliminated the more COOH-terminal LL (2L/2A), both LL sequences (4L/4A), the RxR sequence (2R/2A), or both the LL and RxR sequences (4L2R/6A) (Fig. 4B). Mutation of either the COOH-terminal LL or both LL pairs prevented forward trafficking of mCLCA4 proteins to the cell membrane (Fig. 4C), indicating a potent role of these sequences in forward trafficking. However, mutation of the putative RxR retention sequence also resulted in perinuclear localization of mCLCA4 (Fig. 4C).
Fig. 4.
Forward traffic and ER retention signals. A: alignments of the amino acid sequence of the highly related mCLCAs identifies the dileucine signals in mCLCA4 and the diarginine signals that are unique to these three genes. Similar signals in glutamate-type glial transporter-1 (GLT-1) are shown for comparison. Regions of identity are shown in gray; sequence numbering is shown for mCLCA4. B: alanine mutations of the transport and retention signals are shown in red. C: immunolocalization of the dileucine and diarginine mutants. Note the perinuclear localization of all mutants and an absence of immunoreactive protein at the membrane.
To further understand the role of these motifs, we examined the glycosylation pattern of putatively ER-trapped proteins in lysates from transfected cells. In vitro deglycosylation of tagged proteins from mCLCA4-expressing cells demonstrated that the 125-kDa protein was sensitive to both PNGase F and Endo H (Fig. 5A), indicating that the full-length mClCA4 protein is immaturely glycosylated and resides within the ER compartment, since PNGase F cleaves both mature and immature NH2-linked glycans, whereas Endo H digests high-mannose carbohydrates found in the ER or medial Golgi. Conversely, the 90- and 40-kDa cleaved fragments were sensitive to PNGase F, but markedly less sensitive to Endo H, suggesting that a minor component of these proteins are immaturely glycosylated (Fig. 5A), consistent with export from the ER and mature glycosylation. The 125-kDa full-length protein displayed a deglycosylation pattern consistent with ER trapping (Fig. 5B), and the 90-kDa NH2-terminal fragment was not present in the lysate of 2L/2A and 4L/4A LL mutants. Similarly, mutation of the RxR sequence (2R/2A) resulted in the apparent loss of the 90-kDa fragment in the lysate. To further test whether ER trapping of the export mutants was complete, we immunoprecipitated proteins from the cell media of tagged (NH2-terminal fragment specific) wild-type and ER mutant transfectants. These experiments demonstrated that the ER export mutants (2L/2A and 4L/4A) did not produce the 90-kDa secreted protein, whereas the 90-kDa cleavage product was abundant in the 2R/2A ER retention mutant (Fig. 5C). Our failure to detect the 90-kDa peptide of 2R/2A in cell lysates (Fig. 5B) despite its presence in the media is likely due to the augmented forward trafficking effected by mutation of the retention signal. Unlike GLT-1, the combined mutant (4L2R/6A) did not rescue forward trafficking, since the 90-kDa fragment was not observed in the lysate or in the media (data not shown). Moveover, the mCLCA4-21MycAA and mCLCA4-21MycDD mutants were deglycosylated in a manner consistent with predominately mature glycosylation of the 90- and 40-kDa fragments. The ratio of 90- to 125-kDa protein in the cell lysate and medium for the wild-type, AA mutant, and DD mutant was 81, 60, and 79%, respectively, consistent with less complete processing of the AA mutant. This was not a result of block of secretion of the mutants, since wild-type, AA, and DD proteins were found in the medium (Fig. 5D).
Fig. 5.
