Abstract
Multiple copies of the hexamer TGCATG have been shown to regulate fibronectin pre-mRNA alternative splicing. GCATG repeats also are clustered near the regulated calcitonin-specific 3′ splice site in the rat calcitonin/CGRP gene. Specific mutagenesis of these repeats in calcitonin/CGRP pre-mRNA resulted in the loss of calcitonin-specific splicing, suggesting that the native repeats act to enhance alternative exon inclusion. Mutation of subsets of these elements implies that alternative splicing requires a minimum of two repeats, and that the combination of one intronic and one exonic repeat is necessary for optimal cell-specific splicing. However, multimerized intronic repeats inhibited calcitonin-specific splicing in both the wild-type context and in a transcript lacking endogenous repeats. These results suggest that both the number and distribution of repeats may be important features for the regulation of tissue-specific alternative splicing. Further, RNA containing a single repeat bound cell-specific protein complexes, but tissue-specific differences in protein binding were not detected by using multimerized repeats. Together, these data support a novel model for alternative splicing regulation that requires the cell-specific recognition of multiple, distributed sequence elements.
The search for regulatory sequences in alternatively spliced pre-mRNAs has led to the definition of a variety of sequence elements that are necessary for normal cell-specific splicing (for reviews, see refs. 1–4). These include constitutive sequences necessary for general splicing as well as novel sequences that apparently respond to specific trans-acting factors (5–7). Competition between alternative exons depends on the relative quality of the constitutive splicing signals, a balance that presumably can be shifted by cell-specific factors acting on regulatory sequences. Further, a cell-specific protein complex binding to a regulatory sequence in c-src pre-mRNA was shown to include ubiquitously expressed proteins, raising the possibility that cell-specific modifications in constitutive proteins also may contribute to alternative splicing regulation (7, 8). However, in most cases, the molecular mechanisms underlying tissue-specific alternative splicing remain to be defined.
Recently, a repeated hexanucleotide element, TGCATG, was shown to regulate the tissue-specific splicing of the fibronectin alternative exon EIIIB (9). The EIIIB exon is included in transfected F9 cells, but excluded in transfected HeLa cells. In fibronectin pre-mRNA, TGCATG repeats occur within the intron downstream of EIIIB. Deletion of intron sequences containing these elements caused EIIIB skipping in F9 cells, whereas replacement of these sequences with synthetic TGCATG repeats restored EIIIB inclusion in these cells. These elements may positively activate splicing of the 5′ alternative exon (9); alternatively, they may act negatively by delaying splicing of the 3′ constitutive exon, thus allowing a kinetic advantage for EIIIB recognition.
Extensive mutational analyses to define regulatory sequences in alternative splicing of rat calcitonin/CGRP pre-mRNA have suggested that although constitutive signals play a role in cell-specific splicing, no single element is responsible for the regulation of tissue-specific splicing (10–12). However, these analyses did not rule out a potential regulatory role for multiple redundant sequence elements. In the rat calcitonin/CGRP gene, GCATG repeats occur nine times over a 3.5-kb region, and five repeats are clustered within 490 nucleotides of the regulated calcitonin-specific 3′ splice site. Of the five repeats within the alternatively spliced portion of the gene, three consist of the TGCATG hexamer. The appearance of eight GCATG repeats in the same region of the human gene suggested that these elements may play a role in the control of calcitonin/CGRP alternative splicing (9).
We now show that specific mutagenesis of all five of the endogenous repeats in the alternatively spliced region of the rat calcitonin/CGRP gene resulted in the loss of calcitonin-specific splicing in stably transfected HeLa cells that normally include this exon. Therefore, the endogenous repeats in the wild-type gene serve to enhance the inclusion of the calcitonin-specific exon. Neither single copies of the repeat nor multimerized repeats were able to restore cell-specific splicing of the mutant transcript. Strikingly, the insertion of multimerized repeats in an otherwise wild-type gene also resulted in the loss of calcitonin-specific splicing, suggesting that the distributed arrangement of repeats is a critical aspect of their function. The possibility that single repeats act differently than multimerized repeats is supported by the observation that RNA containing multimerized copies of GCAUG binds similar complexes of nuclear proteins from cells making either calcitonin or CGRP mRNA. However, RNA containing a single GCAUG sequence bound distinct, cell-specific nuclear protein complexes. These results suggest that cell-specific alternative splicing of calcitonin/CGRP pre-mRNA depends on repetitive, dispersed sequence elements that bind cell-specific trans-acting factors.
