Abstract
Recombinational genetic processes are thought to be rare in the uniparentally inherited mitochondrial (mt) DNA molecules of vertebrates and other animals. Here, however, we document extremely rapid concerted microevolution, probably mediated by frequent gene conversion events, of duplicated sequences in the mtDNA control region of mangrove killifishes (Kryptolebias marmoratus). In local populations, genetic distances between paralogous loci within an individual were typically smaller (and often zero) than those between orthologous loci in different specimens. These findings call for the recognition of concerted evolution as a microevolutionary process and gene conversion as a likely recombinational force in animal mtDNA. The previously unsuspected power of these molecular phenomena could greatly impact mtDNA dynamics within germ cell lineages and in local animal populations.
Keywords: duplicated control regions, mangrove killifish, orthologous, paralogous, Kryptolebias marmoratus
1. Introduction
Mitochondrial (mt) DNA is probably the single most popular and powerful molecule for a wide range of phylogenetic, phylogeographic and population genetics studies. It is thus both surprising and lamentable that several features about intracellular genetic processes in animal mtDNA remain poorly understood (Ballard & Whitlock 2004; Ballard & Rand 2005; Kmiec et al. 2006). Nowhere is this more obvious than in ongoing controversies about whether genetic recombination might play some role in the microevolution of this otherwise asexually transmitted genome (McVean 2001). Here, we document extreme concerted microevolution, probably indicative of rapid gene conversion, between two duplicated mtDNA control regions (CR1 and CR2) in local populations of the killifish Kryptolebias marmoratus. The unexpectedly rapid pace of this recombinational process has potential broad implications for intracellular population dynamics and population genetic patterns for animal mtDNA.
Portions of the mtDNA control region are duplicated in many animal taxa (Arndt & Smith 1998; Black & Roehrdanz 1998; Campbell & Barker 1999; Shao & Barker 2003; Ogoh & Ohmiya 2004), and several reports have implicated concerted evolution for those regions at the levels of species or distinct subspecies (Kumazawa et al. 1996; Eberhard et al. 2001; Abbott et al. 2005; Shao et al. 2005; Ogoh & Ohmiya 2007). Lee et al. (2001) first reported duplicated control regions in K. marmoratus, and, based on high sequence similarity between CR1 and CR2 in the single specimen examined, they also noted the possibility of concerted evolution. However, their study could not rule out an alternative hypothesis that the duplication was recent and the two copies had not yet accumulated mutational differences. To demonstrate concerted evolution at the microevolutionary level, it is necessary to (i) identify polymorphic duplicated sequences in a sample of conspecific individuals, preferably within local populations and (ii) show that the genetic differences between paralogous loci in the same individuals are unambiguously smaller than the differences between orthologous loci in different individuals. Here, we fulfil both of these criteria using complete (or nearly so) CR1 and CR2 sequences from 88 specimens.
2. Material and methods
Samples were collected from the following locales (with abbreviations indicated in parentheses): Twin Cays, Belize (TC); Exuma and San Salvador Islands, Bahamas (EI, SS); Everglades National Park near Flamingo in Monroe County, FL (EP); Shark River also in Monroe County, FL (SR); Charlotte County, FL (CC); St Lucie County, FL (SL); Marco Island in Collier County, FL (MI); and No Name Key in the Florida Keys (NK). Additional details on capturing are provided in Mackiewicz et al. (2006).
Following Lee et al. (2001), we refer to CR1 as the 887 bp non-coding region between the 5′ ends of loci coding for tRNA-Pro and tRNA-Phe, and CR2 as a 795 bp non-coding region between the 3′ end of the locus coding for tRNA-Leu(UUR) and the 5′ end of ND1. For each fish specimen, we sequenced two disjunct mtDNA regions: (i) ‘Cytb–trnT–trnP–CR1’, which spans 1243 bp (positions 16 060–17 300 on GenBank Accession AF283503) and (ii) ‘trnL1–CR2–ND1’, which spans 871 bp (positions 2791–3662 on AF283503). A large segment (735 bp) of CR2 displays high overall sequence similarity (greater than 99%) to CR1, and only this region is the focus of our analysis. Region (i) was amplified with 5′-TCGCCTTACTGGCCTCAATTCT-3′ and 5′-TTTAAGCTACACGAGCCCTAAGTTC-3′. Region (ii) was amplified with primers 5′-AACGTCTTGTTAGGGTGGCAGA-3′ and 5′-AGGAAAGCAAGAGCTAAGAGGA-3′. GenBank accession numbers for the nucleotide sequences analysed in this study are EF202348–EF202523.
