Abstract
Whole body protein synthesis is reduced during the fed-to-fasted transition and in cases of chronic dietary restriction; however, less is known about tissue-specific alterations. We have assessed the extent to which protein synthesis in cardiac muscle responds to dietary perturbations compared with liver and skeletal muscle by applying a novel 2H2O tracer method to quantify tissue-specific responses of protein synthesis in vivo. We hypothesized that protein synthesis in cardiac muscle would be unaffected by acute fasting or food restriction, whereas protein synthesis in the liver and gastrocnemius muscle would be reduced when there is a protein-energy deficit. We found that, although protein synthesis in liver and gastrocnemius muscle was significantly reduced by acute fasting, there were no changes in protein synthesis in the left ventricle of the heart for either the total protein pool or in isolated mitochondrial or cytosolic compartments. Likewise, a chronic reduction in calorie intake, induced by food restriction, did not affect protein synthesis in the heart, whereas protein synthesis in skeletal muscle and liver was decreased. The later observations are supported by changes in the phosphorylation state of two critical mediators of protein synthesis (4E-BP1 and eIF2α) in the respective tissues. We conclude that cardiac protein synthesis is maintained in cases of nutritional perturbations, in strong contrast to liver and gastrocnemius muscle, where protein synthesis is decreased by acute fasting or chronic food restriction.
Keywords: protein turnover, nitrogen homeostasis, left ventricular hypertrophy, calorie restriction, stable isotopes
whole body protein synthesis is sensitive to nutritional status (40, 42), with greater rates observed in the fed compared with the fasted state. The synthesis of proteins in liver and skeletal muscle appears to play an important role in trapping exogenous (diet-derived) protein and therein preserving protein homeostasis (10, 12). On the other hand, little is known about the influence of dietary status on myocardial protein dynamics. In particular, since myocardium has a higher metabolic requirement than skeletal muscle or liver, the heart cannot globally downregulate metabolic demands and still maintain normal cardiac output. The ability to modulate myocardial protein synthesis is an important concern in heart disease, with cardiac hypertrophy occurring with hypertension and loss of viable cardiac tissue with end stage heart failure or following myocardial infarction (17). In addition, cardiac failure has been reported in cases when obese patients undergo rapid body mass loss via dietary restriction (38, 39).
Various factors have been implicated in the remodeling heart protein, including nutritional status and insulin. For example, classical studies of Jefferson et al. (16) demonstrated that insulin has a stimulatory effect on protein synthesis in isolated heart muscle. However, tissues were maintained in either the complete absence of insulin or in the presence of insulin (16), whereas in vivo it may be that a relatively low concentration of insulin is sufficient to stimulate normal protein synthesis. In other studies, Sugden and coworkers (26, 27) demonstrated that protein synthesis in heart is affected by nutritional status; however, they compared rates of protein synthesis in fed vs. 3-day-starved animals. Again, the data clearly demonstrate a stimulatory effect of feeding on heart protein synthesis, but the intervention was quite extreme.
It is unclear whether reduced dietary intake leads to reduced protein synthesis in heart compared with liver and skeletal muscle. Therefore, the primary aim of the present study was to determine the role of food intake on cardiac protein synthesis. We aimed to use more physiologically relevant models than the early literature and hypothesized that protein synthesis in cardiac muscle would be unaffected by acute fasting or chronic dietary restriction whereas the liver and gastrocnemius muscle would show reduced protein synthesis when there was a negative energy balance. Two series of experiments were performed. First, we assessed the effect(s) of 20 h of fasting to determine whether a large and sudden shift in protein energy balance would decrease protein synthesis, since digestion/absorption trigger the secretion of insulin and provide amino acid substrates, both of which are required for protein synthesis. Second, we assessed the response to 7 days of food restriction, the rationale being that a more prolonged nutritional imbalance would slow protein turnover and reduce cardiac protein synthesis. Studies were performed in rats by use of a novel 2H2O method to directly quantify protein synthesis in specific tissues (2, 3, 6, 29).
Attention was also directed at studying how chronic food restriction affects protein synthesis at the molecular level. We focused our investigation on the translation initiation factors eIF2α and eIF4E-binding protein-1 (4E-BP1) (18, 30). In particular, eIF2α, GTP, and MettRNAi form the ternary complex that interacts with the 40S ribosomal subunit and the translation preinitiation complex (14, 34), phosphorylation of the α-subunit of eIF2 at Ser51 increases the affinity of interaction of eIF2B with eIF2-GDP and inhibits recycling of the ternary complex, thus blocking a new round of translation initiation. In addition, hyperphosphorylation of 4E-BP1, releases eIF4E, thus increasing cap-dependent translation initiation. Stress conditions (e.g., nutrient limitation) have been associated with the phosphorylation of eIF2 (4, 48) and the dephosphorylation of 4E-BP1, which would lead to a reduction in protein synthesis (19, 24, 31). It isunknown, however, whether/how the molecular control of protein synthesis would respond in different tissues when exposed to the same milieu.