MATERIALS AND METHODS
In Vivo Splicing Assays.
Alternative splicing of calcitonin/CGRP transcripts was assayed as previously described (11). Briefly, an expression plasmid containing a 7.2-kb rat genomic fragment in pSV2neo was stably transfected into HeLa and F9 cells. Mutations in the rat calcitonin/CGRP gene were made either by site-directed mutagenesis or by subcloning annealed oligonucleotide pairs. Total RNA was isolated by guanidine isothiocyanate lysis and assayed by RNase protection (14), by using a chimeric genomic probe consisting of the entire calcitonin-specific exon 4 with 154 nucleotides of intron 3 fused to the entire CGRP-specific exon 5 with 120 nucleotides of intron 4. Probes specific for each mutation were generated by PCR, and protected fragments were quantitated by densitometry of autoradiographic films. Quantitative data are expressed as ++++ = 80–100%, +++ = 60–80%, ++ = 40–60%, + = 20–40%, and − = 0–20%.
Constructs.
Site-directed mutagenesis was used to convert each endogenous GCATG sequence to a restriction site. Because the quality of the constitutive splicing signals are important for alternative calcitonin/CGRP splicing (12), the mutations in the current study were made such that the branchpoint and polypyrimidine stretch were preserved. The repeats at−254,−34,−19, +45, and +236 (relative to the calcitonin-specific 3′ splice site) were changed to SmaI, BamHI, SmaI, BamHI, and ApaI sites, respectively. For insertion of multimerized repeats, the introduced site was used to insert either three copies of GATCCTGCATGACTACTGCTTGCATGCTCGTCATA (R6, originally named T3 in ref. 9), three copies of the mutant sequence GATCCAGTCGTACTACTGCTAGTCGTCTCGTCATA (S6, originally called S3 in ref. 9), or five copies of ACTGCATGGCT, derived from the −254 repeat in the rat calcitonin/CGRP gene (C5). For RNA probes, annealed oligonucleotides representing a single −254 repeat and 13–15 bp of flanking sequences, a mutant repeat in the same context (GCATG to CCCGG), or five copies of the −254 repeat (insert C5) were subcloned into pSP72 (Promega). Full sequence information is available on request.
Gel Mobility-Shift Assays.
RNA gel mobility-shift assays were performed as described (13). Short RNA probes were transcribed from templates consisting of linearized plasmids using a T7 Megashortscript kit (Ambion). Radiolabeled RNA (50,000 cpm, about 10 ng) was incubated with 1 μg yeast tRNA and 1 μl nuclear extract in 12 mM Tris⋅HCl, pH 7.6, 60 mM KCl and 3.2 mM MgCl2 on ice for 10 min. Nuclear extracts were prepared from HeLa, B50 (rat neuronal tumor), and CA (rat thyroid C cell tumor) cell lines (14). Samples were separated on 5% polyacrylamide gels in 50 mM Tris⋅glycine, pH 8.0 at 10 V/cm.
RESULTS
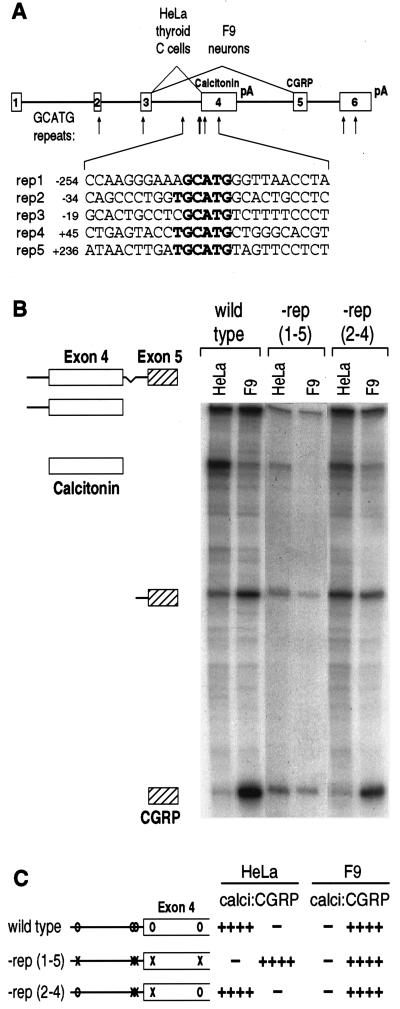
In the rat calcitonin/CGRP gene, there are nine repeats of GCATG, five of them within the alternatively spliced portion of the gene. Of the five repeats surrounding the calcitonin-specific 3′ splice site, three match the fibronectin hexamer, TGCATG, while the others are AGCATG and CGCATG (Fig. 1A). To examine if these repeats are important for calcitonin/CGRP alternative splicing, each of the endogenous repeats was removed individually or in combination by site-directed mutagenesis, changing GCATG to a restriction site. For convenience, we will refer to the repeats at −254, −34, −19, +45, and +236 relative to the calcitonin-specific exon 4 3′ splice site as repeats number 1–5 (Fig. 1A). Mutant transcription units were assayed by stable transfection of F9 and HeLa cells that splice the wild-type gene to form predominantly CGRP and calcitonin mRNA, respectively (11). The quantitative data shown have been corrected for differences in the specific activities of RNase protection probe fragments.