Glycosylation pattern of mutant proteins. A: peptide N-glycosidase F (PNGaseF) or endoglycosidase H (Endo H) deglycosylation of NH2- and COOH-terminal fragment-tagged mCLCA4 proteins indicated that the 125-kDa full-length protein is immaturely glycosylated (sensitive to both enzymes), whereas the 90- and 40-kDa fragments were only partially sensitive to Endo H, indicating more mature glycosylation. B: the dileucine and diarginine (RxR) mutants demonstrate marked ER or early Golgi trapping since there is no observable 90-kDa fragment, and the 125-kDa protein displays an immature glycosylation pattern. C: mutation of the diarginine motif does not result in ER trapping, since the 90-kDa fragment is detected in the media by immunoprecipitation, indicating highly efficient forward trafficking. Conversely, mutation of the dileucine signals results in complete loss of forward trafficking. D: the AA R592ARAPA597 mutant displays a glycosylation pattern consistent with delayed processing, but not ER trapping, since the 90-kDa proteolytic fragment from this mutant, as well as the DD mutant, is resistant to Endo H. Media lanes were by immunoprecipitation.
To more definitively test the function of putative mCLCA4 trafficking signal sequences, we created chimeric proteins in which a 40-amino-acid mCLCA4 peptide containing the wild-type, LL, or R×R mutants was placed immediately COOH-terminal to a secreted EGFP construct (3) (Fig. 6A). Consistent with the mCLCA4 mutational results, 48 h after transfection, chimeric EGFP proteins containing the 4L/4A mutation markedly accumulated within the cell, whereas WT and 2R/2A mutant proteins were poorly retained in cells and in those cases were diffusely present within the cytosol (Fig. 6B). Moreover, immunoprecipitation and immunoblotting indicated substantial WT and 2R/2A protein in the cell media, indicating efficient forward trafficking and secretion, but accumulation in the media of 4L/4A mCLCA4 mutant protein was markedly reduced (Fig. 6C).
Fig. 6.
mCLCA4 dileucine (LL) sequences are required for forward trafficking of secreted proteins. A: schematic of construct used to examine mCLCA4 dileucine and diarginine sequences. The NH2-terminal signal sequence (SS) from human (h) CLCA2 was used to direct EGFP to secretory compartments. A 40-amino-acid fragment of mCLCA4 was positioned COOH-terminal to EGFP, similar to its location with respect to the NH2-terminal mCLCA4 fragment. Dileucine and diarginine (RxR) sequences are shown. B: widefield (above, ×10) and confocal (below, ×40) images of HEK293T cells 48 h after transfection (scaled to equivalent fluorescence intensity). The expression pattern indicates EGFP trapping in the 4L/4A mutant (∼70% of the cells showed EGFP localized to the cytosol, and 30% exhibited a more vesicular staining pattern) and is consistent with rapid secretion of EGFP by wild-type (WT) and RxR mutant cells. Confocal images on bottom show a predominate cytosolic pattern in EGFP, retaining WT and 2R/2A cells, and a vesicular pattern in cells transfected with the 4L/4A mutant. Fluorescence of WT and 2R/2A images was substantially lower and is scaled to show roughly equivalent maximum intensity as 4L/4A. C: immunoprecipitation of enhanced green fluorescent protein (EGFP) from medium 72 h after transfection; quantification of the EGFP band is shown below (representative of 3 experiments). Little or no 4L/4A protein was detected in the media in these experiments, further supporting vesicle trapping, whereas slightly more 2R/2A protein was detected, consistent with rapid secretion of the EGFP constructs in which the retention signal has been mutated. “C” indicates nontransfected control.
These results confirm the function of LL and RxR ER regulatory sequences in mClCA4. No secreted protein was detected in LL mutants, indicating a complete block of forward trafficking, whereas secreted protein was detected in RxR mutants, indicating that the motif functions as an ER retention signal. These observations are consistent with the role of the RxR motif in GLT-1 (11). Moreover, the decrease in processing efficiency observed in the RARSPT mutant mCLCA4-21MycAA likely results from conformational effects on the RxR retention sequence imposed by mutation of the neighboring S595 and/or T597 residues.
DISCUSSION
We have used epitope insertion and immunodetection to examine the processing and fate of mCLCA proteins. Our results indicate that mCLCA4 cDNA encodes a full-length protein of ∼125 kDa that is proteolytically cleaved into large NH2-terminal (90-kDa) and smaller COOH-terminal (40-kDa) fragments. The data are consistent with cleavage within the ER or early in the trans Golgi network, since the full-length mClCA4 protein displays an ER-like immunoreactive pattern, does not traffic to the plasma membrane, and is immaturely glycosylated (Fig. 5D). Both the NH2- and COOH-terminal cleavage products are secreted and found in the media, where they rapidly associate with the plasma membrane, and proteins isolated from cell lysates display a mature glycosylation pattern (Figs. 1B and 5A), further suggesting that only the proteolytic products are forward trafficked beyond the ER.