MATERIALS AND METHODS
Chemicals and Supplies
Unless specified, all chemicals and reagents were purchased from Sigma-Aldrich. 2H2O (99.9 atom percent excess) was purchased from Isotec (Miamisburg, OH). Gas chromatography-mass spectrometry supplies were purchased from Agilent Technologies (Wilmington, DE) and Alltech (Deerfield, IL). Protease and phosphatase inhibitor cocktail tablets were purchased from Roche. Antibodies, rabbit (polyclonal) anti-eIF2α-PSer51 phosphospecific antibodies were purchased from Invitrogen/Biosource (44-728G); mouse anti-eIF2α antibodies were prepared by Quality Controlled Biochemicals; rabbit (polyclonal) anti-4EBP1 antibodies were purchased from Invitrogen/Zymed Laboratories (51-2900).
Animal Protocols
All animal studies were conducted according to Guide for the Care and Use of Laboratory Animals (NIH publication No. 85-23) and approved by the Institutional Animal Care and Use Committee at Case Western Reserve University. Male Wistar rats were purchased from Harlan (∼9–11 wk of age; Indianapolis, IN). Upon arrival, all rats were fed standard lab chow (14% fat, 60% carbohydrate, 26% protein by calories; LabDiet, St. Louis, MO). Animals were weighed and randomized to experimental groups (n = 8–10 per group). Rats were housed in a 12:12-h reverse light-dark cycle with free access to food and water in controlled conditions (23 ± 1°C). Two series of experiments were performed: study 1, fed vs. fasted; study 2, 40% dietary restriction vs. ad libitum fed.
In study 1, rats were fed a standard high-complex-carbohydrate/low-fat diet ad libitum for 7 days (70% carbohydrate, 10% fat, and 20% protein by calories; Research Diets, New Brunswick, NJ), and were then fasted for 20 h (food removed at 6:00 PM) or given free access to food (n = 8 per group). At 8:00 AM, all rats were given an intraperitoneal injection of 99% [2H]saline (8 ml per rat). Animals were then continued either fed or fasted and killed 6 h later.
In study 2, all rats consumed the standard diet for 7 days. Rats were then randomized to a dietary restricted group (DR, n = 8) and fed 60% of the dietary intake of the control group. DR rats were fed twice a day (at 8:00 AM and 11:30 AM) for 7 days (typically rats would consume food until ∼1:30 PM); the control rats (n = 8) were fed ad libitum between 8:00 AM and 1:30 PM for 7 days. After 7 days, all rats were given an intraperitoneal injection of 99% [2H]saline (8 AM) and then killed at ∼1:30 PM. All rats were fed during the 2H2O labeling period, i.e., either ad libitum or dietary restricted.
For tissue harvest, rats were anesthetized with isoflurane, and the heart was exposed by sternotomy. A 1-ml sample of blood was drawn from the inferior vena cava, and plasma was isolated and frozen. The heart was rapidly removed, the right ventricle was sliced away from the left ventricle (LV), both sections were quickly weighed, and ∼300 mg of LV was immediately quick-frozen in liquid nitrogen. Liver and gastrocnemius were also removed, weighed, and stored at −80°C.
Analytic Procedures
Cellular compartment separation.
In study 1, a freshly isolated sample of LV was used for isolation of cardiac subsarcolemmal mitochondria. This was done by placing the tissue in modified Chappell-Perry buffer, CP1 (containing 100 mM KCl, 50 mM MOPS, 1 mM EGTA, 5 mM MgSO4·7H2O, and 1 mM ATP, pH 7.4, at 4°C). The isolation was based on the protocol by Palmer et al. (25), except for the usage of CP1 buffer. Citrate synthase was measured as previously described to determine the purity of the mitochondria (32). The protein concentration in each tissue was measured by spectrophotometer assay kit (Lowry method) using bovine serum albumin as a standard.
2H labeling of body water.
Body water enrichment was measured as previously described by McCabe et al. (20). Briefly, plasma was centrifuged for 10 min at 4,000 rpm, and 20 μl of plasma (or standard) was reacted with 2 μl of 10 N NaOH and 4 μl of a 5% (vol/vol) solution of acetone in acetonitrite overnight at room temperature. Acetone was extracted following the addition of 500 μl of chloroform and 0.5 g of Na2SO4. Each sample was mixed thoroughly, and an aliquot of the chloroform was transferred to a GC-MS vial. 2H labeling of acetone was analyzed using an Agilent 5973N-MSD equipped with an Agilent 6890 GC system. A DB17-MS capillary column (30 m × 0.25 mm × 0.25 μm) was used. The initial program temperature was set at 60°C, increased by 20°C/min to 100°C, increased by 50°C/min to 220°C, and held for 1 min, with a helium flow of 1 ml/min. Acetone elutes at ∼1.5 min. The mass spectrometer was operated in the electron impact mode (70 eV). Selective ion monitoring of m/z 58 and 59 was performed using a dwell time of 10 ms per ion.
2H labeling of protein-bound alanine.
Alanine enrichment was measured as previously described by Dufner et al. (6). Briefly, total protein synthesis in a sample was determined by homogenizing 0.1 g of tissue in 1,000 μl of 6% PCA (wt/vol) and centrifuging for 10 min at 4,000 rpm. The supernatant was then discarded. The pellet was washed with 3% PCA and hydrolyzed for 20 h in 1 ml of 6 N HCl at 100°C.
Rates of mitochondrial and cytosolic protein synthesis were determined as follows. Mitochondria were dissolved in KME [100 mM KCl, 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), internal salt, and 0.5 mM EGTA], and 1,000 μl of 6 N HCl was used to hydrolyze 200 μl of mitochondrial solution. Cytosolic fractions were in CP1 buffer, and 500 μl of 6 N HCl was used to hydrolyze 500 μl of cytosolic solution.