We aligned CR1 and CR2 sequences visually, with assistance from Clustal V (default parameters) as implemented in MegAlign (Lasergene v. 6, DNASTAR, Inc.). We employed the p distance as the measure of sequence divergence (Nei & Kumar 2000). Other distance measures were unnecessary or inappropriate, respectively, because the sequences were so similar and because we also included indels as characters (these would require subjective weight assignments in more complex distance metrics). Gene conversion rates were estimated by the method of Eberhard et al. (2001), which assumes that genetic distances between pairs of sequences scale with time to the most recent gene conversion events. The haplotype network was constructed using the method of Templeton et al. (1992) implemented in software TSC v. 1.21 (Clement et al. 2000).
3. Results and discussion
Alignments of the 735 bp sequences from CR1 and CR2 revealed 27 variable nucleotide positions, including five 1 bp deletions (table 1). A striking result is the near identity of CR1 and CR2 within individuals, despite extensive sequence differences (at as many as nine nucleotide positions) between CR1 and CR2 in different specimens. At nucleotide position 663, for example, most individuals displayed an adenine in both CR1 and CR2, but three individuals displayed a cytosine at both loci and one showed a guanine at both CR1 and CR2. Qualitatively similar outcomes were apparent at each of 26 other polymorphic nucleotide sites surveyed (table 1; figure 1). Overall, this pattern of concerted microevolution is also quantitatively evidenced by a 10-fold higher mean genetic distance (p) between orthologues in different individuals (p=1.9×10−3) vis-à-vis paralogues within individuals (p=1.7×10−4).
Table 1.
Evidence for concerted evolution in CR1 and CR2 sequences. (Shown is the common sequence in most specimens and observed departures from it at the indicated loci in specified individuals at each of 27 polymorphic nucleotide positions. Each letter ‘d’ is a 1 bp deletion. Cases of intra-individual genetic identity for CR1 and CR2 are shown in bold.)
| nucleotides at 27 relevant positions 1112223333334444555567 111450121180000003368034563 014664144541234673765914135 | specimens (and loci) | |
|---|---|---|
| (a) | TACGAATTAAGCCCTTTTGCTCCCGAC | 47 specimens (CR1, CR 2); EI6 (CR1); CC5 (CR1); MI1 (CR1); MI4 (CR1); MI5 (CR1); MI7 (CR1); TC2 (CR2); TC3 (CR2); TC5 (CR2) |
| (b) | –-A–––––––––––- | SR6 (CR1, CR2); SR7 (CR1, CR2); SR9 (CR1, CR2) |
| (c) | ––––––-d––––––- | EI6 (CR2); CC5 (CR2) |
| (d) | –––––––––––––T | TC2 (CR1); TC3 (CR1); TC5 (CR1) |
| (e) | ––––––––--A–––– | MI1 (CR2); MI4 (CR2); MI5 (CR2); MI7 (CR2) |
| (f) | ––––––––––-T––- | SR2 (CR1, CR2); SR3 (CR1, CR2); SR8 (CR1, CR2); SR11 (CR1, CR2) |
| (g) | d––––-A––––T––A– | EP3 (CR1, CR2) |
| (h) | ––––-G––––––T-A– | TC10 (CR1, CR2) |
| (i) | –––C–––––––––– | SR21 (CR1, CR2); SR23 (CR1, CR2); SR30 (CR1, CR2) |
| (j) | ––-G––––––––––- | SS2 (CR1, CR2) |
| (k) | -G––––––d––-C––– | SS1 (CR1, CR2) |
| (l) | ––G––––-d-d––––– | SS3 (CR1, CR2); SS5 (CR1, CR2); SS4 (CR2) |
| (m) | ––G––––-ddd––––– | SS4 (CR1) |
| (n) | ––––––––––––-G- | SR5 (CR1, CR2) |
| (o) | ––––––––––––-C- | SR10 (CR1, CR2) |
| (p) | ––––––T––––––C- | SR22 (CR1, CR2); SR26 (CR1, CR2) |
| (q) | –––-C––––-C––––- | SL4 (CR1) |
| (r) | ––––––––-C––––- | SL3 (CR1, CR2); SL4 (CR2) |
| (s) | –d––-G––––––––– | TC7 (CR1, CR2) |
| (t) | ––––G––––––––– | NK1 (CR1, CR2); NK3 (CR1, CR2); TC9 (CR1, CR2) |
| (u) | –––-C–––––––––- | SL6 (CR1, CR2) |