The topology and cellular fate of CLCA proteins has undergone substantial refinement since the initial modeling indicating multiple transmembrane segments (1, 7, 9). More recent structural analysis indicates a single COOH-terminal transmembrane segment (22) and a conserved NH2-terminal hydrolase domain (19). This prediction has been supported by experimental data indicating the secretion of a CLCA fragment in the extracellular space (3, 6, 14), although differences exist as to whether the NH2-terminal fragment (6) or both the NH2- and COOH-terminal fragments (3) are secreted extracellularly, with the latter data inconsistent with a single transmembrane domain. For mCLCA3, the two proteins are reported to remain physically associated and secreted in the extracellular space (14), whereas the COOH-terminal fragment of the highly related human homolog hCLCA1 does not appear to be associated with the secreted NH2-terminal cleaved peptide and is not found in the extracellular media (6).
Based on results from hCLCA2, Elble et al. (3) have suggested a general model of CLCA processing in which the full-length precursor is transported to the cell surface where it undergoes cleavage by an endopeptidase, resulting in release of the NH2-terminal fragment, whereas the COOH-terminal fragment with its transmembrane segment remains as an integral membrane protein. Our data are in general consistent with this model, although it appears that both proteins remain associated with the plasma membrane and that the full-length precursor of mCLCA4 does not traffic to the plasma membrane, since the 125-kDa fragment is sensitive to Endo H digestion (Fig. 5, A, B, and D). Also, unlike hCLCA2, we were unable to demonstrate tight association between the two fragments by coimmunoprecipitation (data not shown).
We have also extended these observations by determining the luminal motifs that regulate trafficking of mCLCA4. Kalandze et al. (11) described cytosolic RxR retention and LL forward trafficking signals regulating the trafficking of GLT-1, an intrinsic membrane protein. Interestingly, mutation of all of the LL motifs in GLT-1 was required to abolish forward trafficking, since 2L/2A and 4L/4A proteins were still maturely glycosylated and inserted in the plasma membrane. The 6L/6A GLT-1 mutation resulted in ER trapping but also resulted in markedly less full-length protein in the lysate, suggesting misfolding and degradation. By contrast, secretion of the 90-kDa NH2-terminal peptide was completely inhibited in 2L/2A and 4L/4A mutants of the luminal protein mCLCA4, and high amounts of ER-trapped 125-kDa protein were observed (Fig. 5B). Mutation of the RxRs also produced an apparent loss of 90-kDa product in the protein lysate; however, protein was readily detected in the cell medium, suggesting augmented forward trafficking associated with loss of the RxR retention signal. The apparent absence of the 90-kDa peptide from the plasma membrane suggests that the RxR motif may also be required for surface association. Chimeric constructs clearly demonstrated the role of the LL sequences in forward trafficking (Fig. 6, B and C), and the prominent vesicular pattern of EGFP expression in 4L/4A mutants confirms the importance of these sequences as retention signals. Because CLCA proteins appear to undergo distinct processing (3, 6, 14), these motifs may underlie significant functional differences in these family members. Studies designed to determine the function of the separate membrane-associated fragments should provide more clarity with respect to the role of CLCAs in ion transport and secretion.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL-45239 (M. I. Kotlikoff) and by Philip Morris USA and Philip Morris International (R. C. Elble).