An aliquot of the hydrolyzed protein fractions (i.e., total, mitochondrial, or cytosolic) was dried by vacuum centrifuge for 30–60 min. The samples were then reacted to form the “methyl-8” derivative of alanine, made by mixing acetonitrile, methanol, and Methyl-8 Reagent (1:2:3, vol/vol/vol; Pierce) and heating the sample at 75°C for 30 min (35). The sample was transferred to a GC vial and analyzed using the Agilent 5973N-MSD equipped with the Agilent 6890 GC system. A DB17-MS capillary column (30 m × 0.25 mm × 0.25 μm) was used in all assays. The initial temperature program was set at 90°C and held for 5 min, increased by 5°C/min to 130°C, increased by 40°C/min to 240°C, and held for 5 min, with a helium flow of 1 ml/min. Alanine elutes at ∼12 min. The mass spectrometer was operated in the electron impact mode. Selective ion monitoring of m/z 99 and 100 (total 2H labeling of alanine) was performed using a dwell time of 10 ms per ion.
Western Blot Analyses
Tissue was homogenized in buffer containing of 20 mM Tris, pH7.6, 0.1 mM EDTA, 0.5 mM EGTA, 0.1% Triton X, 250 mM sucrose, and protease and phosphatase inhibitors. The protein concentration of total cell lysates was measured with a Bio-Rad Dc protein assay kit. Total cell lysates (40 μg protein) were separated by SDS-PAGE, transferred to PVDF membranes, and probed with antibodies by standard procedures. Western blots were quantified and used to calculate the phosphorylation ratio.
Calculations
Protein synthetic rate.
The rate(s) of protein synthesis was calculated using the formula
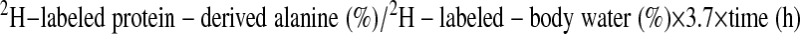 |
(1) |
where the factor 3.7 represents an incomplete exchange of 2H between body water and alanine; i.e., 3.7 of the 4 carbon-bound hydrogens of alanine exchange with water (6, 29). This formula assumes that the 2H labeling in body water equilibrates with free alanine more rapidly than alanine is incorporated into newly made protein and that protein synthesis is linear over the study (6, 43).
The percent change(s) of protein synthesis in various samples was determined by comparing the individual rats in an experimental group (e.g., acutely fasted) against the mean of the respective control group (e.g., ad libitum fed) using the formula
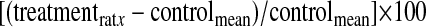 |
(2) |
where treatmentrat x refers to a single rat in the treatment group. The mean ± SE was calculated.
Statistical analysis.
Comparisons were made between control and treatment groups within each study by use of Student's t-test. A one-way ANOVA was run to determine differences in the percent change among tissues within a given experiment, and post hoc testing was done using a Bonferroni test (P < 0.05). Unless noted, all values are presented as means ± SE.
RESULTS
Study 1: Effect of Acute Fasting
Fasting for 20 h decreased body mass (P < 0.01) and caused an ∼30% reduction in protein synthesis in the liver and gastrocnemius muscle (Table 1). There was no difference in either total heart or LV mass, and LV protein synthesis was not altered by fasting (Table 1). Furthermore, when LV tissue was fractionated into mitochondrial and cytosolic compartments, there were no differences in protein synthesis between fed and fasted animals (Table 1). When the change in the rate of protein synthesis between the fed rats was compared with fasted animals, the percent change was significantly greater (P < 0.05, one-way ANOVA) in the liver and skeletal muscle compared with the heart, i.e., −23.2 ± 4.3, −33.1 ± 8.6 and −3.3 ± 4.0%, respectively.
Table 1.
General characteristics and 2H labeling of fed vs. acutely fasted rats
| Study 1 | Fed | Fasted |
|---|---|---|
| Initial body mass, g | 311±4 | 311±3 |
| Terminal body mass, g | 367±6 | 345±3* |
| Heart mass, g | 1.02±0.04 | 0.99±0.02 |
| LV mass, mg | 804±38 | 791±18 |
| LV/tibial length, mg/cm | 189±8 | 184±4 |
| 2H labeling body water, % enrichment | 2.72±0.10 | 2.57±0.09 |
| 2H labeling protein-bound alanine, %enrichment | ||
| LVwhole tissue | 0.37±0.02 | 0.36±0.02 |
| LVmitochondrial compartment | 0.57±0.05 | 0.49±0.07 |
| LVcytosolic compartment | 0.39±0.03 | 0.38±0.02 |
| Liver | 1.56±0.04 | 0.99±0.18* |
| Gastrocnemius | 0.21±0.02 | 0.14±0.02† |
| Protein synthesis rate, %newly made/h | ||
| LVwhole tissue | 0.62±0.04 | 0.58±0.02 |
| LVmitochondrial compartment | 0.98±0.14 | 0.81±0.13 |
| LVcytosolic compartment | 0.65±0.04 | 0.62±0.04 |
| Liver | 2.65±0.10 | 1.96±0.06* |
| Gastrocnemius | 0.35±0.04 | 0.23±0.03† |
Body mass was determined as well as that of various tissues. LV, left ventricle. 2H labeling used to calculate protein synthesis is shown. In all cases, data are shown as means ± SE; n = 8 per group.