| (v) | –––-CG–TT–––––T–- | TC4 (CR1, CR2); TC8 (CR1, CR2) |
| (w) | –––-CG–––––––T–- | TC91 (CR1, CR2); TC92 (CR1, CR2) |
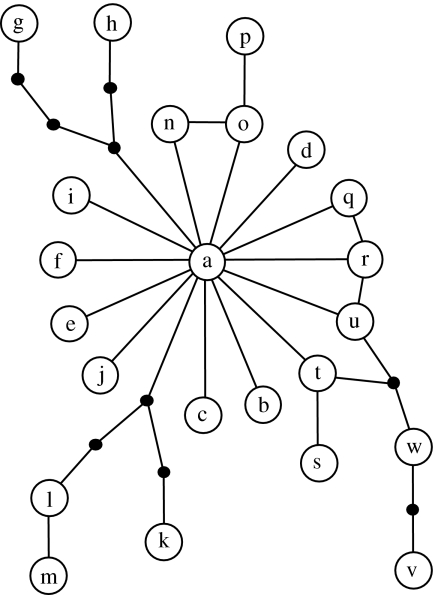
Figure 1.
Haplotype network of CR1 and CR2 sequences. The network is constructed using the method of Templeton et al. (1992) implemented in software TSC v. 1.21 (Clement et al. 2000). Haplotypes and their designations are shown in table 1. Each connecting line represents a single mutation; solid dots are inferred haplotypes.
CR1 and CR2 did differ in sequence within 11 out of 88 specimens, but never by more than one mutation step (table 1). Furthermore, each of these intraindividual sequence discrepancies was confined to a single population, the only possible exception being a 1 bp deletion in CR2 in specimens EI6 and CC5. The finding that most intraspecimen differences between paralogues are confined to a local population further indicates that such discrepancies are short lived due to rapid concerted evolution at these two loci.
Following Eberhard et al. (2001), we provisionally estimated the pace of gene conversion by assuming a standard vertebrate rate for CR evolution (10% change per lineage per million years) as applied to the mean within-individual genetic distance between CR1 and CR2. By this approach, the mean time (±95% CI) between gene conversion events is 850±479 years or about eightfold shorter than the mean time-interval between successive nucleotide substitutions within either CR region. Although the precision of this estimate is debatable, the fact remains that the data evidence a stunning pace of concerted evolution in a molecule that is normally assumed to evolve without appreciable recombinant processes (Wolstenholme 1992; Birky 2001).
A high incidence of concerted evolution in mitochondrial sequences has both practical and conceptual implications. Pragmatically, it means that the two CR regions should be of equal, but redundant, utility in phylogenetic or phylogeographic appraisals (much like the members of any nuclear gene family that experiences strong concerted evolution; Hillis & Dixon 1991). Of potential broader significance, it documents that animal mtDNA can display frequent if not routine recombination. The molecular mechanism itself is not revealed by our data, but all proposed mechanisms of concerted evolution entail genetic recombination of one sort or another: gene turnover via frequent duplications and losses of sequences, or, more likely in this case, some form of gene conversion (non-reciprocal genetic exchange) per se (Kumazawa et al. 1996, 1998).
Previous evidence for recombination in animal mtDNA, e.g. from subtle patterns of linkage disequilibria across the molecule (Awadalla et al. 1999; Eyre-Walker et al. 1999), has been indirect and hotly contested (McVean 2001; Kmiec et al. 2006). Our current evidence for recombinational processes is far more compelling, but, on the other hand, it applies at face value only to the duplicated CR sequences. Perhaps homologous recombination is confined to this small section of mtDNA (one hypothetical mechanism involves recombination between parental and nascent DNA strands during replication of the duplicate loci within the three-stranded D-loop structure; Eberhard et al. 2001). But the mere fact that recombinational machinery in animal mtDNA exists (Thyagarajan et al. 1996) and can apparently function routinely (present study) at least raises the possibility that the phenomenon has much wider ramifications.