Figure 1.
GCATG repeats are necessary for calcitonin-specific splicing. (A) The structure of the rat calcitonin/CGRP gene is diagrammed, indicating the location of GCATG repeats (arrows). The sequence of each repeat and flanking sequences is given below. (B) Total RNAs from stably transfected HeLa and F9 cells were analyzed by RNase protection. Cells were transfected with the wild-type gene, or with mutant genes lacking all five of the repeats indicated in A, −rep (1–5), or the central three repeats, −rep (2–4). Protected fragments are identified at the left. (C) Quantitative results from RNase protections is shown graphically. For ease of identification, mutations in individual repeats are shown by an “X,” whereas the wild-type repeat is indicated by “O.” Note that the quantitative data are corrected for the difference in the specific activities of the calcitonin- and CGRP-specific protected fragments.
Removal of the three central repeats near the calcitonin-specific 3′ splice site had no effect on calcitonin-specific splicing in HeLa cells [Fig. 1 B and C, −rep (2–4)]. However, when all five repeats were removed simultaneously, calcitonin-specific splicing was substantially decreased [Fig. 1 B and C, −rep (1–5)]. In F9 cells, CGRP-specific splicing was unaffected by the removal of three or five GCATG repeats. Although the possibility remains that the mutated sequence itself has unintentional effects on splice choice, we avoided introduction of known constitutive elements, and we had previously established that the elimination of any one of these repeats alone was without effect in either cell type (12). Therefore multiple GCATG repeats are required for calcitonin-specific splicing.
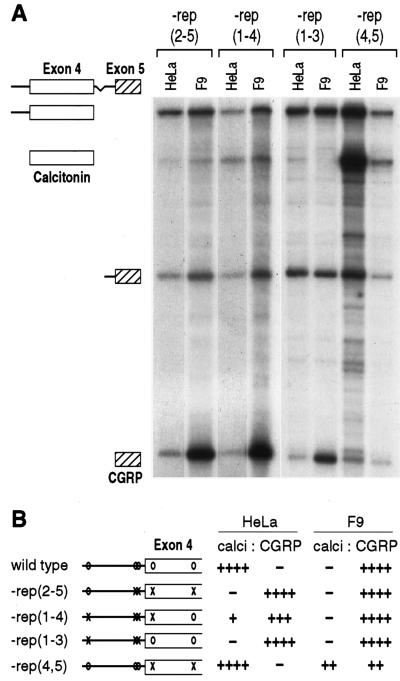
To determine the relative contribution of each repeat to splicing regulation, different combinations of mutations were tested. Removal of four of the five repeats, leaving either the 5′-most or 3′-most element intact [−rep (2–5) and −rep (1–4)], caused the loss of calcitonin-specific splicing in HeLa cells (Fig. 2). These results suggest that a single repeat is not sufficient to direct calcitonin-specific splicing. In addition, the removal of the three intronic repeats [−rep (1–3)] also reduced calcitonin-specific splicing (Fig. 2). In contrast, removal of the two exonic repeats had no effect on calcitonin-specific splicing in HeLa cells [−rep (4, 5), Fig. 2]. Interestingly, with the latter mutation, calcitonin-specific splicing was enhanced in F9 cells that normally exclude this exon. Together, these results suggest that the distribution of the repeated elements is important for alternative exon inclusion and that the exonic repeats function additionally in F9 cells to prevent calcitonin-specific splicing.