P < 0.01,
P < 0.05 vs. fed.
Study 2: Effect of Chronic Dietary Restriction
Dietary restriction lowered whole body and liver mass (P < 0.05) (Table 2). LV mass was lower when expressed in absolute terms; however, there was no significant difference when normalized to tibia length (Table 2). Chronic food restriction significantly reduced protein synthesis in liver and gastrocnemius muscle but did not affect rates of protein synthesis in the heart (Table 2). When the change in the rate of protein synthesis between the ad libitum-fed rats was compared with the food-restricted group (Eq. 2), the percent change was significantly greater (P < 0.05, one-way ANOVA) in the liver and skeletal muscle compared with the heart, i.e., −8.3 ± 3.4, −14.2 ± 3.5, and +2.1 ± 2.3%, respectively.
Table 2.
General characteristics and 2H labeling of control vs. dietary restricted rats
| Study 2 | Ad Libitum Fed | Calorie Restricted |
|---|---|---|
| Initial body mass, g | 356±3 | 355±3 |
| Terminal body mass, g | 355±6 | 333±3† |
| Change in body mass, % | −0.3±0.6 | −6.7±1.1* |
| Food intake, g/day | 17.3±0.4 | 10.9±0.9* |
| Liver mass, g | 11.34±0.38 | 8.27±0.96* |
| Heart mass, g | 0.92±0.04 | 0.84±0.02 |
| LV mass, mg | 769±32 | 688±16† |
| LV/tibial length, mg/cm | 191±6 | 178±6 |
| 2H labeling body water, %enrichment | 3.10±0.07 | 3.40±0.04* |
| 2H labeling protein-bound alanine, %enrichment | ||
| LVwhole tissue | 0.57±0.04 | 0.63±0.02 |
| Liver | 1.41±0.03 | 1.45±0.03 |
| Gastrocnemius | 0.45±0.02 | 0.43±0.02 |
| Protein synthesis rate, %newly made/h | ||
| LVwhole tissue | 0.91±0.06 | 0.92±0.04 |
| Liver | 2.26±0.06 | 2.12±0.03† |
| Gastrocnemius | 0.72±0.02 | 0.62±0.02* |
Body mass was determined as well as that of various tissues. 2H labeling used to calculate protein synthesis is shown. In all cases, data are shown as means ± SE; n = 8 per group.
P < 0.01,
P < 0.05 vs. control.
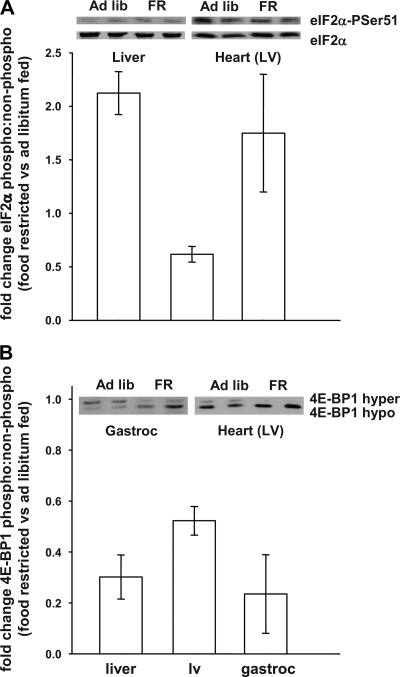
To explain the sustained protein synthesis in the heart compared with the liver and skeletal muscle, we determined the phosphorylation status of two translation initiation factors that are known to regulate protein synthesis rates in a manner that is dependent on nutrient supply (18, 30). The degree of phosphorylation of 4E-BP1 and eIF2α was measured in the three sites (Fig. 1). Those data demonstrate that chronic food restriction (compared with ad libitum feeding) leads to an increase (∼100%) in the phosphorylation of eIF2α in the liver and muscle but a decrease (∼45%) in the heart, consistent with the observed alterations in protein synthesis (Table 2; chronic food restriction leads to a reduction in protein synthesis in liver and muscle with no effect in heart). Figure 1 also demonstrates a reduction in the phosphorylation of 4E-BP1 with chronic food restriction vs. ad libitum feeding in all three tissues, with the largest effects in liver and skeletal muscle compared with heart (∼4-fold vs. ∼2-fold); this is consistent with the changes in protein synthesis (Table 2; protein synthesis is reduced in liver and skeletal muscle but not in heart). In total, these data suggest that the differential effects of chronic food restriction on tissue-specific protein synthesis is (partly) mediated by a combination of molecular mechanisms, including an increased phosphorylation of eIF2α and a more pronounced decrease in the phosphorylation of 4E-BP1 (i.e., in liver and skeletal muscle but not heart).
Fig. 1.
Changes in regulation of protein translation machinery. Immunoblots were probed to examine changes in the phosphorylation state of eukaryotic initiation factor (eIF2)α and eIF4E-binding protein-1 (4E-BP1). A: fold change of eIF2α, expressed as ratio of eIF2αphospho-Ser51 to eIF2α in food-restricted (FR) vs. ad libitum-fed rats for a given tissue. B: fold change in 4E-BP1, expressed as fold change in hyper- to hypophosphorylated protein in food-restricted vs. ad libitum-fed rats for a given tissue. Data are calculated as (food restrictedrat x/ad lib fedmean) and expressed as means ± SE; n = 3. LV, left ventricle.