We wonder, for example, whether concerted microevolution might possibly have any bearing on the phenomenon of homoplasmy. A long-standing puzzle for most animal species is why each individual often displays a single predominant mtDNA genotype (i.e. homoplasmic), despite having developed from a zygote that had many thousands of mtDNA molecules and belonging to a local population of animals in which mtDNA sequence variation is normally extensive (Avise 2000). A traditional explanation for rapid evolutionary transitions through transient heteroplasmic states is that mtDNA numbers undergo population bottlenecks in the germline cells that are precursor to each mtDNA-rich oocyte (Chapman et al. 1982; Laipis et al. 1988). However, direct empirical evidence for such mtDNA bottlenecks in germline cells is meagre, and another hypothetical process (that could operate in conjunction with the first) is rapid, biased gene conversion such that particular mtDNA molecules often convert others to their own sequence within germline cells.
We also wonder whether concerted evolution in mtDNA might have a bearing on mtDNA repair mechanisms, which are generally thought to be deficient compared with those in the cell nucleus. But if gene conversion is widespread and routine, perhaps the underlying mechanisms play a role in mtDNA repair as well. The current data do not address whether the inferred gene conversion events are intramolecular (within one circular mtDNA molecule) or intermolecular (between different mtDNA molecules), but if the latter is true, there are certainly many mtDNA templates within any cell that could, in principle, serve as useful substrates for the recombinational repair of sequence defects.
These possibilities are admittedly highly speculative, but they do highlight the potential significance of the discovery that concerted microevolution does occur and can be extremely rapid in animal mtDNA. Nevertheless, our current findings are unlikely to challenge conventional wisdom about the utility of mtDNA for phylogeographic or phylogenetic inferences about matrilines. For that to be true, routine interparent mtDNA recombination (following paternal leakage) would have to be demonstrated as well, and numerous studies seem to indicate that such processes are at most very rare.
Acknowledgments
We thank Bruce J. Turner, D. Scott Taylor, William P. Davis, William Dunson, John Grizzle and Carole McIvor for sample collections. Walter Fitch made helpful comments on the manuscript. Mark Mackiewicz developed primers for CR2, which he kindly shared. The work was supported by funds from the University of California at Irvine to J.C.A.
References
- Abbott C.L, Double M.C, Trueman J.W.H, Robinson A, Cockburn A. An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses. Mol. Ecol. 2005;14:3605–3613. doi: 10.1111/j.1365-294X.2005.02672.x. doi:10.1111/j.1365-294X.2005.02672.x [DOI] [PubMed] [Google Scholar]
- Arndt A, Smith M.J. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol. Biol. Evol. 1998;15:1009–1016. doi: 10.1093/oxfordjournals.molbev.a025999. [DOI] [PubMed] [Google Scholar]
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Awadalla P, Eyre-Walker A, Maynard Smith J. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science. 1999;286:2524–2525. doi: 10.1126/science.286.5449.2524. doi:10.1126/science.286.5449.2524 [DOI] [PubMed] [Google Scholar]
- Ballard J.W.O, Rand D.M. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 2005;36:621–642. doi:10.1146/annurev.ecolsys.36.091704.175513 [Google Scholar]
- Ballard J.W.O, Whitlock M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. doi:10.1046/j.1365-294X.2003.02063.x [DOI] [PubMed] [Google Scholar]
- Birky C.W. The inheritance of genes in mitochondrial and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. doi:10.1146/annurev.genet.35.102401.090231 [DOI] [PubMed] [Google Scholar]
- Black W.C, Roehrdanz R.L. Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol. Biol. Evol. 1998;15:1772–1785. doi: 10.1093/oxfordjournals.molbev.a025903. [DOI] [PubMed] [Google Scholar]