Figure 2.
Combinations of repeats are required for cell-specific splicing. (A) Total RNAs from cells transfected with mutant genes were analyzed by RNase protection. In −rep (2–5) and −rep (1–4), a single repeat was left intact, at −254 and +236, respectively. All the intronic repeats were removed in −rep (1–3), and the exonic repeats were removed in −rep (4, 5). Protected fragments are identified on the left. (B) Quantitative analysis of the data in A are shown graphically.
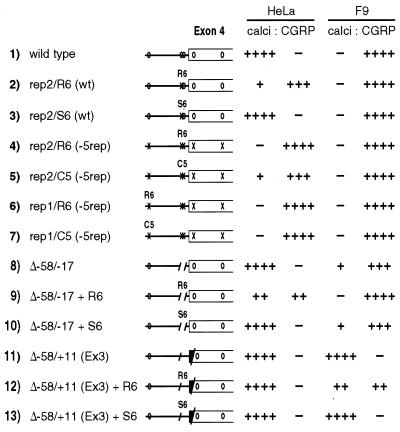
To determine if the re-insertion of GCATG repeats could rescue calcitonin-specific splicing, we replaced repeat number 2 with a synthetic sequence containing six copies of TGCATG (R6, previously termed T3, ref. 9). As a control, a sequence containing six copies of a scrambled hexamer (S6, previously called S3) was tested. For comparison, five copies of the TGCATG repeat as found in the calcitonin/CGRP repeat number 1 (C5) also were introduced. In the absence of all five endogenous repeats, insertion of either the R6 or C5 multimerized repeats did not rescue calcitonin-specific splicing, whether the insertion was made at repeat position 1 or 2 (Fig. 3, lines 4–7). In fact, when multimerized TGCATG repeats were inserted at repeat position 2 in the wild-type gene, calcitonin-specific splicing was substantially reduced, whereas introduction of the scrambled repeats had no effect (Fig. 3, lines 2 and 3). These results suggest that multimerized, closely apposed copies of TGCATG in the intron can suppress the downstream alternative 3′ exon, and have little activating function in this model system.
Figure 3.
Multimerized repeats can suppress downstream exon inclusion. Quantitative data from RNase protections of total RNAs is presented graphically. R6 refers to six copies of the synthetic TGCATG repeat, S6 is a scrambled version of the same sequence, and C5 contains five copies of the calcitonin repeat at −254 relative to exon 4 (see Materials and Methods). rep2 and rep1 indicates the site of multimer insertion, and (wt) and (−5rep) indicate the status of the endogenous repeats.
The negative effect of the TGCATG multimer was further supported by its effect on two other mutations shown previously to stimulate calcitonin-specific splicing in F9 cells (11). The intron deletion Δ−58/−17 removed repeats 2 and 3 and caused a slight increase in exon 4 inclusion in F9 cells, without a change in calcitonin-specific splicing in HeLa cells. Introduction of R6, but not S6, restored CGRP-specific splicing and decreased calcitonin-specific splicing in F9 cells (Fig. 3, lines 9 and 10). Calcitonin-specific splicing was also somewhat decreased in HeLa cells (Fig. 3, Δ−58/−17 + R6). In another activating mutation, the substitution of exon 4 splice acceptor sequences with those from the constitutive exon 3 caused strong calcitonin-specific splicing in both F9 and HeLa cells [ref. 11; Δ−58/+11 (Ex3)]. Insertion of R6 at −42 relative to the 3′ splice site in this substitution mutation caused a substantial decrease in exon 4 inclusion in F9 cells, an effect not observed with S6 insertion. In HeLa cells, the insertion of multimerized repeats did not substantially affect calcitonin-specific splicing in the context of the 3′ splice site substitution (Fig. 3, lines 12 and 13). Together, these results suggest that a multimerized synthetic element inhibits splicing of the downstream alternative exon in both cell types, despite the presence of endogenous repeats, and independently of the sequence context of the 3′ splice site.