DISCUSSION
In the present report, we determined tissue-specific rates of protein synthesis in different models involving nutritional manipulation using a novel 2H2O tracer method (2, 3, 6, 29). This method is based on establishing a precursor-to-product labeling ratio (36, 43); i.e., following the administration of 2H2O, one determines the rate of protein synthesis by measuring the incorporation of [2H]alanine into a protein(s) of interest. We recently demonstrated that, since there is rapid equilibration of 2H between body water and free alanine, 2H2O is well suited for measuring metabolic flexibility during a meal in vivo (6). That observation is consistent with the literature regarding alanine flux measurements (15, 23, 44, 45) and is supported by other recent investigations in which 2H2O was used to quantify protein synthesis in vivo (2, 3). It is important to note that the use of 2H2O was recently validated against the use of a constant infusion of labeled leucine under conditions of a metabolic steady state. However, we suspected that use of a constant infusion of a labeled amino acid (e.g., [13C]leucine) would be problematic in our study since, in addition to requiring catheters, the labeling of the precursor amino acid would be diluted via the influx of cold amino acids during the meal. Thus, the 2H2O method was used since one expects a steady-state 2H labeling of alanine even during extreme conditions (6, 29); i.e., alanine has a rapid turnover (the t1/2 is ∼10 min in humans) (15, 23, 44, 45).
Our overall observations regarding the relative rates of protein synthesis in different tissues are consistent with reports in the literature (10, 11, 26, 28, 33). For example, rates of liver protein synthesis are generally greater than rates of protein synthesis in heart, which are generally greater than rates of protein synthesis in skeletal muscle, e.g., 2.65, 0.62, and 0.35% newly made protein in liver, heart, and skeletal muscle of fed rats, respectively (Table 1, fed rats). Those values are in agreement with studies in which the flooding dose was used to study protein synthesis in rodent models (8, 22). However, an important point to make regarding the use of 2H2O is that the method yields a measurement of protein synthesis over a wide experimental window (∼180–360 min) compared with the flooding dose that is typically used for ∼10–60 min. This is of interest since animals, even when trained to eat via a feeding schedule, require upward of 60 min simply to consume a reasonable amount of food. Furthermore, when using the flooding dose, and studying protein synthesis over a relatively short window, one then extrapolates rates of protein synthesis determined over ∼10–60 min to draw conclusions regarding events that may occur over hours. In that regard, the 2H2O method provides a unique picture of protein dynamics, since one can directly determine rates of protein synthesis over reasonably short intervals (e.g., several hours) or quantify protein synthesis over prolonged periods (e.g., days or weeks) (1–3, 6, 29), therein allowing one to capture the true cumulative effect(s) of temporal changes in the availability of hormones and substrates.
Previous studies found that whole body protein synthesis is reduced by fasting and food restriction (11, 12, 41, 42); however, little was known about the response of the heart to these perturbations. The present study demonstrates that the heart is resistant to the suppression in protein synthesis that occurs in the liver and skeletal muscle in response to a decrease in food intake. For example, although protein synthesis in liver and gastrocnemius muscle was significantly reduced by fasting, there were no changes in protein synthesis in the LV of the heart either for the total protein pool or in isolated mitochondrial or cytosolic compartments (Table 1). Likewise, a chronic reduction in protein energy intake induced by food restriction did not affect protein synthesis in the heart, whereas we observed decreased protein synthesis in skeletal muscle and liver (Table 2). These results clearly demonstrate that cardiac protein synthesis is maintained in response to changes in food intake, in strong contrast to liver and gastrocnemius muscle. It should be emphasized that, in addition to affecting nitrogen calorie balance, our model of dietary restriction also impacts vitamin and mineral intake; i.e., those nutrients were not supplemented to achieve intakes comparable to the ad libitum group. Duffy et al. (5) have demonstrated that reductions in vitamin and mineral intake, in the approximate range as expected in our study, did not affect life-time survival. Therefore, the effects on protein synthesis are presumably related to changes in macronutrient intake, and alterations in micronutrients are not of immediate concern.
We were surprised by the novel finding that cardiac protein synthesis is not affected by acute fasting (Table 1), especially since digestion and absorption trigger the release of insulin and provide amino acid substrates that are needed for the synthesis of new proteins. Nevertheless, our observation agrees with the studies by Young and colleagues (21, 46) in which phenylalanine tracer balance was directly measured across the heart in humans or dogs. Namely, the acute infusion of insulin had no stimulatory effect on protein synthesis; rather, the major effect was an inhibition of protein breakdown. Our observation differs slightly from other work by Young and colleagues, who demonstrated a potential role for branched-chain amino acids in affecting cardiac protein synthesis (47). In a related study (Gilge DA, Bederman IR, Ruszczycky MW, Anderson VE, Ernsberger PE, Loukili O, Rachdaoui B, Previs SF, unpublished observations), we measured protein synthesis via 2H2O labeling in rats following an intraperitoneal bolus of saline (sham) vs. a bolus of glucose plus branched-chain amino acids. Although we observed a stimulation of protein synthesis in skeletal muscle (as expected in the rats given glucose plus branched-chain amino acids), we did not see any effect on protein synthesis in heart. Those findings suggest that the stimulatory effect of branched-chain amino acids on protein synthesis in cardiac tissue may be limited to conditions in which substrates are continuously available in high abundance. In total, our data suggest that attempts to acutely modulate cardiac protein synthesis via an acute nutritional intervention(s) may be of limited efficacy.