- Campbell N.J.H, Barker S.C. The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus: fivefold tandem repetition of a coding region. Mol. Biol. Evol. 1999;16:732–740. doi: 10.1093/oxfordjournals.molbev.a026158. [DOI] [PubMed] [Google Scholar]
- Chapman R.W, Stephens J.C, Lansman R.A, Avise J.C. Models of mitochondrial DNA transmission genetics and evolution in higher eukaryotes. Genet. Res. 1982;40:41–57. doi: 10.1017/s0016672300018899. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Eberhard J.R, Wright T.F, Bermingham E. Duplication and concerted evolution of the mitochondrial control region in the parrot genus Amazona. Mol. Biol. Evol. 2001;18:1330–1342. doi: 10.1093/oxfordjournals.molbev.a003917. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Smith N.H, Maynard Smith J. How clonal are human mitochondria? Proc. R. Soc. B. 1999;266:477–483. doi: 10.1098/rspb.1999.0662. doi:10.1098/rspb.1999.0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis D.M, Dixon M.T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. doi:10.1086/417338 [DOI] [PubMed] [Google Scholar]
- Kmiec B, Woloszynska M, Janska H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 2006;50:149–159. doi: 10.1007/s00294-006-0082-1. doi:10.1007/s00294-006-0082-1 [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol. Biol. Evol. 1996;13:1242–1254. doi: 10.1093/oxfordjournals.molbev.a025690. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. The complete nucleotide sequence of a snake (Dinodon semicarinatus) mitochondrial genome with two identical control regions. Genetics. 1998;150:313–329. doi: 10.1093/genetics/150.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laipis P.J, Van de Walle M.J, Hauswirth W.W. Unequal partitioning of bovine mitochondrial genotypes among siblings. Proc. Natl Acad. Sci. USA. 1988;85:8107–8110. doi: 10.1073/pnas.85.21.8107. doi:10.1073/pnas.85.21.8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-S, Miya M, Lee Y.-S, Kim C.G, Park E.-H, Aoki Y, Nishida M. The complete DNA sequence of the mitochondrial genome of the self-fertilizing fish Rivulus marmoratus (Cyprinodontiformes, Rivulidae) and the first description of duplication of a control region in fish. Gene. 2001;280:1–7. doi: 10.1016/s0378-1119(01)00765-x. doi:10.1016/S0378-1119(01)00765-X [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Taylor D.S, Turner B.J, Avise J.C. Extensive outcrossing and androdioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc. Natl Acad. Sci. USA. 2006;103:9924–9928. doi: 10.1073/pnas.0603847103. doi:10.1073/pnas.0603847103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G.A.T. What do patterns of genetic variability reveal about mitochondrial recombination? Heredity. 2001;87:613–620. doi: 10.1046/j.1365-2540.2001.00965.x. doi:10.1046/j.1365-2540.2001.00965.x [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Oxford University Press; Oxford, UK: 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Ogoh K, Ohmiya Y. Complete mitochondrial DNA sequence of the sea-firefly, Vargula hilgendorfii (Crustacea, Ostracoda) with duplicate control regions. Gene. 2004;327:131–139. doi: 10.1016/j.gene.2003.11.011. doi:10.1016/j.gene.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Ogoh K, Ohmiya Y. Concerted evolution of duplicated control regions within an ostracod mitochondrial genome. Mol. Biol. Evol. 2007;24:74–78. doi: 10.1093/molbev/msl132. doi:10.1093/molbev/msl132 [DOI] [PubMed] [Google Scholar]
- Shao R.F, Barker S.C. The highly rearranged mitochondrial genome of the plague thrips Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. doi:10.1093/molbev/msg045 [DOI] [PubMed] [Google Scholar]
- Shao R.F, Barker S.C, Mitani H, Aoki Y, Fukunaga M. Evolution of duplicate control regions in the mitochondrial genomes of metazoa: a case study with Australasian Ixodes ticks. Mol. Biol. Evol. 2005;22:620–629. doi: 10.1093/molbev/msi047. doi:10.1093/molbev/msi047 [DOI] [PubMed] [Google Scholar]
- Templeton A.R, Crandall K.A, Sing C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Padua R.A, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 1996;271:27 536–27 543. doi: 10.1074/jbc.271.44.27536. doi:10.1074/jbc.271.44.27536 [DOI] [PubMed] [Google Scholar]
- Wolstenholme D.R. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]