To examine if the functional differences between dispersed and multimerized repeats reflected biochemical differences in RNA-protein interactions, short RNAs consisting of a single or multiple repeats were bound to nuclear extract proteins derived from cells making either calcitonin or CGRP mRNA. As shown in Fig. 4, RNA containing a single repeat bound with low efficiency in a few discrete complexes. Protein complex patterns were clearly different between nuclear extracts prepared from HeLa cells (calcitonin-specific splicing), and those made from CA or B50 cells, that, like F9 cells, show strong CGRP-specific splicing (10, 12). Two discrete complexes were observed binding to GCAUG RNA from each extract, and the complexes from each cell type migrated at different rates. No additional complexes were observed when RNA was bound by a mixture of HeLa and B50 nuclear extracts. Binding to the slowest-migrating cell-specific complexes was diminished but not eliminated when a mutant probe was used.
Figure 4.
Cell-specific protein complexes bind to single repeats, but not multimerized repeats. Short radiolabeled RNA probes were bound to nuclear extract proteins derived from cell lines that generate calcitonin mRNA (HeLa), or CGRP mRNA (B50 and CA). Complexes were resolved by gel mobility-shift assays, and the position of bound vs. free RNA is indicated.
In contrast, RNA containing five copies of UGCAUG bound nuclear proteins efficiently, and the protein complexes appeared similar between nuclear extracts from cell types making opposite splice choices (Fig. 4). In the absence of nuclear extract, the multimerized RNA migrated as two separable species, suggesting this RNA may adopt at least two stable secondary structures. Protein complexes appeared indistinct, and the combination of B50 and HeLa nuclear extracts yielded a larger indistinct complex. It is unclear whether the lack of discrete complexes with the multimerized probe reflects the additive effects of single complexes or the recruitment of additional proteins. Nonetheless, the clear difference in cell-specific complexes detected on RNA containing a single repeat contrasts markedly with the pattern detected with a multimerized probe. Therefore, the functional differences between single and multimerized repeats in the regulation of pre-mRNA splicing may reflect biochemical differences in the binding of nuclear proteins.
DISCUSSION
In previous studies, multiple intronic copies of the repeated sequence TGCATG were found to activate the inclusion of the upstream alternative fibronectin exon EIIIB (9). Repeated copies of this sequence also were found to enhance upstream alternative exon inclusion in the heterologous preprotachykinin gene (9). However, in the calcitonin/CGRP context, multimerized repeats inhibited use of the downstream alternative exon. One possible explanation for the positive effect of multimerized repeats on EIIIB inclusion and the negative effect of the same repeats on calcitonin-specific exon inclusion may lie in the relative location of the repeats. The fibronectin and calcitonin/CGRP genes differ both in the location of the native repeats, and in the arrangement of the alternative exons. In the fibronectin gene, the alternative exon occurs between two constitutive exons, and the native GCATG repeats are found eight times in the 1,071-nucleotide downstream intron. In contrast, the alternative exons in the calcitonin/CGRP gene are arranged as competing 3′ splice sites, with different polyadenylation sites, and the native repeats occur five times within 490 nucleotides spanning the upstream alternative 3′ splice site. One model to reconcile the ability of multimerized repeats to enhance alternative exon usage in one context but inhibit it in another might be that multimerized repeats suppress splicing to the downstream exon in both genes. In fibronectin pre-mRNA, a repeat-mediated delay in recognition of the 3′ constitutive exon may allow EIIIB to be recognized more efficiently. Alternatively, trans-acting factors bound by multimerized repeats may function differently when interacting with 5′ vs. 3′ splice sites. In fibronectin pre-mRNA, factors bound by multimerized repeats may interact with splicing components bound at the 5′ splice site of the upstream exon, whereas in calcitonin/CGRP, proteins bound by multimerized repeats may interact with the complex of factors defining the 3′ splice site.
The native arrangement of GCATG repeats in the calcitonin/CGRP gene is not “multimerized,” but distributed over 490 nucleotides, the closest two repeats being spaced by 10 nucleotides. We have not extensively examined the spacing requirements that distinguish positively and negatively acting arrangements of repeats. Further, the role of the 5′ T nucleotide has not been examined in either the fibronectin or the calcitonin/CGRP context. However, it is clear that the precise arrangement of these short repeated sequences is critical for appropriate, cell-specific splicing regulation. In previous studies, we had detected a modest shift toward calcitonin-specific splicing when a deletion was made from −58 to −17 in intron 3. This deletion removed two repeats as well as the branchpoint (11). Subsequent, smaller mutations in the intron did not reveal a single regulatory element in the intron other than the suboptimal branchpoint (12). Therefore, the idea that multiple redundant elements regulate calcitonin/CGRP splicing is entirely consistent with our previous results.