Acknowledgments
We thank Drs. N. Yvonne Tallini and Mark Rishniw for help with the manuscript and figure preparation and Weipeng Mu for tubulin antibody and Western blotting.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem 270: 31016–31026, 1995. [DOI] [PubMed] [Google Scholar]
- 2.Elble RC, Ji G, Nehrke K, DeBiasio J, Kingsley PD, Kotlikoff MI, Pauli BU. Molecular and functional characterization of a murine calcium-activated chloride channel expressed in smooth muscle. J Biol Chem 277: 18586–18591, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU. The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. J Biol Chem 281: 29448–29454, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Elble RC, Widom J, Gruber AD, Abdel-Ghany M, Levine R, Goodwin A, Cheng HC, Pauli BU. Cloning and characterization of lung-endothelial cell adhesion molecule-1 suggest it is an endothelial chloride channel. J Biol Chem 272: 27853–27861, 1997. [DOI] [PubMed] [Google Scholar]
- 5.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, Thompson N. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J Biol Chem 280: 27205–27212, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Gruber AD, Elble RC, Ji HL, Schreur KD, Fuller CM, Pauli BU. Genomic cloning, molecular characterization, and functional analysis of human CLCA1, the first human member of the family of Ca2+-activated Cl- channel proteins. Genomics 54: 200–214, 1998. [DOI] [PubMed] [Google Scholar]
- 8.Gruber AD, Pauli BU. Molecular cloning and biochemical characterization of a truncated, secreted member of the human family of Ca2+-activated Cl- channels. Biochim Biophys Acta 1444: 418–423, 1999. [DOI] [PubMed] [Google Scholar]
- 9.Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU. Molecular cloning and transmembrane structure of hCLCA2 from human lung, trachea, and mammary gland. Am J Physiol Cell Physiol 276: C1261–C1270, 1999. [DOI] [PubMed] [Google Scholar]
- 10.Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol 15: 364–370, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Kalandadze A, Wu Y, Fournier K, Robinson MB. Identification of motifs involved in endoplasmic reticulum retention-forward trafficking of the GLT-1 subtype of glutamate transporter. J Neurosci 24: 5183–5192, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leverkoehne I, Holle H, Anton F, Gruber AD. Differential expression of calcium-activated chloride channels (CLCA) gene family members in the small intestine of cystic fibrosis mouse models. Histochem Cell Biol 126: 239–250, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev 85: 1061–1092, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Mundhenk L, Alfalah M, Elble RC, Pauli BU, Naim HY, Gruber AD. Both cleavage products of the mCLCA3 protein are secreted soluble proteins. J Biol Chem 281: 30072–30080, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, Fujino M. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA 98: 5175–5180, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nara M, Dhulipala PD, Wang YX, Kotlikoff MI. Reconstitution of beta-adrenergic modulation of large conductance, calcium-activated potassium (maxi-K) channels in Xenopus oocytes. Identification of the camp-dependent protein kinase phosphorylation site. J Biol Chem 273: 14920–14924, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Patel AC, Morton JD, Kim EY, Alevy Y, Swanson S, Tucker J, Huang G, Agapov E, Phillips TE, Fuentes ME, Iglesias A, Aud D, Allard JD, Dabbagh K, Peltz G, Holtzman MJ. Genetic segregation of airway disease traits despite redundancy of calcium-activated chloride channel family members. Physiol Genomics 25: 502–513, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, Elble RC. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol 27: 901–905, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Pawlowski K, Lepisto M, Meinander N, Sivars U, Varga M, Wieslander E. Novel conserved hydrolase domain in the CLCA family of alleged calcium-activated chloride channels. Proteins 63: 424–439, 2006. [DOI] [PubMed] [Google Scholar]
- 20.Ritzka M, Stanke F, Jansen S, Gruber AD, Pusch L, Woelfl S, Veeze HJ, Halley DJ, Tummler B. The CLCA gene locus as a modulator of the gastrointestinal basic defect in cystic fibrosis. Hum Genet 115: 483–491, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol 12: 27–54, 1996. [DOI] [PubMed] [Google Scholar]
- 22.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13: 3369–3387, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Dong Q, Louahed J, Dragwa C, Savio D, Huang M, Weiss C, Tomer Y, McLane MP, Nicolaides NC, Levitt RC. Characterization of a calcium-activated chloride channel as a shared target of Th2 cytokine pathways and its potential involvement in asthma. Am J Respir Cell Mol Biol 25: 486–491, 2001. [DOI] [PubMed] [Google Scholar]