Our second study aimed to determine the effect(s) of chronic protein energy restriction on protein synthesis. This appears to be a somewhat novel study, since others have not examined this question, and is of particular importance since sudden cardiac failure has been reported in cases where dietary restriction is used to achieve rapid weight loss (38, 39). Surprisingly, we did not observe any effect on protein synthesis in the heart during chronic dietary restriction (Table 2). In contrast, we did observe reductions in protein synthesis in liver and in skeletal muscle. It is important to note that, although we observed statistically significant reductions in liver and skeletal muscle protein synthesis in chronically food restricted vs. control rats, the magnitude of those changes is relatively small. We suspect that the nature of the dietary restriction may have played a role. For example, rats were maintained via a feeding schedule. Thus, the patterns of protein energy consumption may have affected the outcome. It is likely that the dietary restricted rats received a relatively large bolus of food over the period when protein synthesis was determined; namely, the food-restricted rats and their respective controls were studied in the fed state. Since feeding does stimulate protein synthesis (Table 1), it was reasonable to expect small differences between liver and muscle in study 2.
We were intrigued by the fact that calorie restriction did not influence protein synthesis in heart but did impact liver and skeletal muscle. The differential effects seen in heart vs. skeletal muscle are particularly interesting since, unlike liver, one expects that those tissues are presented with identical concentration profiles of amino acids and hormones after a meal; thus, one would not expect an extracellular signal to explain the effect(s) on protein synthesis. Consequently, we examined components of the intracellular signal transduction system, i.e., the phosphorylation state of 4E-BP1 and eIF2α (Fig. 1). We observed larger reductions in the phosphorylation of 4E-BP1 in liver and skeletal muscle compared with heart (i.e., an ∼4-fold decreases vs. an ∼2-fold decrease, in calorie-restricted vs. ad libitum-fed rats, respectively). The observed changes in the phosphorylation status of 4E-BP1 by food restriction in the three tissues are in agreement with dietary studies in whole animals (7, 9, 24, 31, 37). However, the magnitude and direction of changes in the phosphorylation of eIF2α in liver and skeletal muscle vs. heart (i.e., an ∼2-fold increases vs. an ∼1.5-fold decrease in calorie-restricted vs. ad libitum-fed rats, respectively) were surprisingly different, suggesting that the heart sustains protein synthesis during food restriction via molecular mechanisms that do not increase eIF2α phosphorylation (48). Although these signaling mechanisms were examined in a small subgroup of animals (n = 3), making it difficult to perform statistical measurements, the changes in the phosphorylation status of the two translation factors complement each other (and presumably serve to reinforce the regulation of protein translation). To our knowledge, this novel finding regarding the effect(s) of chronic food restriction on tissue-specific changes in protein synthesis merits further investigation. For example, although these observations suggest some molecular mechanisms that could explain the differential effects of nutritional status on protein synthesis, future studies might consider measuring protein synthesis while 1) quantifying the temporal changes in the response of the signaling pathways following a meal (6) and/or 2) manipulating (tissue-specific) signal transduction. Those types of investigations might suggest the relative importance of the proteins that control translation.
Despite the fact that protein turnover in rat cardiac muscle is fairly active and is characterized by a t1/2 of ∼4.7 days, our observations suggest that protein synthesis in cardiac muscle is preserved over a range of conditions. For example, the normal daily perturbation of eating stimulates protein synthesis in muscle and liver by ∼33 and 23%, respectively, yet there is no effect on protein synthesis in heart. Our data suggest that 1) during periods of reduced dietary intake cardiac protein synthesis may be preserved by the pool of stored amino acids in the body (e.g., skeletal muscle protein), and/or 2) cardiac protein homeostasis is more strongly influenced by changes in protein breakdown (46).
In summary, this study assessed the extent to which protein synthesis in cardiac muscle responds to dietary perturbations, compared with protein synthesis in liver and skeletal muscle, using a novel 2H2O method in rats. The primary aim was to determine whether protein synthesis in cardiac muscle would be affected by dietary manipulation; attention was also directed toward the study of the liver and gastrocnemius muscle. Although protein synthesis in liver and gastrocnemius muscle were significantly reduced by acute fasting, there were no changes in protein synthesis in the left ventricle of the heart for either the total protein pool or in isolated mitochondrial or cytosolic compartments. Likewise, a chronic reduction in calorie intake, induced by food restriction, did not affect protein synthesis in the heart, whereas we observed decreased protein synthesis in skeletal muscle and liver. We conclude that cardiac protein is maintained in response to changes in food intake, in strong contrast to liver and gastrocnemius muscle where protein synthesis is decreased by sudden fasting or chronic food restriction. In addition, our studies have identified two potential intracellular targets that may explain the differential modulation of protein synthesis in vivo in specific tissues.