The regulation of calcitonin/CGRP alternative splicing is a complex process that depends partly on the quality of the constitutive splicing signals surrounding the calcitonin-specific exon (12). Recently, it was reported that an intronic element exists downstream of the calcitonin-specific polyadenylation site that can regulate alternative splicing of the human calcitonin/CGRP pre-mRNA (15). This element resembles both a splice donor and splice acceptor, and may act by recruiting constitutive factors. However, in the rat gene, a deletion in the downstream intron that removed this element was entirely neutral with respect to cell-specific splicing (11). The human gene also differs from the rat gene in that the exon 4 branchpoint nucleotide is different, the extensive CGRP polypyrimidine stretch is twice as large in the rat gene, and there are more GCATG repeats near the calcitonin 3′ splice site in the human gene (9). Therefore, either the deletion that removed the intronic sequence in the rat gene had both positive and negative effects that compensated for each other, or the rat and human genes differ in which sequence elements dominate regulation. In this regard, it would be interesting to examine the role of GCATG repeats in the human gene.
In this report, individual repeats bound protein complexes that differed between nuclear extracts made from cell types that made opposite splice choices. Because CGRP splicing is neuron-specific, the cell-specific binding detected using single repeats resembles that seen in the case of c-src pre-mRNA, which also exhibits neuron-specific splicing. The c-src N1 exon is included in neurons and inclusion depends on flanking intronic sequences (7). As has been noted before (9), one copy of TGCATG occurs within the “downstream control sequence” (DCS) in the intron following the alternative exon N1. The DCS element binds a cell-specific protein complex that includes hnRNP F and a novel protein, KSRP, and the DCS is required for N1 exon inclusion (7, 8). A mutation that lost cell-specific protein binding and functional activity happened to include the two 5′ nucleotides of TGCATG (7), but the role of this specific element in the regulation of N1 splicing has not yet been reported. Although the two genes have different structures, it is tempting to speculate that similar biochemical mechanisms underlie the neural-specific regulation of both c-src and calcitonin/CGRP pre-mRNAs, and that a related mechanism governs fibronectin pre-mRNA regulation.
Finally, the observation that spaced repeats function differently than adjacent repeats suggests that the density and distribution of these repeat elements comprise important aspects of splicing regulation. Because the minimally effective arrangement of repeats consisted of a pair of one intronic and one exonic element, a potential mechanism for their function at the calcitonin-specific 3′ splice site might involve interactions between proteins bound to each repeat that span and thus impose structural constraints on the splice acceptor. A similar mechanism has been proposed for the activity of the RNA binding protein RBP-1 in its binding to both intronic and exonic sites spanning the doublesex alternative 3′ splice site (16). Although the biochemical mechanisms underlying the function of GCATG repeats remain to be defined, the requirement for multiple, redundant, distributed elements in calcitonin/CGRP pre-mRNA splicing suggests that a mechanism combining both biochemical and structural features may direct alternative splicing regulation.
Acknowledgments
We thank J. Bermingham and X.-D. Fu for critical discussions. We also thank P. Myer and C. Nelson for their valuable assistance. M.G.R. and R.O.H. are Investigators with the Howard Hughes Medical Institute. This work was supported by grants from the American Cancer Society to M.G.R. and from the National Cancer Institute to R.O.H.
References
- 1.Green M R. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 2.McKeown M. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- 3.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1–30. [Google Scholar]
- 4.Black D L. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan K J, Cooper T A. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siebel C W, Admon A, Rio D C. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 7.Min H, Chan R C, Black D L. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 8.Min H, Turck C W, Black D L. Genes Dev. 1996;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 9.Huh G S, Hynes R O. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 10.Leff S E, Evans R M, Rosenfeld M G. Cell. 1987;48:517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 11.Emeson R B, Hedjran F, Yeakley J M, Guise J W, Rosenfeld M G. Nature (London) 1989;341:76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- 12.Yeakley J M, Hedjran F, Morfin J-P, Merillat N, Rosenfeld M G, Emeson R B. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeakley J M, Morfin J-P, Rosenfeld M G, Fu X-D. Proc Natl Acad Sci USA. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 15.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrichs V, Baker B S. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]