Acknowledgments
This research was supported by the National Institutes of Health Road Map DK-070291-01, HL-074237, and Training Grant DK-007319 (to D. A. Gilge) and DK-060596 and DK-053307 (to M. Hatzoglou), American Heart Association-Ohio Valley Predoctoral Fellowship (to N. Sharma), and the Mt. Sinai Health Care Foundation, Cleveland, OH.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Bederman IR, Dufner DA, Alexander JC, Previs SF. Novel application of the “doubly labeled” water method: measuring CO2 production and the tissue-specific dynamics of lipid and protein in vivo. Am J Physiol Endocrinol Metab 290: E1048–E1056, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Belloto E, Diraison F, Basset A, Allain G, Abdallah P, Beylot M. Determination of protein replacement rates by deuterated water: validation of underlying assumptions. Am J Physiol Endocrinol Metab 292: E1340–E1347, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Busch R, Kim YK, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta 1760: 730–744, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Carnevalli LS, Pereira CM, Jaqueta CB, Alves VS, Paiva VN, Vattem KM, Wek RC, Mello LE, Castilho BA. Phosphorylation of the alpha subunit of translation initiation factor-2 by PKR mediates protein synthesis inhibition in the mouse brain during status epilepticus. Biochem J 397: 187–194, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy PH, Lewis SM, Mayhugh MA, McCracken A, Thorn BT, Reeves PG, Blakely SA, Casciano DA, Feuers RJ. Effect of the AIN-93M purified diet and dietary restriction on survival in Sprague-Dawley rats: implications for chronic studies. J Nutr 132: 101–107, 2002. [DOI] [PubMed] [Google Scholar]
- 6.Dufner DA, Bederman IR, Brunengraber DZ, Rachdaoui N, Ismail-Beigi F, Siegfried BA, Kimball SR, Previs SF. Using 2H2O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. Am J Physiol Endocrinol Metab 288: E1277–E1283, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Escobar J, Frank JW, Suryawan A, Nguyen HV, Kimball SR, Jefferson LS, Davis TA. Regulation of cardiac and skeletal muscle protein synthesis by individual branched-chain amino acids in neonatal pigs. Am J Physiol Endocrinol Metab 290: E612–E621, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Estornell E, Cabo J, Barber T. Protein synthesis is stimulated in nutritionally obese rats. J Nutr 125: 1309–1315, 1995. [DOI] [PubMed] [Google Scholar]
- 9.Frank JW, Escobar J, Suryawan A, Kimball SR, Nguyen HV, Jefferson LS, Davis TA. Protein synthesis and translation initiation factor activation in neonatal pigs fed increasing levels of dietary protein. J Nutr 135: 1374–1381, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Garlick PJ, McNurlan MA, Essen P, Wernerman J. Measurement of tissue protein synthesis rates in vivo: a critical analysis of contrasting methods. Am J Physiol Endocrinol Metab 266: E287–E297, 1994. [DOI] [PubMed] [Google Scholar]
- 11.Garlick PJ, Millward DJ, James WP. The diurnal response of muscle and liver protein synthesis in vivo in meal-fed rats. Biochem J 136: 935–945, 1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garlick PJ, Wernerman J, McNurlan MA, Heys SD. Organ-specific measurements of protein turnover in man. Proc Nutr Soc 50: 217–225, 1991. [DOI] [PubMed] [Google Scholar]
- 14.Hershey J, Merrick W. The pathway and mechanism of initiation of protein synthesis. In: Translational Control of Gene Expression, edited by Sonenberg N. Cold Spring Harbor, IL: Cold Spring Harbor Laboratory, 2000, p. 3388.
- 15.Hoffer LJ, Yang RD, Matthews DE, Bistrian BR, Bier DM, Young VR. Alanine flux in obese and healthy humans as evaluated by 15N- and 2H3-labeled alanines. Am J Clin Nutr 48: 1010–1014, 1988. [DOI] [PubMed] [Google Scholar]
- 16.Jefferson LS, Rannels DE, Munger BL, Morgan HE. Insulin in the regulation of protein turnover in heart and skeletal muscle. Fed Proc 33: 1098–1104, 1974. [PubMed] [Google Scholar]
- 17.Jessani S, Lip GY. Death or heart failure post acute myocardial infarction? The role of cardiac biomarkers. J Card Fail 12: 641–643, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Kimball SR Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol 26: 155–184, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Kimball SR, Siegfried BA, Jefferson LS. Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J Biol Chem 279: 54103–54109, 2004. [DOI] [PubMed] [Google Scholar]
- 20.McCabe BJ, Bederman IR, Croniger C, Millward C, Norment C, Previs SF. Reproducibility of gas chromatography-mass spectrometry measurements of 2H labeling of water: application for measuring body composition in mice. Anal Biochem 350: 171–176, 2006. [DOI] [PubMed] [Google Scholar]
- 21.McNulty PH, Jacob R, Deckelbaum LI, Young LH. Effect of hyperinsulinemia on myocardial amino acid uptake in patients with coronary artery disease. Metabolism 49: 1365–1369, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Mosoni L, Malmezat T, Valluy MC, Houlier ML, Mirand PP. Muscle and liver protein synthesis adapt efficiently to food deprivation and refeeding in 12-month-old rats. J Nutr 126: 516–522, 1996. [DOI] [PubMed] [Google Scholar]
- 23.Nissim I, Lapidot A. Plasma amino acid turnover rates and pools in rabbits: in vivo studies using stable isotopes. Am J Physiol Endocrinol Metab Gastrointest Physiol 237: E418–E427, 1979. [DOI] [PubMed] [Google Scholar]
- 24.Orellana RA, Jeyapalan A, Escobar J, Frank JW, Nguyen HV, Suryawan A, Davis TA. Amino acids augment muscle protein synthesis in neonatal pigs during acute endotoxemia by stimulating mTOR-dependent translation initiation. Am J Physiol Endocrinol Metab 293: E1416–E1425, 2007. [DOI] [PubMed] [Google Scholar]
- 25.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977. [PubMed] [Google Scholar]
- 26.Preedy VR, Smith DM, Kearney NF, Sugden PH. Rates of protein turnover in vivo and in vitro in ventricular muscle of hearts from fed and starved rats. Biochem J 222: 395–400, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preedy VR, Smith DM, Kearney NF, Sugden PH. Regional variation and differential sensitivity of rat heart protein synthesis in vivo and in vitro. Biochem J 225: 487–492, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preedy VR, Why H, Paice AG, Reilly ME, Ansell H, Patel VB, Richardson PJ. Protein synthesis in the heart in vivo, its measurement and patho-physiological alterations. Int J Cardiol 50: 95–106, 1995. [DOI] [PubMed] [Google Scholar]
- 29.Previs SF, Fatica R, Chandramouli V, Alexander JC, Brunengraber H, Landau BR. Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end-stage renal disease. Am J Physiol Endocrinol Metab 286: E665–E672, 2004. [DOI] [PubMed] [Google Scholar]
- 30.Proud CG Regulation of protein synthesis by insulin. Biochem Soc Trans 34: 213–216, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Reiter AK, Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. Repression of protein synthesis and mTOR signaling in rat liver mediated by the AMPK activator aminoimidazole carboxamide ribonucleoside. Am J Physiol Endocrinol Metab 288: E980–E988, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Srere PA, Matsuoka Y, Mukherjee A. Inhibition studies of rat citrate synthase. J Biol Chem 248: 8031–8035, 1973. [PubMed] [Google Scholar]
- 33.Stein TP, Oram-Smith JC, Leskiw MJ, Wallace HW, Long LC, Leonard JM. Effect of nitrogen and calorie restriction on protein synthesis in the rat. Am J Physiol 230: 1321–1325, 1976. [DOI] [PubMed] [Google Scholar]
- 34.Sudhakar A, Ramachandran A, Ghosh S, Hasnain SE, Kaufman RJ, Ramaiah KV. Phosphorylation of serine 51 in initiation factor 2 alpha (eIF2 alpha) promotes complex formation between eIF2 alpha(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry 39: 12929–12938, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Thenot JP, Horning EC. Amino acid N-dimethylaminomethylene alkyl esters. New derivatives for gas chromatographic and gas chromatographic-mass spectrometric studies. Anal Lett 5: 519–529, 1972. [Google Scholar]
- 36.Ussing HH The rate of protein renewal in mice and rats studied by means of heavy hydrogen. Acta Physiol Scand 2: 209–221, 1941. [Google Scholar]
- 37.Vary TC, Deiter G, Lantry R. Chronic alcohol feeding impairs mTOR(Ser 2448) phosphorylation in rat hearts. Alcohol Clin Exp Res 32: 43–51, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Vertes V, Genuth SM, Hazelton IM. Abuse of the supplemented fast. JAMA 238: 2179, 1977. [PubMed] [Google Scholar]
- 39.Vertes V, Genuth SM, Hazelton IM. Precautions with supplemented fasting. JAMA 238: 2142, 1977. [PubMed] [Google Scholar]
- 40.Waterlow JC Total protein turnover in animals and man. Nutr Rev 28: 115–118, 1970. [DOI] [PubMed] [Google Scholar]
- 41.Waterlow JC Metabolic adaptation to low intakes of energy and protein. Annu Rev Nutr 6: 495–526, 1986. [DOI] [PubMed] [Google Scholar]
- 42.Waterlow JC Whole-body protein turnover in humans—past, present, and future. Annu Rev Nutr 15: 57–92, 1995. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe RR, Chinkes DL. Isotope Tracers in Metabolic Research: Principles and Practice of Kinetic Analyses. New York: Wiley-Liss, 2004.
- 44.Yang RD, Matthews DE, Bier DM, Lo C, Young VR. Alanine kinetics in humans: influence of different isotopic tracers. Am J Physiol Endocrinol Metab 247: E634–E638, 1984. [DOI] [PubMed] [Google Scholar]
- 45.Yang RD, Matthews DE, Bier DM, Wen ZM, Young VR. Response of alanine metabolism in humans to manipulation of dietary protein and energy intakes. Am J Physiol Endocrinol Metab 250: E39–E46, 1986. [DOI] [PubMed] [Google Scholar]
- 46.Young LH, Dahl DM, Rauner D, Barrett EJ. Physiological hyperinsulinemia inhibits myocardial protein degradation in vivo in the canine heart. Circ Res 71: 393–400, 1992. [DOI] [PubMed] [Google Scholar]
- 47.Young LH, McNulty PH, Morgan C, Deckelbaum LI, Zaret BL, Barrett EJ. Myocardial protein turnover in patients with coronary artery disease. Effect of branched chain amino acid infusion. J Clin Invest 87: 554–560, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, Cavener DR. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 22: 6681–6688, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]